Abstract
Avicennia marina forests fulfill essential blue carbon and ecosystem services, including halting coastal erosion and supporting fisheries. Genetic studies of A. marina tissues could yield insight into halophyte adaptive strategies, empowering saline agriculture research. We compare transcriptomes from A. marina pneumatophores, stems, leaves, flowers, seeds, and transcriptomes across four widely divergent environments in the Indo-Pacific (Red Sea, Arabian Gulf, Bay of Bengal, and Red River Delta) to decipher the shared and location-, tissue-, and condition-specific functions. On average, 4.8% of transcripts per tissue were uniquely expressed in that tissue, and 12.2% were shared in all five tissues. Flowers’ transcript expression was the most distinct, with domain-centric gene ontology analysis showing high enrichment for stimulus-responsive processes, as well as genes implicated in flowering (hydroxygeraniol dehydrogenase, TPM = 3687) and floral scent biosynthesis (e.g., benzoyl_coenzyme_A, 2497.2 TPM). Pneumatophores highly expressed antioxidant genes, such as glutathione S-transferase (GST, TPM = 4759) and thioredoxin (TRX, TPM = 936.2), as well as proteins in the GO term ‘Hydroquinone:oxygen oxidoreductase activity’ (enrichment Z = 7.69, FDR-corr. p = 0.000785). Tissue-specific metabolic pathway reconstruction revealed unique processes in the five tissues; for example, seeds showed the most complete expression of lipid biosynthetic and degradation pathways. The leaf transcriptome had the lowest functional diversity among the expressed genes in any tissue, but highly expressed a catalase (TPM = 4181) and was enriched for the GO term ‘transmembrane transporter activity’ (GO:0015238; Z = 11.83; FDR-corr. p = 1.58 × 10−9), underscoring the genes for salt exporters. Metallothioneins (MTs) were the highest-expressed genes in all tissues from the cultivars of all locations; the dominant expression of these metal-binding and oxidative-stress control genes indicates they are essential for A. marina in its natural habitats. Our study yields insight into how A. marina tissue-specific gene expression supports halotolerance and other coastal adaptative strategies in this halophytic angiosperm.
1. Introduction
Mangroves support coastal ecosystems despite salinity, oxygen, and temperature extremes [1,2,3,4,5]. They are comprised of distantly related tree species that have converged to dwell on saline shorelines [6,7]. Among mangroves, Avicennia species are particularly hardy in the face of environmental extremes. However, they have relatively shallow genetic diversity and are vulnerable to environmental perturbations from anthropogenic climate changes [8,9,10,11].
Avicennia forests provide blue carbon and ecosystem services for their surrounding populations [4]. Their ability to halt coastal erosion, support fisheries, and sequester atmospheric carbon make them outstanding contributors to global environmental sustainability. These services are enabled by the preservation and propagation of crucial species, such as A. marina [12]. Further genome-level studies will reinforce afforestation efforts. Establishing additional omics resources for A. marina, including genomics, transcriptomics, metabolomics, and subsequent comparative analyses, will inform critical decisions regarding cultivar choices, suitable afforestation locations, and possible transgenic species implementation.
The unique ecophysiology of A. marina has facilitated its survival in a wide variety of Indo-Pacific coastal biomes [8,10,13,14,15,16,17,18,19,20,21,22]. It has colonized coasts from Africa and Asia to Zealandia since the early Holocene [23]. Habitat environments range from hypo- to hypersaline, and annual average, low, and high temperatures in A. marina-colonized locations range from Africa to New Zealand. The species has adapted to survive in a wide range of intertidal biome types. Furthermore, unrelated mangrove lineages have undergone convergent evolution towards functionally similar adaptations [6,7]. Factors advantageous in a seafaring lifestyle include seed survival across ocean voyages, secondary growth through successive cambia formation [20,24], halotolerance [25,26,27,28,29], and multifunctional pneumatophores that cope with a vast array of abiotic stressors [15,30,31].
A. marina inhabits the coastline of the Red Sea [32], a region occasionally characterized by extreme heat and salinity [13,14,19,21,22,33]. Cultivars from this region are suitable models to study the resilience of these angiosperms in the face of intense abiotic stress. The mechanisms behind A. marina’s resilient biology, especially in areas such as the Red Sea, can inform translational agronomical research. Oxidative stress results from salt toxicity that prohibits saline agriculture of most commercial crops; increasing resistance will allow productive transgenic species to thrive using readily available saline irrigation. The implementation of seawater irrigation would promote food security in the face of anticipated shortages in freshwater supply [34,35,36,37], as well as expand saline agriculture into inarable regions [28,38,39,40,41,42].
The mechanisms used by A. marina leaves to maintain photosynthesis in the face of high salt levels is the subject of intensive research, as salt is toxic to photosystems [43,44,45,46,47,48] yet accumulates at high levels in A. marina leaves [26]. A recent study examined Avicennia officinalis leaves’ response to salt stress and found 1404 genes expressed after salt treatment [30]. Salt stress can be mitigated by osmolyte accumulation [45,49,50,51] and active transport [42,52,53,54,55,56,57]. Membrane integral active transporters are essential for A. marina survival and responsible for the large salt crystals that commonly form on the surface of A. marina leaves [58]. Despite intensive research into Avicennia’s leaves and pneumatophores, research into its other tissues, including flowers, seeds, and developing shoots, is lacking. Insight into how these tissues support Avicennia’s seafaring lifestyle could be obtained by examining their transcriptomic expression profiles.
Here, we highlight the expression of genes involved in processes essential for these understudied tissues. We use A. marina transcriptomes from different tissues and geographies to find commonalities of intrinsic gene expression patterns responsible for its ability to survive in conditions that would be prohibitive for other angiosperms. This information will be useful in understanding what core genotypes can promote successful saline agronomy endeavors.
2. Materials and Methods
2.1. Sequence Sources
Our five-tissue transcriptome dataset consisted of tissue samples we previously isolated and used to annotate a high-quality A. marina reference [59]. We used the high-quality reference genome recently published by the NYUAD Marine Biology Laboratory [59] for chromosome-level read mapping (see Figure S1). Reference transcripts for expression analyses were from He et al. (2020) [7]. Finally, we used public RNAseq data from A. marina cultivars from the Red Sea (NCBI BioProject: PRJNA591919), Vietnam (NCBI Bioproject: PRJDB6605, Department of Biotechnology, Graduate School of Engineering, Osaka University, Japan), India (NCBI Bioproject: PRJNA283781, M.S. Swaminathan Research Foundation, Chennai, India), and China (NCBI Bioproject: PRJNA350568, Lingnan Normal University, Zhanjiang, China) to further relate our tissue-specific transcriptomes to natural gene expression patterns.
2.2. Sequence Processing and Alignment
Adapters and filter low-quality bases (<Q20) were removed with the CLC Genomics Workbench (v11.0; Qiagen, Hilden, DE) trim tool. Filtered reads were used for mapping to the reference transcriptome [7] to determine the differentially expressed genes (DEGs, see Table S1) using the CLC Genomics Workbench (v11.0; Qiagen, Hilden, DE). Mapping of RNAseq reads to the reference genome [59] was done in HISAT2 [60], similarly to [61], to observe the global expression patterns (see also Dataset S1). Mapping to the reference transcripts was done in the CLC Genomics Workbench (v11.0; Qiagen, Hilden, DE) in the same manner as [62]. Briefly, genes were extracted from the reference genome [59] (using annotations of the type gene). Other annotations on the gene sequences were preserved (e.g., coding sequences, introns, exons, and untranslated regions). Then, the annotated transcripts (using annotations of type mRNA) were extracted. If there were several annotated splice variants, they were all extracted. The mRNA annotation type was used for deciphering exon–exon boundaries. Reads were mapped against all the transcripts plus the entire gene, so all the exon–exon junctions were joined in the extracted transcript. Mapped reads were categorized and assigned to the genes (elaborated later in this section), and expression values for each gene and transcript were calculated. Then, putative exons were identified, following guidelines [63,64,65]. The default parameters for alignment were used, including no more than two mismatches per aligned read, ten maximum hits for a read, a 90% minimum length fraction mapped, and a minimum similarity fraction of 0.8.
Differential expression was calculated for each gene using transcripts-per-kilobase million (TPM) for accurate comparison between samples (TPM = (R * 106)/(T * L), where T is the sum of all length-normalized transcript counts, R is the number of reads mapped to a transcript, and L is the length of the transcript) [66,67,68,69]. Transcripts with a TPM value of ten or higher were classified as actively expressed unless stated otherwise, and those with TPM values of less than ten were deemed unexpressed. This filtering strategy for a classification-based feature selection resolved 400–600 ‘uniquely’ expressed genes in each tissue. A caveat of this study was the lack of triplicate samples for each condition; thus, qualitative comparative analyses comprised methods used here. Expression values for TPM, FPKM, RPKM, and raw values are in Table S1 (Red Sea, five tissues) and Table S2 (samples from various geographies).
2.3. Uniform Manifold Approximation Projection
The DEG TPMs in each tissue informed the Uniform manifold approximation projection (UMAP, [70]) in TensorFlow (tensorflow.org; https://projector.tensorflow.org/, accessed on 12 March 2021) to cluster different annotated genes by their tissue-specific expression profiles. Although UMAP is primarily used to decipher single-cell RNAseq expression data, its adept embedding clustering algorithm lends itself to a wide variety of applications. Briefly, UMAP clusters data points from variables in multiple dimensions using fuzzy simplices to preserve local relationship information while approximating their embedding. For the projection shown in Figure S1, the ‘3D’ visualization mode was selected, and the 15 nearest neighbors for each data point were used to calculate the approximations. Graphs were manually optimized for visual clarity. Data points represented individual transcripts and were colored according to the tissue showing the highest expression in TPM.
2.4. Hierarchical Clustering
Hierarchical clustering was done using Pearson’s correlation coefficient or 1-cosine similarity (Figure S3) distance metrics and by using an average agglomeration method [71] in Morpheus (https://software.broadinstitute.org/morpheus, accessed on 12 March 2021).
2.5. Intersection Analyses
2.6. Domain-Centric GO Enrichment Analysis
The tissue-specific enrichment analysis was carried out using Domain-centric Gene Ontology (dcGO, [74]). Preliminary HMMsearch (hmmer.org) was performed on translated reference transcripts with the command ‘hmmsearch –noali -E 0.000000001 –cpu 28 –domtblout $OUT $IN.aa.fa’. The software dcGO statistically infers associations between GO terms and combinations of PFAM domains. Unique transcripts were used as inputs for each tissue; the discovered GO terms shown are non-unique. An additional dcGO analysis with unique GO terms in each tissue from uniquely expressed genes (doubly-unique) is shown in Figure S4. A false discovery rate (FDR) threshold of 0.001 was used for detection of enrichment.
2.7. GO Network
The baseline GO network was constructed from the full GO list downloaded from http://geneontology.org/docs/download-ontology/ (accessed on 5 September 2020). The ‘core ontology’ file (go.obo http://purl.obolibrary.org/obo/go.obo, accessed on 3 July 2022; Core ontology (OBO Format)) was used to create the network infrastructure in Cytoscape (cytoscape.org, accessed on 3 July 2022). Expression data were mapped to the nodes on the network corresponding to the GO terms.
2.8. Metabolic Map Reconstruction
Metabolic maps were reconstructed using the Interactive Pathways Explorer (iPath) (a web-based tool for the visualization, analysis, and customization of various pathway maps; https://pathways.embl.de/, accessed on 23 November 2021). Briefly, KEGG ortholog (KO) designation lists corresponding to genes expressed at >10 TPM were uploaded for each tissue (see also Table S3).
2.9. Manhattan and Violin Plots
Sequence reads from public RNAseq data from A. marina cultivars in Vietnam (NCBI Bioproject: PRJDB6605, Department of Biotechnology, Graduate School of Engineering, Osaka University, Japan), India (NCBI Bioproject: PRJNA283781, M.S. Swaminathan Research Foundation, Chennai, India), and China (NCBI Bioproject: PRJNA350568, Lingnan Normal University, Zhanjiang, China) were mapped to reference transcripts [7], to obtain the TPMs, and plotted in Plotly (https://plotly.com/, accessed on 23 November 2021, px.scatter, y=violin), to show the highest-expressed transcripts and overall expression distributions2.10. Graphics
Figure 1B was provided by NASA. The International Space Station Program supports the laboratory as part of the ISS National Lab to help astronauts take pictures of Earth that will be of great value to scientists and the public, and to make those images freely available on the Internet. Astronaut photograph ISS052-E-15927 was acquired on 14 July 2017, with a Nikon D4 digital camera using an 1150 mm lens and is provided by the ISS Crew Earth Observations Facility and the Earth Science and Remote Sensing Unit, Johnson Space Center. The image was taken by a member of the Expedition 52 crew. The image has been cropped and enhanced to improve contrast, and lens artifacts have been removed. Caption by Andi Hollier, Hx5, JETS Contract at NASA-JSC.
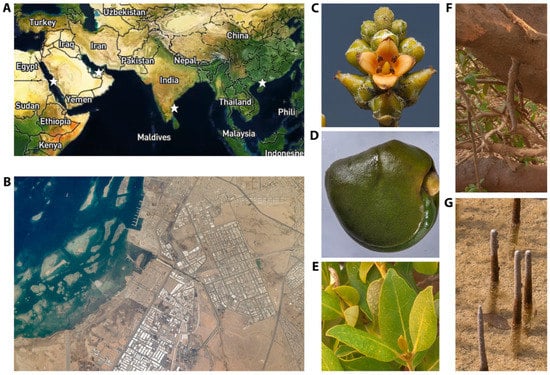
Figure 1.
Overview of the RNAseq data source tissues and geographies. (A) Locations (stars) of the A. marina RNAseq samples. Map and location visualized in Plotly. (B) Jeddah Port, showing the industrialization near the Red Sea mangrove sampling site. Image courtesy of NASA. (C) A. marina flowers. (D) A. marina seeds. (E) A. marina leaves. (F) A. marina stems. (G) A. marina pneumatophores. Mangrove tissue photos reproduced with permission from Dr. Alexey Sergeev.
Automatic generation of images was carried out in Plotly (Plotly express; https://plotly.com/python/plotly-express/, accessed on 23 November 2021) and R (https://www.r-project.org, accessed on 23 November 2021), ggplot2 (https://ggplot2.tidyverse.org, accessed on 23 November 2021), and processed with Adobe Illustrator (v25.2.3) and Adobe Photoshop CC (v20.0.6) for clarity and annotation. Circos (http://circos.ca/, accessed on 4 April 2020 [75]) was used to plot the TPM expression values of the RNAseq from various tissues mapped to the reference chromosomes for Figure S1 [59] with HISAT2 [60]. Hierarchical clustering was done in Morpheus (https://software.broadinstitute.org/morpheus, accessed on 19 May 2021). The GO network was created in Cytoscape (cytoscape.org, accessed on 3 July 2022).
3. Results and Discussion
We sampled sequencing reads obtained from A. marina in various locations in the Indo-Pacific region and collated them into a database. These reads were the basis for transcriptome comparisons between tissue types and geographies (Figure 1). Our characterization of the expressed functions was facilitated by the reference genome and coding sequence annotations [7]. In addition, we used the high-quality reference genome recently published by the NYUAD Marine Biology Laboratory [59] for mapping reads to chromosomes (Figure S1; Dataset S1). Finally, we used public RNAseq data from A. marina cultivars in Vietnam (NCBI Bioproject: PRJDB6605, Department of Biotechnology, Graduate School of Engineering, Osaka University, Japan), India (NCBI Bioproject: PRJNA283781, M.S. Swaminathan Research Foundation, Chennai, India), and China (NCBI Bioproject: PRJNA350568, Lingnan Normal University, Zhanjiang, China) to further relate our tissue-specific transcriptomes to natural gene expression patterns in A. marina. Together, these datasets provide a basis to study organ-specific transcriptomes in A. marina.
3.1. A. marina Coding Potential and Transcriptome
A. marina can survive extreme conditions because of the genes transcribed by its relatively compact, unchanging genome. We examined de novo-predicted genes, de novo-assembled transcriptomes, and evaluated expression profiles from two published reference sequence sources. Compared to other angiosperms, all A. marina transcriptomes published so far show high expression of a collection of genes implicated in heat and drought tolerance. Thus, their transcriptomic profiles contain the blueprints for angiosperm survival in harsh conditions.
Here, we analyze five separate transcriptome profiles from pneumatophore, leaf, flower, seed, and shoot tissue samples of A. marina. We compare these profiles with each other as well as transcriptomes from various tissues and locations throughout the Indo-Pacific, including India, Vietnam, and China. Investigation into these transcriptomic profiles yielded insight into the functional roles of each tissue and how they support A. marina’s seafaring lifestyle.
One of the most remarkable findings from our studies was that genes for metallothionein showed the highest expression regardless of tissue type or cultivar location (see Table S1). This finding provides strong evidence that metallothioneins are key components for A. marina antioxidant systems regardless of location, climate, or environment. Other significant findings include arrays of desiccation-related genes, especially in seawater-immersed tissues, and flower-specific genes involved in pollinator attraction processes. Below, we detail the features of the sampled transcriptomes. Tissue-specific transcriptomes are described in the order of their expression track rings in Figure S1.
3.2. A. marina Tissue-Specific Transcript Expression
Expressed genes were determined by having a coverage depth of >10 transcripts per kilobase million (TPM) unless stated otherwise. Downstream analyses on defined gene sets were uniform manifold approximation projection (UMAP [70]; Figure S1), hierarchical bi-clustering (Figure 2), Hidden Markov Model (HMM) alignment to known protein families (PFAMs [76,77]), and gene ontology (GO) enrichment analyses by tissue and location. The five tissues shared major chromosomal regions of high expression, with notable exceptions (Figure S1). These regions were in the outer arms; centromeric regions displayed canonical low transcript expression (Figure S1; Dataset S1).
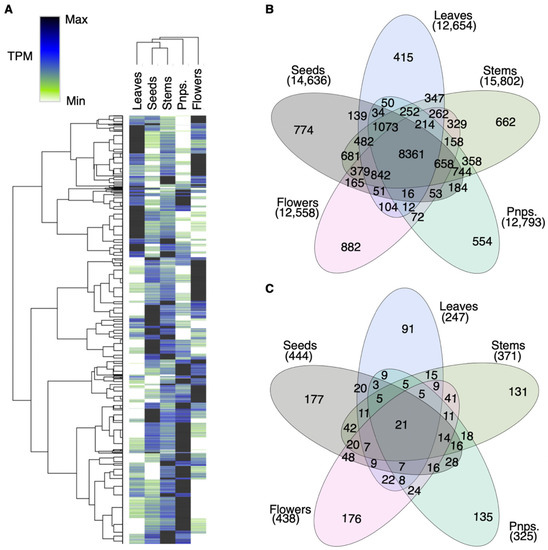
Figure 2.
Segregation of Red Sea A. marina gene clusters based on tissue-specific expression patterns. Pnps. = Pneumatophores. (A) Hierarchical bi-clustering of transcripts-per-million (TPM) normalized read counts from the five mangrove tissues using Pearson’s correlation (see Table S1). The y-axis indicates transcripts. Darker clusters in each tissue correspond with GO enrichment networks. (B) Venn diagram showing reference transcripts >10 TPM from five tissues. Leaves had the smallest unique set, while stems expressed the most transcripts >10 TPM. Figure S2 shows additional intersections not shown here. (C) Unique and shared KEGG Orthology (KO) in the unique gene sets from (B). Approximately 60% of KOs from the uniquely expressed genes in each tissue are shared with one or more other tissues. Nearly one-third of (C) KOs in any given tissue are unique to that tissue, while the ratio of unique (B) transcripts is 1/15th the expressed transcript count. This discrepancy suggests a high redundancy in function coded for by A. marina transcripts.
Hierarchical bi-clustering of DEGs highlighted tissue-specific gene clusters (Figure 2). Uniquely-expressed (>10 TPM) clusters of DEGs (Table S1) for each tissue are described in the following sections, with implications for A. marina’s abiotic stress resistance. For insight into the relationship of different A. marina gene expressions at a snapshot of the Red Sea cultivar, Uniform Manifold Approximation Projection (UMAP; [70]) was applied (Figure S1) to the TPM profiles from five tissues (Table S1). Root-specific genes clustered near the seed and leaf genes; flowers and stems clustered tightly but were isolated from other tissues. Leaf-dominant genes were centrally projected, implying a median expression level for highly differentially expressed genes between the flower-stem and pneumatophore-seed groups. Roots and seeds are the only two A. marina tissues with prolonged seawater exposure; thus, similar gene expression networks may be required by these tissues to survive in these conditions.
We used unique KEGG KO terms (see References [6,7]) as input for each tissue, with red lines representing unique pathways for flowers, leaves, pneumatophores, seeds, and stems (see Figure 3). The reference transcripts were used as queries in HMMsearch to detect PFAMs (E < 1 × 10−9). These PFAMs were used to discover domain-centric gene ontology (dcGO; [74]) functional enrichment using either one or two exclusion filters. The first filtering step removed transcripts expressed in more than one tissue; uniquely expressed transcripts were used for dcGO [74] enrichment (Figure 3). The second filter removed any GO terms shared amongst tissues.
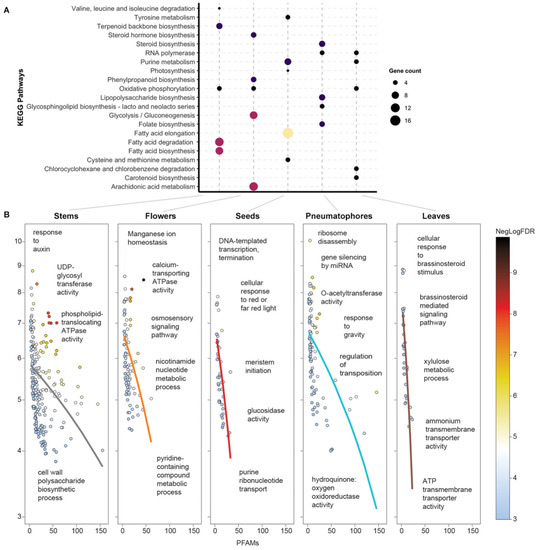
Figure 3.
Tissue-specific KEGG pathways and GO term enrichment in A. marina. (A) Top five KEGG pathways with regard to uniquely expressed transcripts for each tissue. The y-axis represents the pathway name. The size of the bubble indicates the number of transcripts in the KEGG pathway. The color represents the gene count, with yellow representing a higher value and black representing a lower value. Transcripts expressed at >10 TPM (see Table S1) in each tissue were used to retrieve the KEGG orthologs. (B) Domain-centric gene ontology (dcGO; [74]) enrichment in uniquely expressed transcripts in the Red Sea cultivar.
3.3. Flower-Specific Transcript Expression
A. marina flowers accumulate relatively large amounts of sweet nectar and are pollinated by insects. Pollination by long-distance-traveling insects is crucial for species stability. Honeybees are the dominant pollinator of temperate A. marina, and pollinator-mediated competition has been reported [78]. Recent studies have hinted at a hidden world of mangrove insect interactions, with up to 160 distinct species per sampled site [79]. Insect-flower interactions can be interrogated at the molecular level in A. marina by examining the genes involved in nectar formation and chemical signaling. We examined the expressed genes involved in these processes.
Flower-specific genes yielded 5398 PFAMs, which contributed to enrichment in 577 GO terms (FDR corr. p = 0.001). Gene ontology analyses of flower-specific genes showed enrichment for stimulus-responsive processes; for example, the term ‘response to hormone’ (GO:0009725) was highly enriched in flowers (Z = 14.81; FDR-corr. p = 3.85 × 10−25; see also Table S3).
A large portion of flower-specific transcripts coded for proteins with known roles in flowering timing, such as cytochromes [44,80,81,82,83,84,85]. Putative cytochromes and ferredoxins were uniquely expressed in the flower transcriptome (Table S1), corresponding to the proposed roles of these enzymes in season-responsive flower developmental timing [86,87,88,89]. We note that multi-copper-containing glycoproteins (e.g., laccases; EC 1.10.3.2; i.e., p-diphenol: dioxygen oxidoreductase) were uniquely expressed in flowers. Mutations in Arabidopsis homologs of these genes have shortened the time to flowering [90]. An 8-hydroxygeraniol dehydrogenase (HGD; AM_02435) was also highly expressed in flowers (TPM = 3687). Gene ontology analysis with flower-specific transcripts revealed enrichment in processes related to the aforementioned redox reactions (e.g., ‘cation homeostasis’ (GO:0055080; Z = 8.83; FDR-corr. p = 7.36 × 10−11) and ‘response to metal ion’ (GO:0010038; Z = 8.96; FDR-corr. p = 3.72 × 10−10) (see also Table S3).
The flower RNAseq evidence is consistent with a pollinator-dependent flowering strategy in A. marina. Genes in pathways for nectar formation were uniquely expressed in flowers. Polygalacturonases (AM_00332 and AM_0031), three pectate lyases (AM_01234, AM_03511, AM_07809), and a pectinesterase (AM_01717) were uniquely expressed in flowers. These genes are implicated in nectar formation to attract pollinators, including honeybees [78]. Cheminformatic research has established molecular links between phytochemical signaling interventions and the regulation of the multi-enzyme arachidonic acid (AA) metabolic network [91]. The cysteine-rich peptide family gibberellic acid (GA)-stimulated Arabidopsis (GASA)/GA-stimulated transcript (GAST) plays critical roles in plant growth, development, and responses to abiotic and biotic stresses [92,93,94]. The GA-stimulated transcript (GAST1) was found in Arabidopsis roots and shoots, whereas our unique transcripts (AM_24369) were found in the flowers.
Pollination is a crucial yet understudied aspect of A. marina’s dispersal. Volatile organic carbon compounds produced by flowers attract and regulate pollinator visitations [95]. Insight into the genetic mechanisms governing A. marina’s pollination preferences would assist conservation and afforestation endeavors. Here, we show the uniquely expressed genes in flowers with putative roles in floral scent biosynthesis and nectar formation. Benzoyl_coenzyme_A (benzyl_alcohol_benzoyl_transferase, AM_23483, TPM = 2497.2) was one of the highest-expressed flower genes. This enzyme is responsible for the floral scent in a wide variety of ornamental plants (REFs). Thus, its role in Avicennia is likely to attract pollinators. 8-Hydroxygeraniol dehydrogenase (HGD; AM_02435) was highly expressed in flowers (TPM = 3687). In plants, monoterpene 8-hydroxygeraniol is an intermediate between geraniol and secologanin, among other floral compounds [96]. Secologanin is a branchpoint monoterpene in terpene indole alkaloid biosynthesis; activity through this pathway produces bioactive alkaloid secondary metabolites that sources the cornucopia of volatile organic carbons to attract pollinators [97,98,99,100,101,102].
A. marina has a short flowering period; the high expression of HGD is consistent with a burst of floral volatile organic carbon (VOC) production, which could signal migratory insects for pollination. Pollination by migratory insects is especially prevalent in desert regions, where resources are scarce or are only seasonally available. The sampling site and flanking areas (i.e., MENA countries and territories) occupy the largest deserts on earth; migratory pollinators in these areas likely provide significant benefit for A. marina propagation and, perhaps, evolution of the species. Finally, an MIP-family (NIP) aquaporin (AM_01631) was uniquely expressed in flowers (TPM = 4.3). Although its expression was lower than the 10 TPM cutoff used in this study, this aquaporin may have a flower-specific role, with implications for flower stability (i.e., wilt prevention) in high-heat conditions.
3.4. Seed-Specific Transcript Expression
Avicennia species have less capacity for local dispersion by clonal propagation compared to other mangrove species (e.g., Rhizophora [103]) but propagate to distant shores through ocean currents [104] with resilient, cryptoviviparic seeds. This shift in energy expenditure investment from clonal propagation to seed potency has given them an evolutionary edge to live in otherwise inhabitable shorelines. Seeds expressed 14,636 genes with TPM >10; 774 of these were unique. Seed-expressed genes had putative roles in development, including coordinating the timing for senescence and ripening. Unique seed-expressed genes included an egg cell-secreted protein (AM_33231). Orthologs of this gene are known to mediate gamete interactions during fertilization in Arabidopsis [105,106]. We found an ethylene_receptor_isoform_1 unique transcription factor (AM_06004) in seeds. Ethylene is a simple two-carbon gas that is required for a variety of plant processes, such as seed germination, seedling growth, leaf and petal abscission, organ senescence, and pathogen responses. Genes for Jumonji (AM_23304) and Myb (AM_16379), among other (AM_18056, AM_08631) transcription factors, were uniquely expressed (>10 TPM) in seeds. These genes play roles in chromatin remodeling and temporal- and temperature-dependent transcription orchestration [107,108,109], which are processes implicated in seed senescence and ripening timing [110].
A type 2 metallothionein (MT-2; AM_26466) was expressed at relatively low levels (96 TPM) in seeds; however, its expression was high in the other four tissues from the Red Sea cultivar: leaves (35,065 TPM), stems (30,600 TPM), pneumatophores (18,035 TPM), and flowers (9652 TPM; also see Table S1). An MT-3 protein had similarly high expression in all tissues except flowers, consistent with tissue-specific suppression [111]. Other genes with potential roles in ripening include the uniquely expressed isoform of an ethylene receptor (AM_06004; TPM = 6237 in seeds) and a metalloendopeptidase (AM_05873) in seeds. This transcript was expressed at 194 TPM in seeds but less than 10 TPM in other tissues. This digestive enzyme may promote ripening in A. marina seeds. Leaf and seed expression profiles clustered together based on Pearson correlation scores (Figure 2A); transcript expression similarity between these two tissues is seen in other plant species [112]. A G3P dehydrogenase was highly expressed in seeds; this was one of the rare examples of a primary metabolic gene with an expression like leaves (TPM = 2614 in seeds, TPM = 3082 in leaves; expressed as AM_02817 in both tissues). Fatty acid (FA) biosynthesis and degradation pathways showed higher activity in seeds and pneumatophores.
3.5. Leaf-Specific Transcript Expression
Leaves had less overall diversity of transcript expression; however, photosynthesis-associated genes were highly expressed in leaves. For example, rubisco subunits were highly expressed (TPM 15,000–20,000; see Table S1). The expression of transcripts involved in photosynthetic processes was an order of magnitude higher in leaf tissue, which can be attributed to their abundant chloroplasts [113]. Transcript expression was suppressed on several leaf chromosomes (e.g., right arms of Chr 21 and Chr 12), and leaves had the lowest number of expressed CDSs with annotated PFAMs. Overall, 12,645 genes were expressed at TPM >10 in leaves, and 415 of these were unique to leaves (Table S1). The leaf transcript with the highest expression was AM_26466 (35,065 TPM in leaves), annotated as an MT-2 [18]. Leaves had relatively high expression of central metabolism genes. The AM_26466 transcript was also expressed at high levels in the other four tissue and likely plays a critical role in A. marina survival. Glyceraldehyde-3-phosphate dehydrogenase was expressed at 1911 TPM in leaves but at less than 400 TPM in the other tissues, and fructose-bisphosphate aldolase was expressed at 5404 TPM in leaves but at less than 1000 TPM in other tissues (Table S1). These results are consistent with high metabolic activity in leaves, exemplifying the capacity for energy generation in these chloroplast-rich tissues. Reconstructed GO networks also display strong enrichment for ‘plastid’ and related processes in leaves (Figure 4).
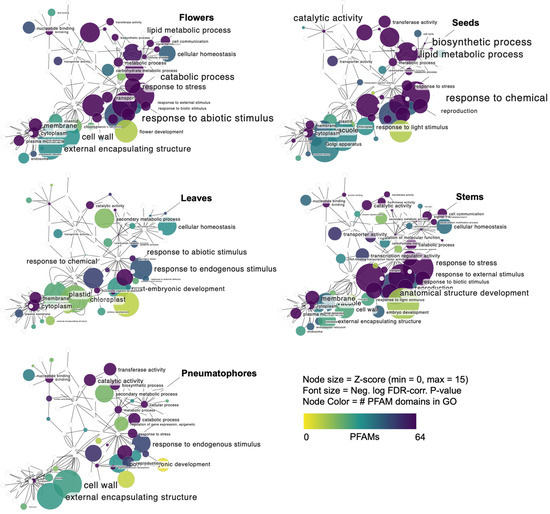
Figure 4.
Domain-centric gene ontology (dcGO, [74]) enrichment in uniquely expressed transcripts in the Red Sea cultivar visualized in Cytoscape using the base GO core ontology association network. Panels show GO enrichment networks from uniquely expressed genes in each of five A. marina tissues. Hypergeometric distributions were the null hypotheses; false discovery rate (FDR, [114])-corrected p values (colorscale) are from Fischer’s exact tests and Benjamini–Hochberg correction for multiple hypotheses against the hypergeometric null background. Z-scores indicate enrichment and are calculated by subtracting matches and dividing by the standard deviation with all UniProt entries following a true-path rule. A least-squares regression trendline is overlaid on each plot. Leaves showed a sharp bias in Z-score compared to the PFAM counts, indicating specialization. Z-score indicates enrichment or distance of the overlapping PFAM group with the constituents of the GO term. All p-values shown are < 0.001. Protein family domain counts in each GO term are shown in the color scale. The node size corresponds to the Z-score from dcGO [74] enrichment analysis.
Active salt efflux relies on ATP and other intracellular energy stores. Avicennia modulates salt influx and efflux through leaf pores; rhythmic fluctuations in salt secretion rates correspond to environmental salinity [29]. Candidate salt gland genes were expressed in concordance with [27]. The leaf-specific expression of ATPases may assist salt efflux in this tissue. The ATPc subunit was uniquely expressed in leaves at >10 TPM. This subunit has a rapid turnover rate in vivo [115,116], explaining its high expression in leaves. The GO term ‘transmembrane transporter activity’ (GO:0015238) was highly enriched in leaves (Z = 11.83; FDR-corr. p = 1.58 × 10−9, 13 PFAMs in this GO term), highlighting a possible role for the constituent genes in salt efflux.
Dehydrins have characteristically high expression in salt glands [27]; we observed the CDSs AM_1543 (Dehydrin, TPM = 67.2) and AM_31570 (Dehydrin_DHN1, TPM = 1504) expressed in leaves. Stems (TPM = 2701) and seeds (TPM = 2099) also expressed this dehydrin isoform at high levels, indicating that salt glands may be present and active in these non-leaf A. marina tissues.
Although the Red Sea samples were collected in the winter, we observed the high expression of genes involved in photorespiration in leaves. For example, a photorespiration enzyme, glycolate oxidase (GLO; AM_06008), was highly expressed in leaves (TPM = 1603). Although GLO isoforms have divergent roles [48], the overexpression of at least one GLO isoform has been reported to confer improved photosynthetic capacity under high heat and light conditions [117].
We searched for the expression of genes involved in cuticle formation. This type of secondary growth may be essential for A. marina’s desiccation tolerance, especially in periods of high temperatures, such as in the summers in the Middle East and Africa. A variety of wax and lipid biosynthetic genes are responsible for cuticle formation, although this process has not been well-studied in A. marina. Leaves expressed 39 lipid metabolism CDSs at TPM >10, many with putative roles in cuticle synthesis (see Figure S3). However, the only wax synthase we observed, ‘wax synthase isoform 3′ (AM_21438), was highly expressed in flowers (TPM = 79.3) rather than in the leaves (TPM = 6.1). Hierarchal clustering of extracted lipid metabolic genes revealed two clusters of genes uniquely and highly expressed in leaves (Figure S3). Genes in the leaf-specific clusters included a non-specific lipid-transfer protein-like protein (AM_27366), plastid lipid-associated proteins (AM_10546, AM_04326), a lipid-binding protein (AM_04326), a StAR-related lipid transfer protein (AM_18353), calcium lipid-binding proteins (AM_28011, AM_13767), a galactolipid galactosyltransferase (AM_07797), and a constitutive plastid-lipid associated protein (AM_04709; see also Figure S3).
A type-2 MT, together with a collection of photosynthetic genes, was expressed far more than other leaf-expressed genes (Table S1). Type-2 MTs respond to oxidative stress (Mir, 2004), bind metals [118], and mitigate oxidative stress in Avicennia [119] (Babaei-Bondarti and Shahpiri, 2020); however, MTs are less studied than their mammalian counterparts [120] (Freisinger, 2008). Transgenic E. coli expressing a recombinant type-2 MT (GST-OsMTI-2b) accumulated more Pb2+, Ni2+, Cd2+, Zn2+, and Cu2+ than the controls and had a higher tolerance to these metals [121]. Metallothioneins mitigate metal-induced oxidative stress [118,120,122,123,124], but recent studies have shown that MT isoforms can mitigate salt-induced oxidative stress as well. For example, a salt-responsive, date palm metallothionein 2A (PdMT2A) conferred salt tolerance in transgenic experiments on Saccharomyces cerevisiae and Arabidopsis thaliana [125].
Over the last decade, several strategies for improving photosynthesis and crop yield have been extensively discussed, including leaf morphology and light interception. Leaf morphology and physiology are inextricably linked to light capture efficiency and temperature regulation. The leaf-specific transcription is related to alpha-form rubisco activase (AM 33275). Increased rubisco activase levels may thus improve photosynthetic efficiency by increasing the amount of rubisco activated for CO2 fixation.
The antioxidant capacity of A. marina leaves must be sufficient to defend cellular integrity, especially of cellular membranes, in the face of seawater immersion from tidal fluxes. Visibly dehydrated salts from tidal fluctuations are regularly observed on A. marina leaves, as well as large salt crystals from cellular efflux from salt glands. A transcript coding for a catalase was highly expressed in leaves (AM_25336; TPM = 4181). Similar catalases facilitate radical detoxification (e.g., H2O2 removal [46,126,127,128,129]). A ferric-chelate reductase expressed at high levels (AM_22465; leaf TPM = 2578) may also be involved in maintaining redox homeostasis in leaves.
3.6. Stem-Specific Transcript Expression
Stems had the highest number of genes expressed at >10 TPM (n = 15,802 transcripts). Gene ontologies with high counts of contributing PFAMs were seen, indicating that A. marina stems retain a generalist expression profile. The expression profile in stem tissue represented an intermediate state between leaf and flower expression profiles (also see Figure S1). The correlation of expression patterns in these three tissues may be due to their relatively dry lifestyle; in contrast, pneumatophores and seeds endure long seawater immersion periods. Metabolic pathways related to the mitochondrial beta-oxidation pathway were prominently featured among the unique KEGG terms. This process is a major source of energy from the degradation of fatty acids, which are abundant in stems and seeds.
Many highly expressed, stem-specific transcripts had putative antioxidant functions (Table S1). Although metallothioneins (MTs) were among the most highly expressed genes in all five tissues, stems had the widest variety of MT types expressed. The highest-expressed stem transcript, an MT-2 (AM_26466; TPM = 30,600), had more than twice as high the expression of the next-ranked gene (Uncharacterized protein; AM_04606; TPM = 4708). A BlastP search with the 262 amino acid protein from AM_04606 yielded a match with a homolog in Olea europaea var. sylvestris (E-value = 9e × 10−77, 47.04% identity) that was annotated as an acid phosphatase. An Interproscan (ebi.ac.uk/interpro) search revealed that AM_04606 was a haloacid dehydrogenase (HAD). The HAD superfamily includes a variety of detoxification-related proteins [130]. These proteins respond to heavy metal (e.g., cadmium [131]) and salinity and alkaline [37] stress in other angiosperms, and their high expression here indicates that the HAD salinity and alkaline stress response mechanisms are conserved and amplified in A. marina. The implementation of HAD in transgenic, phosphate-deficient wild-type rice (Oryza sativa) [132] led to phosphate-accumulating rice able to overcome nutrient shortages.
Dehydrins are essential for plant response and adaptation to abiotic stresses. Dehydrins are highly hydrophilic, thermostable proteins that accumulate in vegetative plant tissues during drought or salt stress, or in seeds during maturation and drying [133]. A hydrophilic, glycine-rich dehydrin (AM_31570; DHN1; TPM = 2700) was highly expressed in stem tissue. These short (~100 amino acid) proteins are induced in response to dehydration, elevated salt, and low temperature [134]. Our data suggest that they preferentially occupy hardy stem tissue in A. marina. Although two lignin-forming anionic peroxidases were unique to stems (AM_07165, AM_07166), lignin glucosyltransferases (AM_31811, AM_31812) were uniquely expressed in pneumatophores. The high expression of HAD, MT, and other detoxification and antioxidant transcripts in stems (e.g., HSP70 (AM_04996; TPM = 1107), gamma-thionin/defensin (AM_11821; TPM = 1204), and thioredoxin (AM_24260; TPM = 1870.9)) outline the genetic mechanisms for abiotic stress resistance in A. marina.
3.7. Pneumatophore-Specific Transcript Expression
The pneumatophores of A. marina develop in oxygen-poor soils and filter out salt while absorbing water, essential nutrients, and oxygen [135]. They grow as pencil-width tubes to penetrate dense, low-oxygen mud (Figure 1). We found 554 genes uniquely expressed (>10 TPM) in A. marina pneumatophores compared to the other four tissues from the Red Sea cultivar, BioProject: PRJNA591919. Transcripts with putative roles in lignin biosynthesis outline a route to hardy, successful pneumatophore formation necessary for roots in low-oxygen soils. The pneumatophore transcriptome had the unique expression of several genes with putative roles in halogen degradation, indicating a possible mechanism to degrade otherwise harmful environmental xenobiotics.
A. marina must cope with salt stress while absorbing water and nutrients from the surroundings. A key mechanism for absorbing water while repelling excess salt is ultrafiltration, a process carried out by aquaporins and similar integral membrane transporters [42,136,137,138,139,140]. Six transcripts coding for aquaporins (NIP1.1) were expressed in pneumatophores: AM_29290 and AM_02471 were expressed at >10 TPM; AM_29291, AM_24990, AM_32021, and AM_01631 were expressed at sub-threshold levels. These aquaporins likely contribute to A. marina’s uptake of water from hypersaline waters, although their full ecophysiology is yet to be elucidated. Various transporter proteins, including H+ -ATPases, ATP-binding cassette transporters, and aquaporins, were found in proteomic studies of A. officinalis membranes [141]. NIP1.1 aquaporin expression was found to contribute to hydrogen peroxide sensitivity in Arabidopsis [142]; thus, A. marina may tightly regulate the expression of its NIP1.1 isoforms to maintain osmotic balance. The low expression of four NIP1.1 genes in A. marina may reflect this delicate balance between water uptake and membrane lipid peroxidation mitigation. Aquaporins are essential for water filtration in A. marina [29,42,57,136,137,138,139,140,141,142,143]; our data suggest that low levels of expression are sufficient in natural conditions.
The Avicennia–seawater interface is dominated by thick mucilages, comprising the pneumatophore extra cellular matrix (ECM). Avicennia ECMs support the maintenance of osmotic balance between the seawater and inner tissues. A xyloglucan endo-transglycosylase (AM_08706) was uniquely expressed in pneumatophores (TPM = 134.97). These enzymes remodel cell wall hemicellulose molecules and likely help to generate salt-resistant ECMs for Avicennia.
The chemistry of mangrove root environments is highly reducing and rich in hydrogen sulfides and other toxic molecules [31,144]. Furthermore, they are highly anaerobic compared with the root environments of most other angiosperms that are waterlogged in their natural environments. For example, poplar has a robust short-term waterlogging response, invoking hydrological conductance detours in a shift to fermentative metabolism [145]. A. marina root environments are hostile to most angiosperms and necessitate robust molecular defenses by plant roots. Here, we investigated the pneumatophore-specific expression for responsible genes.
The A. marina rhizosphere is an assemblage of microorganisms exchanging nutrient and waste molecules and providing physical, stabilizing interactions for the A. marina pneumatophore system [14]. The A. marina rhizosphere is scantly studied, although microbiome interactions, and potential fauna interactions, have critical roles in A. marina pneumatophores and their greater mangrove ecosystems. The assimilation of other nutrients, especially organic molecules that may require processing for a bioactive form, is likely facilitated by halotolerant pneumatophore microbiome community members [3,14,15]. Uniquely expressed transcripts in pneumatophores included genes predicted with roles in A. marina’s rhizosphere interactions (as detailed in Reference [3], e.g., using purine (nitrogen) transporters and cytokines). An aspartate-semialdehyde dehydrogenase (ASD, AM_32125, EC 1.2.1.11) was uniquely expressed in pneumatophores (TPM = 11.7). The ASD enzyme is not well-conserved in higher plants [146,147,148,149,150,151]; but, its presence indicates potential rhizosphere feeding strategies. The ASD reaction represents a critical branch point in nitrogenous amino acid synthesis with possible involvement in nitrogenous compound transformation in relationship to surrounding microbial producers. It forms an early branch point in the biosynthetic pathway to lysine, methionine, leucine, and isoleucine from aspartate. The ASD also synthesizes microbial cell wall constituents, including diaminopimelate. Neighboring microbes may feed A. marina nitrogenous compounds with a payback of more complex molecular building blocks for specialized cell walls as in other plants [152,153,154,155]. Additionally, the biosynthesis of steroids and steroid precursor molecules was also prominently highlighted in our root-specific KEGG analyses, and these may form the building blocks for microbe–host communication in A. marina’s rhizosphere.
Protein-L-isoaspartate O-methyltransferase (AM_26147; EC:2.1.1.77) was highly expressed in pneumatophores (TPM = 110.9); PRIM proteins in Arabidopsis thaliana respond to salt and other abiotic stress [156]. Arabidopsis PRIMs promote seed longevity and germination vigor; however, a similar role in A. marina was not supported by our transcriptomic evidence. MTs are crucial components of the cellular antioxidant network [18,157]. Transgenic experiments using A. marina MTs in E. coli have considerably boosted its antioxidant capacity [119]. Our data suggest that the expression of these proteins should be considered the primary defense mechanism of A. marina against oxidative stress in each of the five tissues we sampled and in leaf tissues from cultivars in the Indian and Pacific oceans.
Although MTs were shared, high-expression outliers in most samples, a variety of other putative antioxidant genes were also highly expressed. A glutathione S-transferase (GST) transcript had relatively high expression (TPM = 4759) in pneumatophores. We previously reported the relatively high copy number of GSTs in coastal photosynthetic microbes [158]; the duplication and increased expression of GSTs may also facilitate coastal habitation. Protein families in the GO term ‘Hydroquinone:oxygen oxidoreductase activity’ were uniquely enriched in pneumatophores (Z-score = 7.69, FDR-corr. p = 0.00079). Quinone cofactors are crucial components of redox cycling; many quinone derivatives can form a redox cycle in conjunction with glutathione to generate oxygen [159]. Salt accelerates oxidative stress, and the salinity in the Red Sea can reach 40.5–41.8 ppt [13]. Strong redox buffer cycles, such as the quinone regeneration processed detected in our enrichment analysis, are necessary for oxidative stability at the tide-delimited border between anoxia and hyperoxia in A. marina pneumatophores.
3.8. Comparison of A. marina Transcriptomes across Geographies and Climates
Transcripts with the highest expression were common in A. marina samples from different geographic locations, solidifying the role of these critical proteins as supportive of A. marina growth and survival in diverse habitats (Figure 5). For example, AM_26466, a metallothionein (Huang and Wang, 2010), was highly expressed in nearly every sample. Interproscan searches with this small (72 amino acids) protein matched IPR000347, which has a class II metallothionein protein family domain (PF01439) and the GO molecular function ’metal ion binding’ (GO:0046872). A. marina leaf samples from the Red Sea, India, and Vietnam showed higher expression of AM_26466 than heavy-metal-treated lab cultivars, indicating that the high expression of this gene is inherent to A. marina’s natural transcriptome. Another short protein (AM_31570, 203 amino acids) was predicted to function as a water-responsive dehydrin. The only protein observed to be highly expressed specifically in treated (either PEG or heavy metals) A. marina leaves was AM_20119 (GO terms: ‘glycolytic process’ (GO:0006096), ‘fructose-bisphosphate aldolase activity’ (GO:0004332), and ‘catalytic activity’ (GO:0003824)). The only high-molecular-weight (HMW) protein (1662 amino acids) with considerable expression (920.5 TPM) was annotated as a multifunctional redox/glutamate synthase, expressed exclusively in Red Sea leaves. A larger protein (AM_33275) was highly expressed in Red Sea A. marina leaves as well as heavy-metal and control leaves from Shanghai. The AM_33275 protein was predicted to have ATPase activity (IPR003959, PF00004) and membrane localization (SignalP-TM).
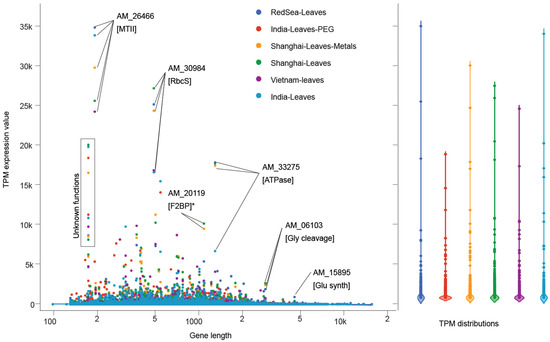
Figure 5.
Comparison of the A. marina expression profiles in distant Indo-Pacific samples. Expression values for A. marina leaves from the Red Sea and India, Shanghai, and Vietnam coasts, as well as their distributions, are shown along the y-axis. High expression outliers are noted with annotation names and functions (MTII = type-2 metallothionein; RbcS = rubisco small subunit; F2BP = fructose 2,6-bisphospatase; Gly cleavage = glycine cleavage; Glu synth = glutamine synthesis). Metallothioneins and rubisco subunits had the highest expression in most samples, indicating that an antioxidant defense system fueled by high carbon input is characteristic of A. marina independent of geography or climate.
Transcripts coding for MTs were highly expressed in A. marina leaves from distant geographies and habitats. These proteins regulate heavy metal homeostasis in the human immune system [123] and act to chelate toxic metals in plants and their microbial community members [118,120,122,124,160]. Several studies have highlighted the potential for bioaccumulation of pollutants, especially heavy metals, in the Red Sea [161,162,163,164]. The toxicity of heavy metals near the Jeddah coast has been documented in fish [161,162,163,164]. Heavy metals cause damaging oxidative stress to cells through redox interactions to form free radicals [165,166,167]. Various biomolecules, including MTs [118,120,168], function to protect cells from damaging free radicals. The ubiquitous high expression of MTs in A. marina transcriptomes from various sources highlights its importance for survival across a broad range of abiotic stressors.
We sought to discover the genetic bases for the unique phenotypes of A. marina’s organs. Overall, the RNAseq data yields insight into how tissue-specific expression forms the basis for the stress resistance and broad propagation of A. marina. Genes such as dehydrins and metallothioneins form the backbone of Avicennia’s environmental resilience, and our data shows that their expression is high across a variety of tissues and geographical locations. Our analyses showed quite divergent expression profiles in the different tissues, and this reveals that many collections of expressed genes are missed when sampling single tissues for organismal analysis. Sampling multiple tissues will need to become a more prevalent practice in mangrove studies, to establish a comprehensive overview of these model halophytes.
4. Conclusions
We investigated the transcriptomic profiles of A. marina tissues and from different ecotypes to understand the genetic basis for its ability to survive a seafaring lifestyle. The expression of genes such as MTs, dehydrins, salt efflux pumps, cytochromes, HADs, and lignin glucosyltransferases were prominent features in the tissues implicated in sustaining growth in the face of seawater immersion. The foremost expression of MTs in all samples suggests their primary role in halotolerance. Although many of the A. marina genes we described have biotechnology potential for saline agriculture, optimizing the use of transgenic MTs should provide a strong backbone for non-halotolerant plants to tolerate seawater irrigation. In conclusion, we demonstrated the distinct transcripts and functions that can be used for future research into the use of A. marina, to determine their potential application in improving plant stress tolerance. Our analyses of tissue-specific analyses of A. marina transcriptomes will constitute a valuable resource for researching specific processes, functional descriptions, and pathways.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12092030/s1, Figure S1: Circos and UMAP plots of tissue-specific transcriptomes. Figure S2: UpsetR [73] plot highlighting shared genes expressed at >10 TPM in the five tissues from the Red Sea cultivar. Figure S3: Expression patterns of lipid metabolic genes in the five Red Sea tissues. Table S1: Differentially-expressed genes mapped to the reference CDSs. Table S2: Expression values (TPM) for the mapping of external datasets (.fastqs) downloaded from NCBI. Table S3: dcGO enrichment from PFAMs in tissue-specific, uniquely-expressed transcripts (≥10 TPM). And Supplementary datasets for “Tissue-specific transcriptomes outline halophyte adaptive strategies in the gray mangrove, Avicennia marina” are available at https://zenodo.org/record/6793097#.YsJ7xexBxew (accessed on 3 July 2022).
Author Contributions
Conceptualization, K.S.-A., B.K. and D.R.N.; methodology, A.C., D.R.N., A.C. and K.S.-A.; software, A.C. and D.R.N.; formal analysis, D.R.N. and K.M.H.; investigation, D.R.N. and K.M.H.; resources, K.S.-A., J.A.B. and K.M.A.A.; data curation, A.J. and D.R.N.; writing—original draft preparation, A.A. and D.R.N.; writing—review and editing, J.A.B., G.F., D.R.N. and K.S.-A.; visualization, D.R.N.; supervision, K.S.-A., K.M.A.A. and J.A.B.; project administration, K.S.-A., J.A.B.; D.R.N. and A.C. contributed equally to this paper. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by KAUST Laboratory Baseline Funds and by New York University Abu Dhabi (NYUAD) Faculty Research fund AD060 and NYUAD Research Institute grant 73 71210 CGSB9. JB was supported by the NYUAD Water Research Center grant CG007, while GF was supported by the NYUAD-CGSB grant CGSB5.
Data Availability Statement
Supplementary Dataset S1 are available at Zenodo (10.5281/zenodo.6793097). The underlying RNAseq datasets used for the analyses in this study are publicly available. We used public RNAseq data from A. marina specimens from the Red Sea (NCBI BioProject: PRJNA591919), Vietnam (NCBI Bioproject: PRJDB6605, Department of Biotechnology, Graduate School of Engineering, Osaka University, Japan), India (NCBI Bioproject: PRJNA283781, M.S. Swaminathan Research Foundation, Chennai, India), and China (NCBI Bioproject: PRJNA350568, Lingnan Normal University, Zhanjiang, China).
Acknowledgments
We thank the New York University Abu Dhabi High-Performance Computing Facility for computational resources. We thank Alexey Sergeev for permission to use the Avicennia photographs and NASA for the Jeddah satellite image. This manuscript is dedicated to the memory of Basel Khraiwesh.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cavanaugh, K.C.; Dangremond, E.M.; Doughty, C.L.; Williams, A.P.; Parker, J.D.; Hayes, M.A.; Rodriguez, W.; Feller, I.C. Climate-driven regime shifts in a mangrove-salt marsh ecotone over the past 250 years. Proc. Natl. Acad. Sci. USA 2019, 116, 21602–21608. [Google Scholar] [CrossRef]
- Charles, S.P.; Kominoski, J.S.; Armitage, A.R.; Guo, H.; Weaver, C.A.; Pennings, S.C. Quantifying how changing mangrove cover affects ecosystem carbon storage in coastal wetlands. Ecology 2019, 101, e02916. [Google Scholar] [CrossRef] [PubMed]
- Inoue, T.; Shimono, A.; Akaji, Y.; Baba, S.; Takenaka, A.; Tuck Chan, H. Mangrove-diazotroph relationships at the root, tree and forest scales: Diazotrophic communities create high soil nitrogenase activities in Rhizophora stylosa rhizospheres. Ann. Bot. 2019, 125, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Sasmito, S.D.; Taillardat, P.; Clendenning, J.N.; Cameron, C.; Friess, D.A.; Murdiyarso, D.; Hutley, L.B. Effect of land-use and land-cover change on mangrove blue carbon: A systematic review. Glob. Change Biol. 2019, 25, 4291–4302. [Google Scholar] [CrossRef] [PubMed]
- Theuerkauff, D.; Rivera-Ingraham, G.A.; Lambert, S.; Mercky, Y.; Lejeune, M.; Lignot, J.H.; Sucre, E. Wastewater bioremediation by mangrove ecosystems impacts crab ecophysiology: In-situ caging experiment. Aquat. Toxicol. 2019, 218, 105358. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Li, X.; Yang, M.; Wang, X.; Zhong, C.; Duke, N.C.; Wu, C.-I.; Shi, S. Speciation with gene flow via cycles of isolation and migration: Insights from multiple mangrove taxa. Natl. Sci. Rev. 2019, 6, 275–288. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Xu, S.; Zhang, Z.; Guo, W.; Lyu, H.; Zhong, C.; Boufford, D.E.; Duke, N.C.; Shi, S.; The International Mangrove Consortium. Convergent adaptation of the genomes of woody plants at the land–sea interface. Natl. Sci. Rev. 2020, 7, 978–993. [Google Scholar] [CrossRef]
- Arnaud-Haond, S.; Teixeira, S.; Massa, S.I.; Billot, C.; Saenger, P.; Coupland, G.; Duarte, C.M.; Serrão, E.A. Genetic structure at range edge: Low diversity and high inbreeding in Southeast Asian mangrove (Avicennia marina) populations. Mol. Ecol. 2006, 15, 3515–3525. [Google Scholar] [CrossRef]
- Dodd, R.S.; Afzal-Rafii, Z.; Kashani, N.; Budrick, J. Land barriers and open oceans: Effects on gene diversity and population structure in Avicennia germinans L. (Avicenniaceae). Mol. Ecol. 2002, 11, 1327–1338. [Google Scholar] [CrossRef]
- Hazarika, D.; Thangaraj, M.; Sahu, S.K.; Kathiresan, K. Genetic diversity in three populations of Avicennia marina along the eastcoast of India by RAPD markers. J. Environ. Biol. 2013, 34, 663–666. [Google Scholar]
- Maguire, T.L.; Peakall, R.; Saenger, P. Comparative analysis of genetic diversity in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae) detected by AFLPs and SSRs. Theor. Appl. Genet. 2002, 104, 388–398. [Google Scholar] [CrossRef]
- Friis, G.; Burt, J.A. Evolution of mangrove research in an extreme environment: Historical trends and future opportunities in Arabia. Ocean Coast. Manag. 2020, 195, 105288. [Google Scholar] [CrossRef]
- Almahasheer, H.; Duarte, C.M.; Irigoien, X. Phenology and Growth dynamics of Avicennia marina in the Central Red Sea. Sci. Rep. 2016, 6, 37785. [Google Scholar] [CrossRef]
- Alzubaidy, H.; Essack, M.; Malas, T.B.; Bokhari, A.; Motwalli, O.; Kamanu, F.K.; Jamhor, S.A.; Mokhtar, N.A.; Antunes, A.; Simoes, M.F.; et al. Rhizosphere microbiome metagenomics of gray mangroves (Avicennia marina) in the Red Sea. Gene 2016, 576, 626–636. [Google Scholar] [CrossRef]
- Chen, C.; Han, S.; Zhu, Z.; Fu, G.; Wang, R.; Zhang, Q.; Ye, Y.; Ren, Y.; Yan, C.; Xu, L.; et al. Idiomarina mangrovi sp. nov., isolated from rhizosphere soil of a mangrove Avicennia marina forest. Int. J. Syst. Evol. Microbiol. 2019, 69, 1662–1668. [Google Scholar] [CrossRef]
- Clarke, P.J.; Allaway, W.G. The regeneration niche of the grey mangrove (Avicennia marina): Effects of salinity, light and sediment factors on establishment, growth and survival in the field. Oecologia 1993, 93, 548–556. [Google Scholar] [CrossRef]
- Ganesan, G.; Sankararamasubramanian, H.; Narayanan, J.M.; Sivaprakash, K.; Parida, A. Transcript level characterization of a cDNA encoding stress regulated NAC transcription factor in the mangrove plant Avicennia marina. Plant Physiol. Biochem. 2008, 46, 928–934. [Google Scholar] [CrossRef]
- Huang, G.Y.; Wang, Y.S. Expression and characterization analysis of type 2 metallothionein from grey mangrove species (Avicennia marina) in response to metal stress. Aquat. Toxicol. 2010, 99, 86–92. [Google Scholar] [CrossRef]
- Khraiwesh, B.; Pugalenthi, G.; Fedoroff, N.V. Identification and analysis of red sea mangrove (Avicennia marina) microRNAs by high-throughput sequencing and their association with stress responses. PloS ONE 2013, 8, e60774. [Google Scholar] [CrossRef]
- Schmitz, N.; Robert, E.M.; Verheyden, A.; Kairo, J.G.; Beeckman, H.; Koedam, N. A patchy growth via successive and simultaneous cambia: Key to success of the most widespread mangrove species Avicennia marina? Ann. Bot. 2008, 101, 49–58. [Google Scholar] [CrossRef]
- Simoes, M.F.; Antunes, A.; Ottoni, C.A.; Amini, M.S.; Alam, I.; Alzubaidy, H.; Mokhtar, N.A.; Archer, J.A.; Bajic, V.B. Soil and Rhizosphere Associated Fungi in Gray Mangroves (Avicennia marina) from the Red Sea--A Metagenomic Approach. Genom. Proteom. Bioinform. 2015, 13, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Usman, A.R.; Alkredaa, R.S.; Al-Wabel, M.I. Heavy metal contamination in sediments and mangroves from the coast of Red Sea: Avicennia marina as potential metal bioaccumulator. Ecotoxicol. Environ. Saf. 2013, 97, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Mildenhall, D.C.; Brown, L.J. An early Holocene occurrence of the mangrove Avicennia marina in Poverty Bay, North Island, New Zealand: Its climatic and geological implications. N. Z. J. Bot. 1987, 25, 281–294. [Google Scholar] [CrossRef]
- Schmitz, N.; Verheyden, A.; Kairo, J.G.; Beeckman, H.; Koedam, N. Successive cambia development in Avicennia marina (Forssk.) Vierh. is not climatically driven in the seasonal climate at Gazi Bay, Kenya. Dendrochronologia 2007, 25, 87–96. [Google Scholar] [CrossRef]
- Cheng, B.; Li, H. Impact of climate change and human activities on economic values produced by ecosystem service functions of rivers in water shortage area of Northwest China. Environ. Sci. Pollut. Res. Int. 2020, 27, 26570–26578. [Google Scholar] [CrossRef]
- Cheng, H.; Inyang, A.; Li, C.D.; Fei, J.; Zhou, Y.W.; Wang, Y.S. Salt tolerance and exclusion in the mangrove plant Avicennia marina in relation to root apoplastic barriers. Ecotoxicology 2020, 29, 676–683. [Google Scholar] [CrossRef]
- Jyothi-Prakash, P.A.; Mohanty, B.; Wijaya, E.; Lim, T.-M.; Lin, Q.; Loh, C.-S.; Kumar, P.P. Identification of salt gland-associated genes and characterization of a dehydrin from the salt secretor mangrove Avicennia officinalis. BMC Plant Biol. 2014, 14, 291. [Google Scholar] [CrossRef]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef]
- Tan, W.-K.; Lin, Q.; Lim, T.-M.; Kumar, P.; Loh, C.-S. Dynamic secretion changes in the salt glands of the mangrove tree species Avicennia officinalis in response to a changing saline environment. Plant Cell Environ. 2013, 36, 1410–1422. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Mohanty, B.; Wijaya, E.; Lee, D.Y.; Lim, T.M.; Lin, Q.; Xu, J.; Loh, C.S.; Kumar, P.P. Transcriptomics analysis of salt stress tolerance in the roots of the mangrove Avicennia officinalis. Sci. Rep. 2017, 7, 10031. [Google Scholar] [CrossRef]
- Kryger, L.; Lee, S.K. Effects of Mangrove Soil Ageing on the Accumulation of Hydrogen Sulphide in Roots of Avicennia spp. Biogeochemistry 1996, 35, 367–375. [Google Scholar] [CrossRef]
- Arshad, M.; Eid, E.M.; Hasan, M. Mangrove health along the hyper-arid southern Red Sea coast of Saudi Arabia. Environ. Monit. Assess. 2020, 192, 189. [Google Scholar] [CrossRef]
- Masoud, M.S.; Abdel-Halim, A.M.; El Ashmawy, A.A. Seasonal variation of nutrient salts and heavy metals in mangrove (Avicennia marina) environment, Red Sea, Egypt. Environ. Monit. Assess. 2019, 191, 425. [Google Scholar] [CrossRef]
- Gao, X.; Liu, Y.; Sun, B. Water shortage risk assessment considering large-scale regional transfers: A copula-based uncertainty case study in Lunan, China. Environ. Sci. Pollut. Res. Int. 2018, 25, 23328–23341. [Google Scholar] [CrossRef]
- Kummu, M.; Guillaume, J.H.; de Moel, H.; Eisner, S.; Florke, M.; Porkka, M.; Siebert, S.; Veldkamp, T.I.; Ward, P.J. The world’s road to water scarcity: Shortage and stress in the 20th century and pathways towards sustainability. Sci. Rep. 2016, 6, 38495. [Google Scholar] [CrossRef]
- Nabi, G.; Ali, M.; Khan, S.; Kumar, S. The crisis of water shortage and pollution in Pakistan: Risk to public health, biodiversity, and ecosystem. Environ. Sci. Pollut. Res. Int. 2019, 26, 10443–10445. [Google Scholar] [CrossRef]
- Xu, Z.; Cai, X.; Yin, X.; Su, M.; Wu, Y.; Yang, Z. Is water shortage risk decreased at the expense of deteriorating water quality in a large water supply reservoir? Water Res. 2019, 165, 114984. [Google Scholar] [CrossRef]
- Hayat, K.; Zhou, Y.; Menhas, S.; Bundschuh, J.; Hayat, S.; Ullah, A.; Wang, J.; Chen, X.; Zhang, D.; Zhou, P. Pennisetum giganteum: An emerging salt accumulating/tolerant non-conventional crop for sustainable saline agriculture and simultaneous phytoremediation. Environ. Pollut. 2020, 265, 114876. [Google Scholar] [CrossRef]
- Illera-Vives, M.; Lopez-Mosquera, M.E.; Salas-Sanjuan Mdel, C.; Lopez-Fabal, A. Leaching techniques for saline wastes composts used as growing media in organic agriculture: Assessment and modelling. Environ. Sci. Pollut. Res. Int. 2015, 22, 6854–6863. [Google Scholar] [CrossRef]
- Jin, Y.; Weining, S.; Nevo, E. A MAPK gene from Dead Sea fungus confers stress tolerance to lithium salt and freezing-thawing: Prospects for saline agriculture. Proc. Natl. Acad. Sci. USA 2005, 102, 18992–18997. [Google Scholar] [CrossRef]
- Qadir, M.; Oster, J.D. Crop and irrigation management strategies for saline-sodic soils and waters aimed at environmentally sustainable agriculture. Sci. Total Environ. 2004, 323, 1–19. [Google Scholar] [CrossRef]
- Qin, S.; Liu, Y.; Han, Y.; Xu, G.; Wan, S.; Cui, F.; Li, G. Aquaporins and their function in root water transport under salt stress conditions in Eutrema salsugineum. Plant Sci. 2019, 287, 110199. [Google Scholar] [CrossRef]
- Guo, R.Z.; Yan, H.Y.; Li, X.X.; Zou, X.X.; Zhang, X.J.; Yu, X.N.; Ci, D.W.; Wang, Y.F.; Si, T. Green leaf volatile (Z)-3-hexeny-1-yl acetate reduces salt stress in peanut by affecting photosynthesis and cellular redox homeostasis. Physiol. Plant 2020, 10, 785. [Google Scholar] [CrossRef]
- Lekklar, C.; Suriya-Arunroj, D.; Pongpanich, M.; Comai, L.; Kositsup, B.; Chadchawan, S.; Buaboocha, T. Comparative genomic analysis of rice with contrasting photosynthesis and grain production under salt stress. Genes 2019, 10, 562. [Google Scholar] [CrossRef]
- Nounjan, N.; Chansongkrow, P.; Charoensawan, V.; Siangliw, J.L.; Toojinda, T.; Chadchawan, S.; Theerakulpisut, P. High Performance of Photosynthesis and Osmotic Adjustment Are Associated With Salt Tolerance Ability in Rice Carrying Drought Tolerance QTL: Physiological and Co-expression Network Analysis. Front. Plant Sci. 2018, 9, 1135. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, H.; Ramesh, A.; Roberts, L.J., 2nd; Zhou, L.; Lin, X.; Zhao, Y.; Guo, Z. Overexpression of Cu/Zn-superoxide dismutase and/or catalase accelerates benzo(a)pyrene detoxification by upregulation of the aryl hydrocarbon receptor in mouse endothelial cells. Free Radic. Biol. Med. 2009, 47, 1221–1229. [Google Scholar] [CrossRef]
- Wasai, S.; Kanno, N.; Matsuura, K.; Haruta, S. Increase of Salt Tolerance in Carbon-Starved Cells of Rhodopseudomonas palustris Depending on Photosynthesis or Respiration. Microorganisms 2018, 6, 4. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, X.; Cui, L.; Meng, S.; Ye, N.; Peng, X. Catalytic and functional aspects of different isozymes of glycolate oxidase in rice. BMC Plant Biol. 2017, 17, 135. [Google Scholar] [CrossRef]
- Chang, B.; Yang, L.; Cong, W.; Zu, Y.; Tang, Z. The improved resistance to high salinity induced by trehalose is associated with ionic regulation and osmotic adjustment in Catharanthus roseus. Plant Physiol. Biochem. 2014, 77, 140–148. [Google Scholar] [CrossRef]
- Frank, W.; Ratnadewi, D.; Reski, R. Physcomitrella patens is highly tolerant against drought, salt and osmotic stress. Planta 2005, 220, 384–394. [Google Scholar] [CrossRef]
- Pires, R.M.O.; Avila, M.A.B.; Leite, D.G.; Santos, H.O.; Souza, G.A.; Von Pinho, E.V.R. Physiological and enzymatic alterations in sesame seeds submitted to different osmotic potentials. Genet. Mol. Res. 2017, 16, gmr16039425. [Google Scholar] [CrossRef] [PubMed]
- Empitu, M.A.; Kadariswantiningsih, I.N. Modelling salt transport disorders of human kidney in zebrafish: The grain of salt. J. Physiol. 2019, 597, 5529–5530. [Google Scholar] [CrossRef] [PubMed]
- Grosell, M.; Heuer, R.M.; Wu, N.C.; Cramp, R.L.; Wang, Y.; Mager, E.M.; Dwyer, R.G.; Franklin, C.E. Salt-water acclimation of the estuarine crocodile Crocodylus porosus involves enhanced ion transport properties of the urodaeum and rectum. J. Exp. Biol. 2020, 223, jeb210732. [Google Scholar] [CrossRef] [PubMed]
- Stolting, G.; Fahlke, C. Chloride channels in renal salt and water transport. Acta Physiol. 2017, 219, 11–13. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z. The Importance of Cl− Exclusion and Vacuolar Cl− Sequestration: Revisiting the Role of Cl− Transport in Plant Salt Tolerance. Front. Plant Sci. 2019, 10, 1418. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ Balance and Transport Regulatory Mechanisms in Weedy and Cultivated Rice (Oryza sativa L.) Under Salt Stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, J.; Ahammed, G.J.; Li, J.; Yang, Y. Genome-wide identification and expression analysis of aquaporin gene family related to abiotic stress in watermelon. Genome 2019, 62, 643–656. [Google Scholar] [CrossRef]
- Severi, E.; Hosie, A.H.; Hawkhead, J.A.; Thomas, G.H. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol. Lett. 2010, 304, 47–54. [Google Scholar] [CrossRef]
- Friis, G.; Vizueta, J.; Smith, E.G.; Nelson, D.R.; Khraiwesh, B.; Qudeimat, E.; Salehi-Ashtiani, K.; Ortega, A.; Marshell, A.; Duarte, C.M.; et al. A high-quality genome assembly and annotation of the gray mangrove, Avicennia marina. G3 Genes Genomes Genet. 2021, 11, jkaa025. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Meth. 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szczesniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Lee, S.; Seo, C.H.; Lim, B.; Yang, J.O.; Oh, J.; Kim, M.; Lee, S.; Lee, B.; Kang, C.; Lee, S. Accurate quantification of transcriptome from RNA-Seq data by effective length normalization. Nucleic Acids Res. 2011, 39, e9. [Google Scholar] [CrossRef]
- Li, P.; Piao, Y.; Shon, H.S.; Ryu, K.H. Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data. BMC Bioinform. 2015, 16, 347. [Google Scholar] [CrossRef]
- Wu, P.Y.; Phan, J.H.; Zhou, F.; Wang, M.D. Evaluation of Normalization Methods for RNA-Seq Gene Expression Estimation. In Proceedings of the 2011 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), Atlanta, GA, USA, 12–15 November 2011; pp. 50–57. [Google Scholar] [CrossRef]
- Zyprych-Walczak, J.; Szabelska, A.; Handschuh, L.; Gorczak, K.; Klamecka, K.; Figlerowicz, M.; Siatkowski, I. The Impact of Normalization Methods on RNA-Seq Data Analysis. Biomed. Res. Int. 2015, 2015, 621690. [Google Scholar] [CrossRef]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, A.; Moeller, S. gplots: Various R Programming Tools for Plotting Data. R Package Version 2009, 2. Available online: https://scholar.google.com/citations?view_op=view_citation&hl=en&user=o7GsfJwAAAAJ&citation_for_view=o7GsfJwAAAAJ:u-x6o8ySG0sC (accessed on 3 July 2022).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef]
- Fang, H.; Gough, J. A domain-centric solution to functional genomics via dcGO Predictor. BMC Bioinform. 2013, 14 (Suppl. S3), S9. [Google Scholar] [CrossRef]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Sonnhammer, E.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Landry, C.L. Pollinator-mediated competition between two co-flowering Neotropical mangrove species, Avicennia germinans (Avicenniaceae) and Laguncularia racemosa (Combretaceae). Ann. Bot. 2013, 111, 207–214. [Google Scholar] [CrossRef]
- Liu, L.; Guo, Z.; Zhong, C.; Shi, S. DNA barcoding reveals insect diversity in the mangrove ecosystems of Hainan Island, China. Genome 2018, 61, 797–806. [Google Scholar] [CrossRef]
- De Vetten, N.; ter Horst, J.; van Schaik, H.P.; de Boer, A.; Mol, J.; Koes, R. A cytochrome b5 is required for full activity of flavonoid 3', 5'-hydroxylase, a cytochrome P450 involved in the formation of blue flower colors. Proc. Natl. Acad. Sci. USA 1999, 96, 778–783. [Google Scholar] [CrossRef]
- Holton, T.A.; Brugliera, F.; Lester, D.R.; Tanaka, Y.; Hyland, C.D.; Menting, J.G.; Lu, C.Y.; Farcy, E.; Stevenson, T.W.; Cornish, E.C. Cloning and expression of cytochrome P450 genes controlling flower colour. Nature 1993, 366, 276–279. [Google Scholar] [CrossRef]
- Liu, Z.; Boachon, B.; Lugan, R.; Tavares, R.; Erhardt, M.; Mutterer, J.; Demais, V.; Pateyron, S.; Brunaud, V.; Ohnishi, T.; et al. A Conserved Cytochrome P450 Evolved in Seed Plants Regulates Flower Maturation. Mol. Plant. 2015, 8, 1751–1765. [Google Scholar] [CrossRef]
- Martsinkovskaya, A.I.; Poghosyan, Z.P.; Haralampidis, K.; Murphy, D.J.; Hatzopoulos, P. Temporal and spatial gene expression of cytochrome B5 during flower and fruit development in olives. Plant Mol. Biol. 1999, 40, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Petkova-Andonova, M.; Imaishi, H.; Ohkawa, H. CYP92B1, A cytochrome P450, expressed in petunia flower buds, that catalyzes monooxidation of long-chain fatty acids. Biosci. Biotechnol. Biochem. 2002, 66, 1819–1828. [Google Scholar] [CrossRef] [PubMed]
- Shahak, Y.; Posner, H.B.; Avron, M. Evidence for a Block between Plastoquinone and Cytochrome f in a Photosynthetic Mutant of Lemna with Abnormal Flowering Behavior. Plant Physiol. 1976, 57, 577–579. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ito, A.; Saito, T.; Nishijima, T.; Moriguchi, T. Effect of extending the photoperiod with low-intensity red or far-red light on the timing of shoot elongation and flower-bud formation of 1-year-old Japanese pear (Pyrus pyrifolia). Tree Physiol. 2014, 34, 534–546. [Google Scholar] [CrossRef]
- Koornneef, M. Plant development: Timing when to flower. Curr. Biol. 1997, 7, R651–652. [Google Scholar] [CrossRef]
- Lord, E.M.; Crone, W.; Hill, J.P. Timing of events during flower organogenesis: Arabidopsis as a model system. Curr. Top Dev. Biol. 1994, 29, 325–356. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D. Flower morphogenesis: Timing is key. Dev. Cell. 2009, 16, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Davis, E.J.; Ballif, J.; Liang, M.; Bushman, E.; Haroldsen, V.; Torabinejad, J.; Wu, Y. Mutant identification and characterization of the laccase gene family in Arabidopsis. J. Exp. Bot. 2006, 57, 2563–2569. [Google Scholar] [CrossRef] [PubMed]
- Salekeen, R.; Mou, S.N.; Islam, M.E.; Ahmed, A.; Billah, M.M.; Rahman, S.M.M.; Islam, K.M.D. Predicting multi-enzyme inhibition in the arachidonic acid metabolic network by Heritiera fomes extracts. J. Biomol. Struct. Dyn. 2022, 40, 4259–4272. [Google Scholar] [CrossRef]
- Aubert, D.; Chevillard, M.; Dorne, A.M.; Arlaud, G.; Herzog, M. Expression patterns of GASA genes in Arabidopsis thaliana: The GASA4 gene is up-regulated by gibberellins in meristematic regions. Plant Mol. Biol. 1998, 36, 871–883. [Google Scholar] [CrossRef]
- Roxrud, I.; Lid, S.E.; Fletcher, J.C.; Schmidt, E.D.; Opsahl-Sorteberg, H.G. GASA4, one of the 14-member Arabidopsis GASA family of small polypeptides, regulates flowering and seed development. Plant Cell Physiol. 2007, 48, 471–483. [Google Scholar] [CrossRef]
- Rubinovich, L.; Weiss, D. The Arabidopsis cysteine-rich protein GASA4 promotes GA responses and exhibits redox activity in bacteria and in planta. Plant J. Cell Mol. Biol. 2010, 64, 1018–1027. [Google Scholar] [CrossRef]
- Carvalho, A.T.; Dotterl, S.; Schlindwein, C. An aromatic volatile attracts oligolectic bee pollinators in an interdependent bee-plant relationship. J. Chem. Ecol. 2014, 40, 1126–1134. [Google Scholar] [CrossRef]
- Ilc, T.; Parage, C.; Boachon, B.; Navrot, N.; Werck-Reichhart, D. Monoterpenol Oxidative Metabolism: Role in Plant Adaptation and Potential Applications. Front. Plant Sci. 2016, 7, 509. [Google Scholar] [CrossRef]
- Awadasseid, A.; Li, W.; Liu, Z.; Qiao, C.; Pang, J.; Zhang, G.; Luo, Y. Characterization of Camptotheca acuminata 10-Hydroxygeraniol oxidoreductase and iridoid synthase and their application in biological preparation of nepetalactol in Escherichia coli featuring NADP+-NADPH cofactors recycling. Int. J. Biol. Macromol. 2020, 162, 1076–1085. [Google Scholar] [CrossRef]
- Carbonezi, C.A.; Martins, D.; Young, M.C.; Lopes, M.N.; Furlan, M.; Rodriguez Filho, E.; Bolzani Vda, S. Iridoid and seco-iridoid glucosides from Chioccoca alba (Rubiaceae). Phytochemistry 1999, 51, 781–785. [Google Scholar] [CrossRef]
- Hofer, R.; Dong, L.; Andre, F.; Ginglinger, J.F.; Lugan, R.; Gavira, C.; Grec, S.; Lang, G.; Memelink, J.; Van der Krol, S.; et al. Geraniol hydroxylase and hydroxygeraniol oxidase activities of the CYP76 family of cytochrome P450 enzymes and potential for engineering the early steps of the (seco)iridoid pathway. Metab. Eng. 2013, 20, 221–232. [Google Scholar] [CrossRef]
- Miettinen, K.; Dong, L.; Navrot, N.; Schneider, T.; Burlat, V.; Pollier, J.; Woittiez, L.; van der Krol, S.; Lugan, R.; Ilc, T.; et al. The seco-iridoid pathway from Catharanthus roseus. Nat. Commun. 2014, 5, 3606. [Google Scholar] [CrossRef] [PubMed]
- Valletta, A.; Trainotti, L.; Santamaria, A.R.; Pasqua, G. Cell-specific expression of tryptophan decarboxylase and 10-hydroxygeraniol oxidoreductase, key genes involved in camptothecin biosynthesis in Camptotheca acuminata Decne (Nyssaceae). BMC Plant Biol. 2010, 10, 69. [Google Scholar] [CrossRef]
- Willems, M. Quantitative Determination of Seco-Iridoid Glucosides from the Fruits of Ligustrum vulgare by HPLC. Planta Med. 1988, 54, 66–68. [Google Scholar] [CrossRef]
- Do, B.T.N.; Koedam, N.; Triest, L. Avicennia marina maintains genetic structure whereas Rhizophora stylosa connects mangroves in a flooded, former inner sea (Vietnam). Estuar. Coast. Shelf Sci. 2019, 222, 195–204. [Google Scholar] [CrossRef]
- Geng, Q.; Wang, Z.; Tao, J.; Kimura, M.K.; Liu, H.; Hogetsu, T.; Lian, C. Ocean Currents Drove Genetic Structure of Seven Dominant Mangrove Species Along the Coastlines of Southern China. Front. Genet. 2021, 12, 615911. [Google Scholar] [CrossRef] [PubMed]
- Rademacher, S.; Sprunck, S. Downregulation of egg cell-secreted EC1 is accompanied with delayed gamete fusion and polytubey. Plant Signal. Behav. 2013, 8, e27377. [Google Scholar] [CrossRef] [PubMed]
- Sprunck, S.; Rademacher, S.; Vogler, F.; Gheyselinck, J.; Grossniklaus, U.; Dresselhaus, T. Egg cell-secreted EC1 triggers sperm cell activation during double fertilization. Science 2012, 338, 1093–1097. [Google Scholar] [CrossRef] [PubMed]
- Deveson, I.W.; Holleley, C.E.; Blackburn, J.; Marshall Graves, J.A.; Mattick, J.S.; Waters, P.D.; Georges, A. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 2017, 3, e1700731. [Google Scholar] [CrossRef] [PubMed]
- Gan, E.S.; Xu, Y.; Wong, J.Y.; Goh, J.G.; Sun, B.; Wee, W.Y.; Huang, J.; Ito, T. Jumonji demethylases moderate precocious flowering at elevated temperature via regulation of FLC in Arabidopsis. Nat. Commun. 2014, 5, 5098. [Google Scholar] [CrossRef]
- Zheng, S.; Hu, H.; Ren, H.; Yang, Z.; Qiu, Q.; Qi, W.; Liu, X.; Chen, X.; Cui, X.; Li, S.; et al. The Arabidopsis H3K27me3 demethylase JUMONJI 13 is a temperature and photoperiod dependent flowering repressor. Nat Commun 2019, 10, 1303. [Google Scholar] [CrossRef]
- Leon, K.E.; Aird, K.M. Jumonji C Demethylases in Cellular Senescence. Genes 2019, 10, 33. [Google Scholar] [CrossRef]
- Bentz, A.B.; Thomas, G.W.C.; Rusch, D.B.; Rosvall, K.A. Tissue-specific expression profiles and positive selection analysis in the tree swallow (Tachycineta bicolor) using a de novo transcriptome assembly. Sci. Rep. 2019, 9, 15849. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Penin, A.A. Gene Expression Maps in Plants: Current State and Prospects. Plants 2019, 8, 309. [Google Scholar] [CrossRef]
- Kubínová, Z.; Janáček, J.; Lhotáková, Z.; Kubínová, L.; Albrechtová, J. Unbiased estimation of chloroplast number in mesophyll cells: Advantage of a genuine three-dimensional approach. J. Exp. Bot. 2014, 65, 609–620. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Lunde, C.; Drew, D.P.; Jacobs, A.K.; Tester, M. Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress. Plant Physiol. 2007, 144, 1786–1796. [Google Scholar] [CrossRef][Green Version]
- Qudeimat, E.; Frank, W. Ca2+ signatures: The role of Ca2+-ATPases. Plant Signal. Behav. 2009, 4, 350–352. [Google Scholar] [CrossRef][Green Version]
- Cui, L.-L.; Lu, Y.-S.; Li, Y.; Yang, C.; Peng, X.-X. Overexpression of Glycolate Oxidase Confers Improved Photosynthesis under High Light and High Temperature in Rice. Front. Plant Sci. 2016, 7, 1165. [Google Scholar] [CrossRef]
- Fernandez, L.R.; Vandenbussche, G.; Roosens, N.; Govaerts, C.; Goormaghtigh, E.; Verbruggen, N. Metal binding properties and structure of a type III metallothionein from the metal hyperaccumulator plant Noccaea caerulescens. Biochim. Biophys. Acta 2012, 1824, 1016–1023. [Google Scholar] [CrossRef]
- Babaei-Bondarti, Z.; Shahpiri, A. A metallothionein type 2 from Avicennia marina binds to iron and mediates hydrogen peroxide balance by activation of enzyme catalase. Phytochemistry 2020, 176, 112396. [Google Scholar] [CrossRef]
- Freisinger, E. Plant MTs-long neglected members of the metallothionein superfamily. Dalton. Trans. 2008, 6663–6675. [Google Scholar] [CrossRef]
- Pirzadeh, S.; Shahpiri, A. Functional characterization of a type 2 metallothionein isoform (OsMTI-2b) from rice. Int. J. Biol. Macromol. 2016, 88, 491–496. [Google Scholar] [CrossRef]
- Cabral, A.C.S.; Jakovleska, J.; Deb, A.; Penner-Hahn, J.E.; Pecoraro, V.L.; Freisinger, E. Further insights into the metal ion binding abilities and the metalation pathway of a plant metallothionein from Musa acuminata. J. Biol. Inorg. Chem. 2018, 23, 91–107. [Google Scholar] [CrossRef]
- Rice, J.M.; Zweifach, A.; Lynes, M.A. Metallothionein regulates intracellular zinc signaling during CD4+ T cell activation. BMC Immunol. 2016, 17, 13. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Tripathi, S.; Shanker, K.; Sharma, A. Cadmium-induced conformational changes in type 2 metallothionein of medicinal plant Coptis japonica: Insights from molecular dynamics studies of apo, partially and fully metalated forms. J. Biomol. Struct. Dyn. 2019, 37, 1520–1533. [Google Scholar] [CrossRef] [PubMed]
- Patankar, H.V.; Al-Harrasi, I.; Al Kharusi, L.; Jana, G.A.; Al-Yahyai, R.; Sunkar, R.; Yaish, M.W. Overexpression of a Metallothionein 2A Gene from Date Palm Confers Abiotic Stress Tolerance to Yeast and Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 2871. [Google Scholar] [CrossRef] [PubMed]
- Hamelet, J.; Seltzer, V.; Petit, E.; Noll, C.; Andreau, K.; Delabar, J.M.; Janel, N. Cystathionine beta synthase deficiency induces catalase-mediated hydrogen peroxide detoxification in mice liver. Biochim. Biophys. Acta 2008, 1782, 482–488. [Google Scholar] [CrossRef] [PubMed]
- Pheomphun, P.; Treesubsuntorn, C.; Jitareerat, P.; Thiravetyan, P. Contribution of Bacillus cereus ERBP in ozone detoxification by Zamioculcas zamiifolia plants: Effect of ascorbate peroxidase, catalase and total flavonoid contents for ozone detoxification. Ecotoxicol. Environ. Saf. 2019, 171, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Sahoo, R.; Bhattacharjee, A.; Majumdar, U.; Ray, S.S.; Dutta, T.; Ghosh, S. A novel role of catalase in detoxification of peroxynitrite in S. cerevisiae. Biochem. Biophys. Res. Commun. 2009, 385, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Stührwohldt, N.; Bühler, E.; Sauter, M.; Schaller, A. Phytosulfokine (PSK) precursor processing by subtilase SBT3.8 and PSK signaling improve drought stress tolerance in Arabidopsis. J. Exp. Bot. 2021, 72, 3427–3440. [Google Scholar] [CrossRef]
- Koonin, E.V.; Tatusov, R.L. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J. Mol. Biol. 1994, 244, 125–132. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, X.; He, X.; Lü, Q.; Qian, X.; Xiao, Q.; Lin, R. Effects of Pseudomonas TCd-1 on rice (Oryza sativa) cadmium uptake, rhizosphere soils enzyme activities and cadmium bioavailability under cadmium contamination. Ecotoxicol. Environ. Saf. 2021, 218, 112249. [Google Scholar] [CrossRef]
- Pandey, B.K.; Mehra, P.; Verma, L.; Bhadouria, J.; Giri, J. OsHAD1, a Haloacid Dehalogenase-Like APase, Enhances Phosphate Accumulation. Plant Physiol. 2017, 174, 2316–2332. [Google Scholar] [CrossRef]
- Hanin, M.; Brini, F.; Ebel, C.; Toda, Y.; Takeda, S.; Masmoudi, K. Plant dehydrins and stress tolerance: Versatile proteins for complex mechanisms. Plant Signal. Behav. 2011, 6, 1503–1509. [Google Scholar] [CrossRef]
- Hinniger, C.; Caillet, V.; Michoux, F.; Ben Amor, M.; Tanksley, S.; Lin, C.; McCarthy, J. Isolation and characterization of cDNA encoding three dehydrins expressed during Coffea canephora (Robusta) grain development. Ann. Bot. 2006, 97, 755–765. [Google Scholar] [CrossRef][Green Version]
- Nath, B.; Birch, G.; Chaudhuri, P. Assessment of sediment quality in Avicennia marina-dominated embayments of Sydney Estuary: The potential use of pneumatophores (aerial roots) as a bio-indicator of trace metal contamination. Sci. Total Environ. 2014, 472, 1010–1022. [Google Scholar] [CrossRef]
- Alishahi, M.; Kamali, R. Forced diffusion of water molecules through aquaporin-5 biomembrane; a molecular dynamics study. Biophys. Physicobiol. 2018, 15, 255–262. [Google Scholar] [CrossRef]
- Merlaen, B.; De Keyser, E.; Ding, L.; Leroux, O.; Chaumont, F.; Van Labeke, M.C. Physiological responses and aquaporin expression upon drought and osmotic stress in a conservative vs prodigal Fragaria x ananassa cultivar. Plant Physiol. Biochem. 2019, 145, 95–106. [Google Scholar] [CrossRef]
- Sahebi, S.; Fadaie, N.; Mirshekar, L.; Kamarehie, B.; Mohammadi, T. Assessing biomimetic aquaporin membrane for forward osmosis desalination process: A dataset. Data Brief 2019, 26, 104482. [Google Scholar] [CrossRef]
- Wei, X.; Jin, X.; Ndayambaza, B.; Min, X.; Zhang, Z.; Wang, Y.; Liu, W. Transcriptome-wide characterization and functional identification of the aquaporin gene family during drought stress in common Vetch. DNA Cell Biol. 2019, 38, 374–384. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, J. Genome-wide comparison reveals divergence of cassava and rubber aquaporin family genes after the recent whole-genome duplication. BMC Genom. 2019, 20, 380. [Google Scholar] [CrossRef]
- Krishnamurthy, P.; Tan, X.F.; Lim, T.K.; Lim, T.-M.; Kumar, P.P.; Loh, C.-S.; Lin, Q. Proteomic analysis of plasma membrane and tonoplast from the leaves of mangrove plant Avicennia officinalis. Proteomics 2014, 14, 2545–2557. [Google Scholar] [CrossRef]
- Sadhukhan, A.; Kobayashi, Y.; Nakano, Y.; Iuchi, S.; Kobayashi, M.; Sahoo, L.; Koyama, H. Genome-wide Association Study Reveals that the Aquaporin NIP1;1 Contributes to Variation in Hydrogen Peroxide Sensitivity in Arabidopsis thaliana. Mol. Plant 2017, 10, 1082–1094. [Google Scholar] [CrossRef]
- Yusupov, M.; Razzokov, J.; Cordeiro, R.M.; Bogaerts, A. Transport of reactive oxygen and nitrogen species across aquaporin: A molecular level picture. Oxid. Med. Cell Longev. 2019, 2019, 2930504. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, N.H.; Thibodeau, F.R. Association between Pore Water Sulfide Concentrations and the Distribution of Mangroves. Biogeochemistry 1985, 1, 183–192. [Google Scholar] [CrossRef]
- Kreuzwieser, J.; Rennenberg, H. Molecular and physiological responses of trees to waterlogging stress: Responses of tree to waterlogging. Plant Cell Environ. 2014, 37, 2245–2259. [Google Scholar] [CrossRef] [PubMed]
- Amala, M.; Rajamanikandan, S.; Prabhu, D.; Surekha, K.; Jeyakanthan, J. Identification of anti-filarial leads against aspartate semialdehyde dehydrogenase of Wolbachia endosymbiont of Brugia malayi: Combined molecular docking and molecular dynamics approaches. J. Biomol. Struct. Dyn. 2019, 37, 394–410. [Google Scholar] [CrossRef]
- Cohen, G.N. Aspartate-semialdehyde dehydrogenase from Escherichia coli. Methods Enzymol. 1985, 113, 600–602. [Google Scholar] [CrossRef]
- Cox, R.J.; Gibson, J.S.; Hadfield, A.T. Design, synthesis and analysis of inhibitors of bacterial aspartate semialdehyde dehydrogenase. Chembiochem 2005, 6, 2255–2260. [Google Scholar] [CrossRef]
- Dahal, G.P.; Viola, R.E. Structural insights into inhibitor binding to a fungal ortholog of aspartate semialdehyde dehydrogenase. Biochem. Biophys. Res. Commun. 2018, 503, 2848–2854. [Google Scholar] [CrossRef]
- Evitt, A.S.; Cox, R.J. Synthesis and evaluation of conformationally restricted inhibitors of aspartate semialdehyde dehydrogenase. Mol. Biosyst. 2011, 7, 1564–1575. [Google Scholar] [CrossRef]
- Gayler, K.R.; Morgan, W.R. An NADP-dependent Glutamate Dehydrogenase in Chloroplasts from the Marine Green Alga Caulerpa simpliciuscula. Plant Physiol. 1976, 58, 283–287. [Google Scholar] [CrossRef]
- Born, T.L.; Blanchard, J.S. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall biosynthesis. Curr. Opin. Chem. Biol. 1999, 3, 607–613. [Google Scholar] [CrossRef]
- Mengin-Lecreulx, D.; van Heijenoort, J.; Park, J.T. Identification of the mpl gene encoding UDP-N-acetylmuramate: L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase in Escherichia coli and its role in recycling of cell wall peptidoglycan. J. Bacteriol. 1996, 178, 5347–5352. [Google Scholar] [CrossRef]
- Park, J.T.; Raychaudhuri, D.; Li, H.; Normark, S.; Mengin-Lecreulx, D. MppA, a periplasmic binding protein essential for import of the bacterial cell wall peptide L-alanyl-gamma-D-glutamyl-meso-diaminopimelate. J. Bacteriol. 1998, 180, 1215–1223. [Google Scholar] [CrossRef]
- Wehrmann, A.; Phillipp, B.; Sahm, H.; Eggeling, L. Different modes of diaminopimelate synthesis and their role in cell wall integrity: A study with Corynebacterium glutamicum. J. Bacteriol. 1998, 180, 3159–3165. [Google Scholar] [CrossRef]
- Villa, S.T.; Xu, Q.; Downie, A.B.; Clarke, S.G. Arabidopsis protein repair l-isoaspartyl methyltransferases: Predominant activities at lethal temperatures. Physiol. Plant. 2006, 128, 581–592. [Google Scholar] [CrossRef]
- Gonzalez-Mendoza, D.; Moreno, A.Q.; Zapata-Perez, O. Coordinated responses of phytochelatin synthase and metallothionein genes in black mangrove, Avicennia germinans, exposed to cadmium and copper. Aquat. Toxicol. 2007, 83, 306–314. [Google Scholar] [CrossRef]
- Nelson, D.R.; Chaiboonchoe, A.; Fu, W.; Hazzouri, K.M.; Huang, Z.; Jaiswal, A.; Daakour, S.; Mystikou, A.; Arnoux, M.; Sultana, M.; et al. Potential for Heightened Sulfur-Metabolic Capacity in Coastal Subtropical Microalgae. iScience 2019, 11, 450–465. [Google Scholar] [CrossRef]
- Cotelle, N.; Moreau, S.; Cotelle, P.; Catteau, J.P.; Bernier, J.L.; Henichart, J.P. Generation of free radicals by simple prenylated hydroquinone derivatives, natural antitumor agents from the marine urochordate Aplidium californicum. Chem. Res. Toxicol. 1991, 4, 300–305. [Google Scholar] [CrossRef]
- Naik, M.M.; Pandey, A.; Dubey, S.K. Pseudomonas aeruginosa strain WI-1 from Mandovi estuary possesses metallothionein to alleviate lead toxicity and promotes plant growth. Ecotoxicol. Environ. Saf. 2012, 79, 129–133. [Google Scholar] [CrossRef]
- Attia, O.E.; Ghrefat, H. Assessing heavy metal pollution in the recent bottom sediments of Mabahiss Bay, North Hurghada, Red Sea, Egypt. Environ. Monit. Assess. 2013, 185, 9925–9934. [Google Scholar] [CrossRef]
- Idris, A.M.; Said, T.O.; Omran, A.A.; Fawy, K.F. Combining multivariate analysis and human risk indices for assessing heavy metal contents in muscle tissues of commercially fish from Southern Red Sea, Saudi Arabia. Environ. Sci. Pollut. Res. Int. 2015, 22, 17012–17021. [Google Scholar] [CrossRef]
- Mohamed, S.A.; Elshal, M.F.; Kumosani, T.A.; Mal, A.O.; Ahmed, Y.M.; Almulaiky, Y.Q.; Asseri, A.H.; Zamzami, M.A. Heavy Metal Accumulation is Associated with Molecular and Pathological Perturbations in Liver of Variola louti from the Jeddah Coast of Red Sea. Int. J. Environ. Res. Public Health 2016, 13, 342. [Google Scholar] [CrossRef]
- Saleh, Y.S.; Marie, M.A. Assessment of metal contamination in water, sediment, and tissues of Arius thalassinus fish from the Red Sea coast of Yemen and the potential human risk assessment. Environ. Sci. Pollut. Res. Int. 2015, 22, 5481–5490. [Google Scholar] [CrossRef]
- Euceda, N.; Jahnke, J.; Espinal, A.; Louis, M.F.; Bashkin, E.; Roccanova, P.; Espaillat, A.; Fuentes, G.V.; Nieto, F.; Gao, R. Thioguanine restoration through type I photosensitization-superoxide oxidation-glutathione reduction cycles. Physical. Chem. Chem. Phys. 2021, 23, 5069–5073. [Google Scholar] [CrossRef]
- Simunkova, M.; Barbierikova, Z.; Jomova, K.; Hudecova, L.; Lauro, P.; Alwasel, S.H.; Alhazza, I.; Rhodes, C.J.; Valko, M. Antioxidant vs. Prooxidant Properties of the Flavonoid, Kaempferol, in the Presence of Cu(II) Ions: A ROS-Scavenging Activity, Fenton Reaction and DNA Damage Study. Int. J. Mol. Sci. 2021, 22, 1619. [Google Scholar] [CrossRef]
- Tsang, T.; Davis, C.I.; Brady, D.C. Copper biology. Curr. Biol. 2021, 31, R421–R427. [Google Scholar] [CrossRef]
- Mir, G. A plant type 2 metallothionein (MT) from cork tissue responds to oxidative stress. J. Exp. Bot. 2004, 55, 2483–2493. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).