Molecular Phylogenetic Analysis of Salt-Tolerance-Related Genes in Root-Nodule Bacteria Species Sinorhizobium meliloti
Abstract
1. Introduction
2. Materials and Methods
2.1. Complete Genomes
2.2. Sequence Similarity Analysis
2.3. Phylogenetic Analysis
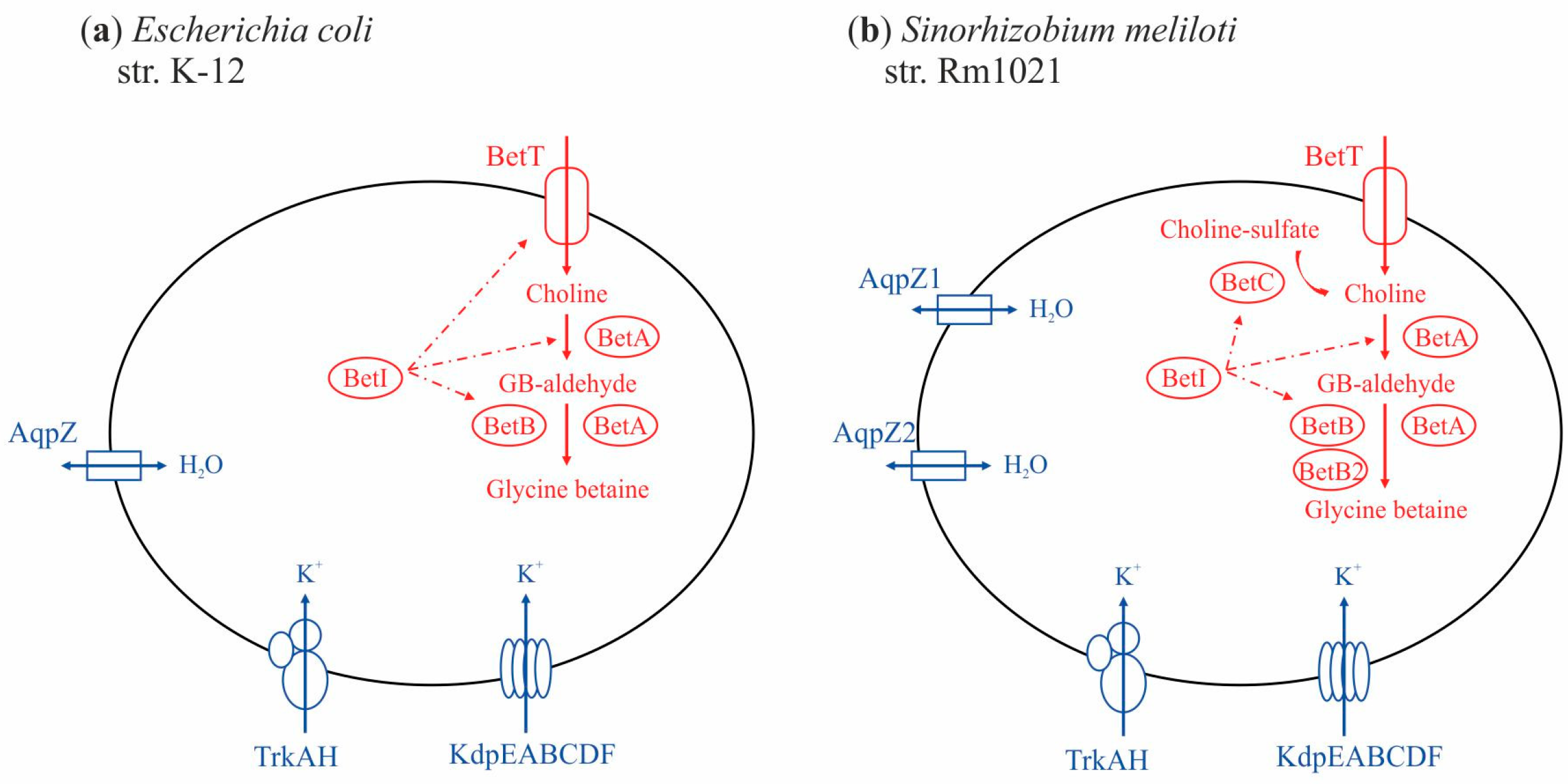
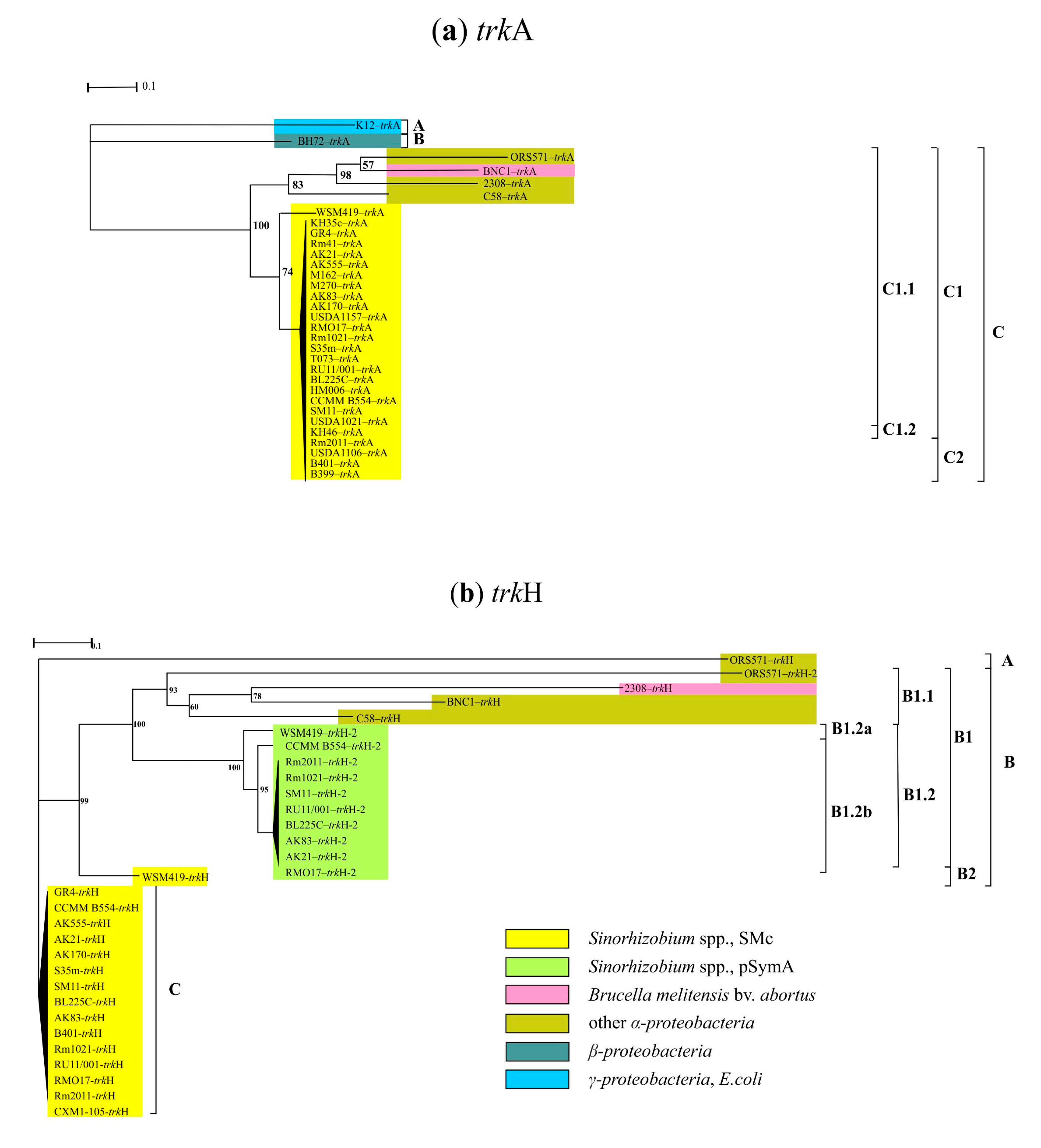
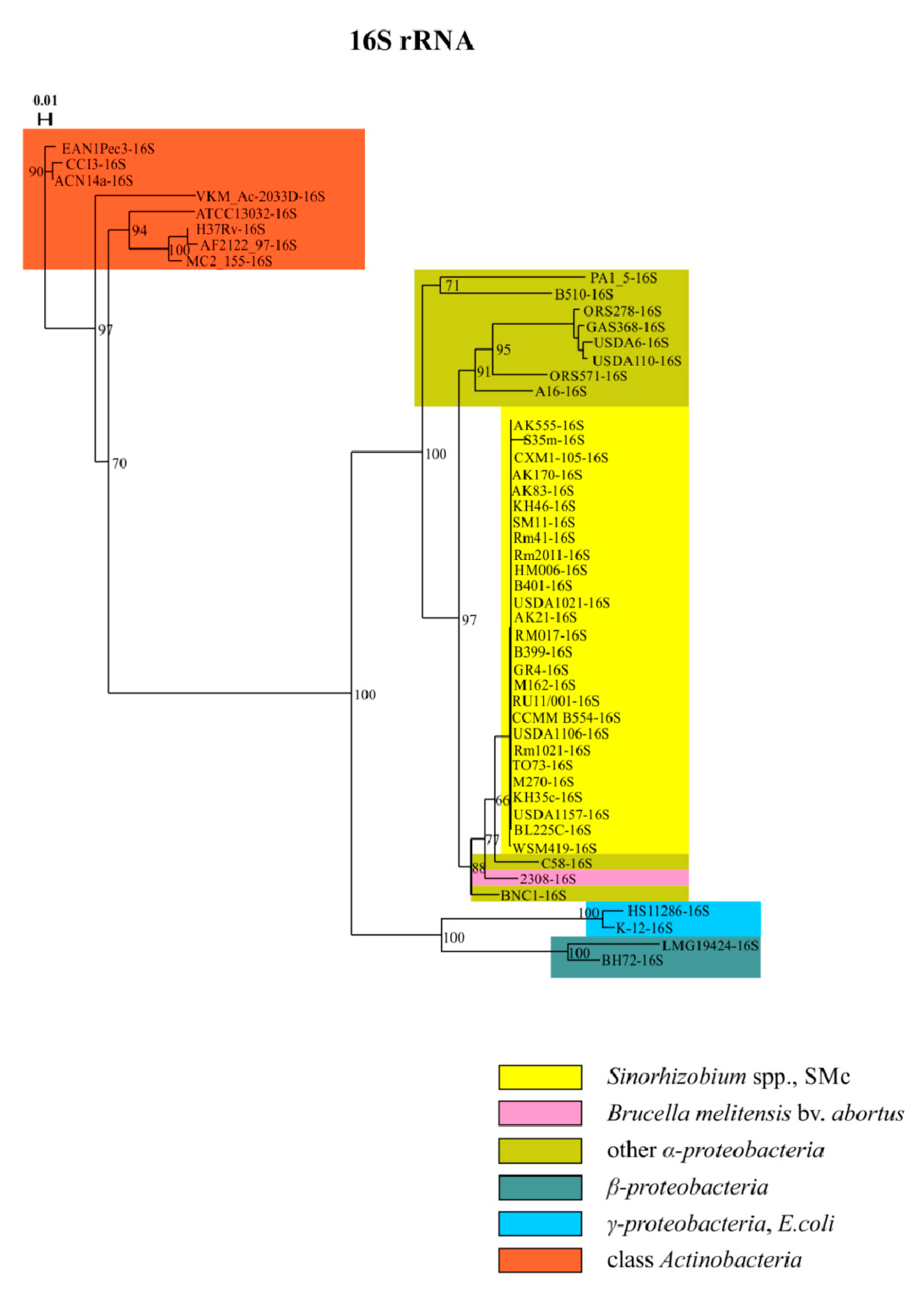
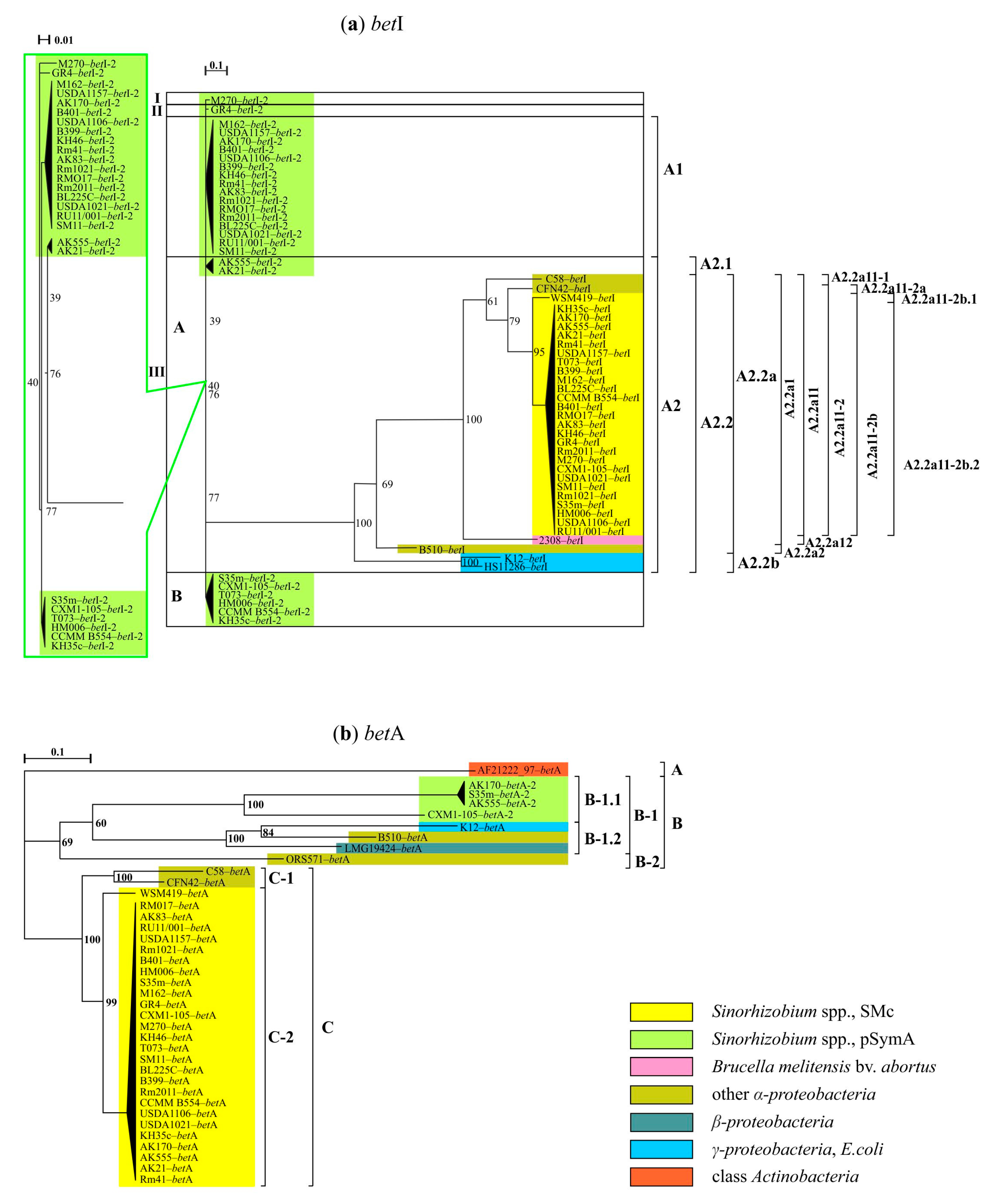
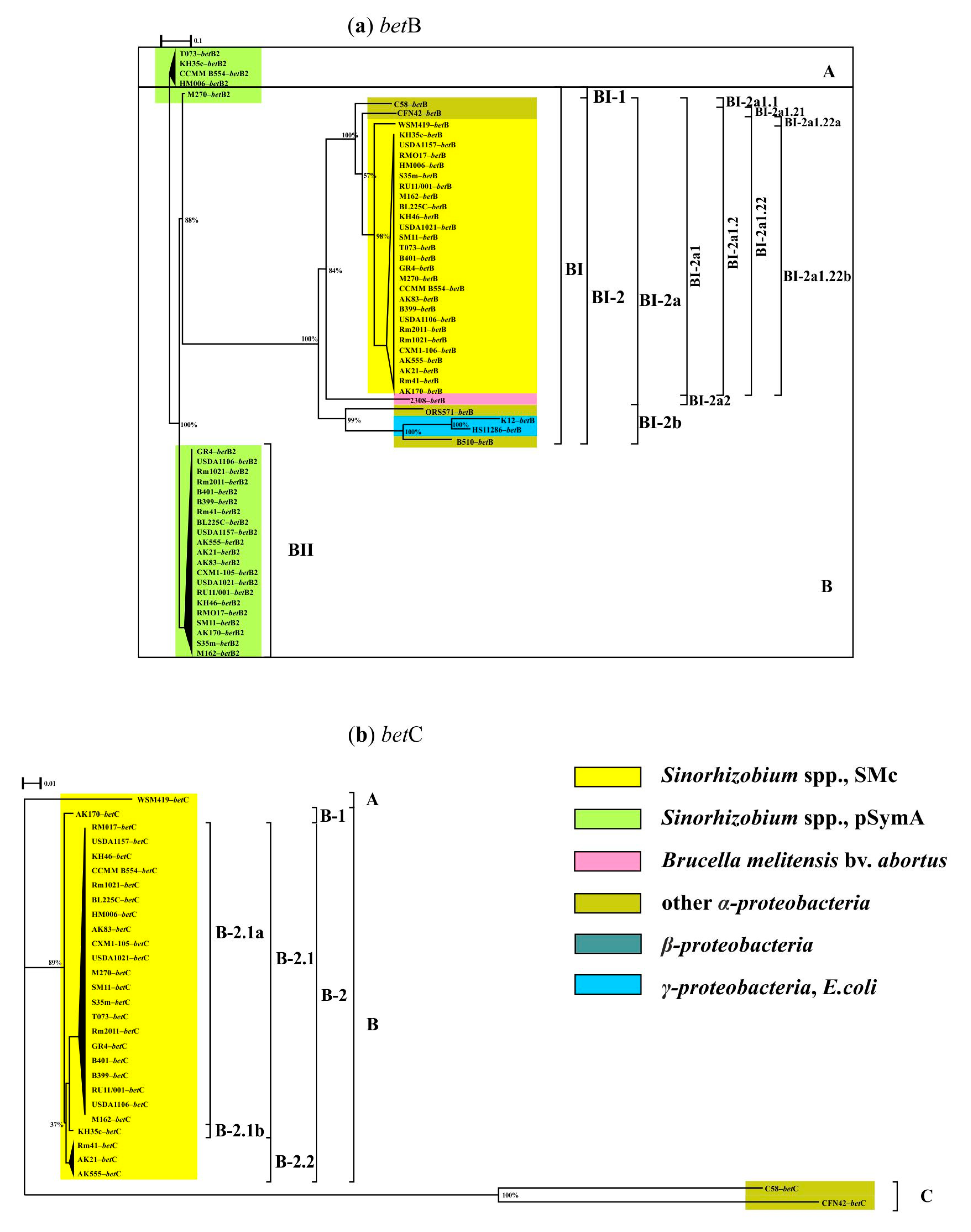
3. Results
3.1. Genes of the First Stage of the Response to Salt Stress
3.2. Genes of the Second Stage of the Response to Salt Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Arzani, A.; Ashraf, M. Smart Engineering of Genetic Resources for Enhanced Salinity Tolerance in Crop Plants. Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Zahran, H.H. Legume-Microbe Interactions Under Stressed Environments. In Microbes for Legume Improvement; Zaidi, A., Khan, M.S., Musarrat, J., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 301–339. ISBN 978-3-319-59173-5. [Google Scholar]
- Roumiantseva, M.L.; Saksaganskaia, A.S.; Muntyan, V.S.; Cherkasova, M.E.; Simarov, B.V. Structural Polymorphism of Sinorhizobium meliloti Genes Related to Virulence and Salt Tolerance. Russ. J. Genet. 2018, 54, 525–535. [Google Scholar] [CrossRef]
- Ermilova, E.V. Molecular Aspects of Prokaryote Adaptation, 2nd ed.; Khimizdat: St. Petersburg, Russia, 2012; p. 128. (In Russian) [Google Scholar]
- Kempf, B.; Bremer, E. Uptake and Synthesis of Compatible Solutes as Microbial Stress Responses to High-Osmolality Environments. Arch. Microbiol. 1998, 170, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Epstein, W. The Roles and Regulation of Potassium in Bacteria. Prog. Nucleic Acid Res. Mol. Biol. 2003, 75, 293–320. [Google Scholar] [CrossRef]
- Dietz, S.; von Bülow, J.; Beitz, E.; Nehls, U. The Aquaporin Gene Family of the Ectomycorrhizal Fungus Laccaria bicolor: Lessons for Symbiotic Functions. New Phytol. 2011, 190, 927–940. [Google Scholar] [CrossRef]
- Li, T.; Hu, Y.; Hao, Z.; Li, H.; Wang, Y.; Chen, B. First Cloning and Characterization of Two Functional Aquaporin Genes from an Arbuscular Mycorrhizal Fungus Glomus intraradices. New Phytol. 2013, 197, 617–630. [Google Scholar] [CrossRef]
- An, B.; Li, B.; Li, H.; Zhang, Z.; Qin, G.; Tian, S. Aquaporin8 Regulates Cellular Development and Reactive Oxygen Species Production, a Critical Component of Virulence in Botrytis cinerea. New Phytol. 2016, 209, 1668–1680. [Google Scholar] [CrossRef]
- Saha, P.; Sade, N.; Arzani, A.; Rubio Wilhelmi, M.D.M.; Coe, K.M.; Li, B.; Blumwald, E. Effects of Abiotic Stress on Physiological Plasticity and Water Use of Setaria viridis (L.). Plant Sci. 2016, 251, 128–138. [Google Scholar] [CrossRef]
- Yamada, S.; Katsuhara, M.; Kelly, W.B.; Michalowski, C.B.; Bohnert, H.J. A Family of Transcripts Encoding Water Channel Proteins: Tissue-Specific Expression in the Common Ice Plant. Plant Cell 1995, 7, 1129–1142. [Google Scholar] [CrossRef]
- Kaldenhoff, R.; Kölling, A.; Richter, G. Regulation of the Arabidopsis Thaliana Aquaporin Gene AthH2 (PIP1b). J. Photochem. Photobiol. B 1996, 36, 351–354. [Google Scholar] [CrossRef]
- Calamita, G.; Kempf, B.; Bonhivers, M.; Bishai, W.R.; Bremer, E.; Agre, P. Regulation of the Escherichia coli Water Channel Gene AqpZ. Proc. Natl. Acad. Sci. USA 1998, 95, 3627–3631. [Google Scholar] [CrossRef] [PubMed]
- Calamita, G. The Escherichia Coli Aquaporin-Z Water Channel. Mol. Microbiol. 2000, 37, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Castro, R.; Rodríguez, M.C.; Seoane, A.; García Lobo, J.M. The Aquaporin Gene aqpX of Brucella abortus Is Induced in Hyperosmotic Conditions. Microbiology 2003, 149, 3185–3192. [Google Scholar] [CrossRef][Green Version]
- Rivers, R.L.; Dean, R.M.; Chandy, G.; Hall, J.E.; Roberts, D.M.; Zeidel, M.L. Functional Analysis of Nodulin 26, an Aquaporin in Soybean Root Nodule Symbiosomes. J. Biol. Chem. 1997, 272, 16256–16261. [Google Scholar] [CrossRef] [PubMed]
- Gavrin, A.; Kaiser, B.N.; Geiger, D.; Tyerman, S.D.; Wen, Z.; Bisseling, T.; Fedorova, E.E. Adjustment of Host Cells for Accommodation of Symbiotic Bacteria: Vacuole Defunctionalization, HOPS Suppression, and TIP1g Retargeting in Medicago. Plant Cell 2014, 26, 3809–3822. [Google Scholar] [CrossRef]
- Rodríguez, M.C.; Froger, A.; Rolland, J.-P.; Thomas, D.; Agüero, J.; Delamarche, C.; García-Lobo, J.M. A Functional Water Channel Protein in the Pathogenic Bacterium Brucella abortus The GenBank Accession Number for the Nucleotide Sequence Reported in This Paper Is AF148066. Microbiology 2000, 146, 3251–3257. [Google Scholar] [CrossRef][Green Version]
- Verbavatz, J.M.; Brown, D.; Sabolić, I.; Valenti, G.; Ausiello, D.A.; Van Hoek, A.N.; Ma, T.; Verkman, A.S. Tetrameric Assembly of CHIP28 Water Channels in Liposomes and Cell Membranes: A Freeze-Fracture Study. J. Cell Biol. 1993, 123, 605–618. [Google Scholar] [CrossRef]
- Cheng, A.; van Hoek, A.N.; Yeager, M.; Verkman, A.S.; Mitra, A.K. Three-Dimensional Organization of a Human Water Channel. Nature 1997, 387, 627–630. [Google Scholar] [CrossRef]
- Eskandari, S.; Wright, E.M.; Kreman, M.; Starace, D.M.; Zampighi, G.A. Structural Analysis of Cloned Plasma Membrane Proteins by Freeze-Fracture Electron Microscopy. Proc. Natl. Acad. Sci. USA 1998, 95, 11235–11240. [Google Scholar] [CrossRef]
- Meinild, A.-K.; Klaerke, D.A.; Zeuthen, T. Bidirectional Water Fluxes and Specificity for Small Hydrophilic Molecules in Aquaporins 0–5. J. Biol. Chem. 1998, 273, 32446–32451. [Google Scholar] [CrossRef]
- Hohmann, S.; Bill, R.M.; Kayingo, G.; Prior, B.A. Microbial MIP Channels. Trends Microbiol. 2000, 8, 33–38. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic Osmolytes as Compatible, Metabolic and Counteracting Cytoprotectants in High Osmolarity and Other Stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef] [PubMed]
- Burg, M.B.; Ferraris, J.D. Intracellular Organic Osmolytes: Function and Regulation. J. Biol. Chem. 2008, 283, 7309–7313. [Google Scholar] [CrossRef]
- Tkachenko, A.G. Molecular Mechanisms of Stress Responses in Microorganisms; Ural Branch of the Russian Academy of Sciences: Ekaterinburg, Russia, 2012; p. 181. (In Russian) [Google Scholar]
- Kempf, B.; Bremer, E. Stress Responses of Bacillus subtilis to High Osmolarity Environments: Uptake and Synthesis of Osmoprotectants. J. Biosci. 1998, 23, 447–455. [Google Scholar] [CrossRef]
- Deole, R.; Hoff, W.D. A Potassium Chloride to Glycine Betaine Osmoprotectant Switch in the Extreme Halophile Halorhodospira halophila. Sci. Rep. 2020, 10, 3383. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.; Ali, M.; Kubra, G.; Fareed, A.; Hasan, H.; Khursheed, A.; Gul, A.; Amir, R.; Fatima, N.; Khan, S.U. Role of Osmoprotectants in Salinity Tolerance in Wheat. In Climate Change and Food Security with Emphasis on Wheat; Elsevier: Amsterdam, The Netherlands, 2020; pp. 93–106. ISBN 978-0-12-819527-7. [Google Scholar]
- Pinevich, A.V. Microbiology. In Biology of Prokaryotes; Publishing House of St. Petersburg State University: St. Petersburg, Russia, 2009; Volume 3, p. 361. (In Russian) [Google Scholar]
- Perullini, M.; Amoura, M.; Roux, C.; Coradin, T.; Livage, J.; Japas, M.L.; Jobbágy, M.; Bilmes, S.A. Improving Silica Matrices for Encapsulation of Escherichia coli Using Osmoprotectors. J. Mater. Chem. 2011, 21, 4546–4552. [Google Scholar] [CrossRef]
- Stecca, J.D.L.; Martin, T.N.; Lúcio, A.D.; Deak, E.A.; Fipke, G.M.; Bruning, L.A. Inoculation of Soybean Seeds Coated with Osmoprotector in Diferents Soil PH’s. Acta Sci. Agron. 2018, 41, 39482. [Google Scholar] [CrossRef]
- Talibart, R.; Jebbar, M.; Gouffi, K.; Pichereau, V.; Gouesbet, G.; Blanco, C.; Bernard, T.; Pocard, J. Transient Accumulation of Glycine Betaine and Dynamics of Endogenous Osmolytes in Salt-Stressed Cultures of Sinorhizobium meliloti. Appl. Environ. Microbiol. 1997, 63, 4657–4663. [Google Scholar] [CrossRef]
- Kappes, R.M.; Kempf, B.; Bremer, E. Three Transport Systems for the Osmoprotectant Glycine Betaine Operate in Bacillus subtilis: Characterization of OpuD. J. Bacteriol. 1996, 178, 5071–5079. [Google Scholar] [CrossRef]
- Gouffi, K.; Pica, N.; Pichereau, V.; Blanco, C. Disaccharides as a New Class of Nonaccumulated Osmoprotectants for Sinorhizobium meliloti. Appl. Environ. Microbiol. 1999, 65, 1491–1500. [Google Scholar] [CrossRef]
- Domínguez-Ferreras, A.; Soto, M.J.; Pérez-Arnedo, R.; Olivares, J.; Sanjuán, J. Importance of Trehalose Biosynthesis for Sinorhizobium meliloti Osmotolerance and Nodulation of Alfalfa Roots. J. Bacteriol. 2009, 191, 7490–7499. [Google Scholar] [CrossRef] [PubMed]
- Østerås, M.; Boncompagni, E.; Vincent, N.; Poggi, M.-C.; Le Rudulier, D. Presence of a Gene Encoding Choline Sulfatase in Sinorhizobium meliloti bet Operon: Choline-O-Sulfate Is Metabolized into Glycine Betaine. Proc. Natl. Acad. Sci. USA 1998, 95, 11394–11399. [Google Scholar] [CrossRef] [PubMed]
- Tani, Y.; Mori, N.; Ogata, K.; Yamada, H. Production and Purification of Choline Oxidase from Cylindrocarpon didymum M-1. Agric. Biol. Chem. 1979, 43, 815–820. [Google Scholar] [CrossRef]
- Rozwadowski, K.L.; Khachatourians, G.G.; Selvaraj, G. Choline Oxidase, a Catabolic Enzyme in Arthrobacter Pascens, Facilitates Adaptation to Osmotic Stress in Escherichia coli. J. Bacteriol. 1991, 173, 472–478. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.L.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete Genome Sequence of Pseudomonas Aeruginosa PAO1, an Opportunistic Pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- Mao, F.; Dam, P.; Chou, J.; Olman, V.; Xu, Y. DOOR: A Database for Prokaryotic Operons. Nucleic Acids Res. 2009, 37, D459–D463. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced Annotations and Features for Comparing Thousands of Pseudomonas Genomes in the Pseudomonas Genome Database. Nucleic Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef]
- Cregut, M.; Durand, M.-J.; Thouand, G. The Diversity and Functions of Choline Sulphatases in Microorganisms. Microb. Ecol. 2014, 67, 350–357. [Google Scholar] [CrossRef]
- Mandon, K.; Østerås, M.; Boncompagni, E.; Trinchant, J.C.; Spennato, G.; Poggi, M.C.; Le Rudulier, D. The Sinorhizobium meliloti Glycine Betaine Biosynthetic Genes ( BetICBA ) Are Induced by Choline and Highly Expressed in Bacteroids. Mol. Plant-Microbe Interact. 2003, 16, 709–719. [Google Scholar] [CrossRef]
- Boscari, A.; Van de Sype, G.; Le Rudulier, D.; Mandon, K. Overexpression of BetS, a Sinorhizobium meliloti High-Affinity Betaine Transporter, in Bacteroids from Medicago sativa Nodules Sustains Nitrogen Fixation During Early Salt Stress Adaptation. Mol. Plant-Microbe Interact. 2006, 19, 896–903. [Google Scholar] [CrossRef][Green Version]
- Yurgel, S.N.; Rice, J.; Mulder, M.; Kahn, M.L.; Belova, V.S.; Roumiantseva, M.L. Truncated betB2-144 Plays a Critical Role in Sinorhizobium meliloti Rm2011 Osmoprotection and Glycine-Betaine Catabolism. Eur. J. Soil Biol. 2013, 54, 48–55. [Google Scholar] [CrossRef]
- Shimada, T.; Ogasawara, H.; Ishihama, A. Single-target regulators form a minor group of transcription factors in Escherichia coli K-12. Nucleic Acids Res. 2018, 46, 3921–3936. [Google Scholar] [CrossRef] [PubMed]
- Angiuoli, S.V.; Gussman, A.; Klimke, W.; Cochrane, G.; Field, D.; Garrity, G.M.; Kodira, C.D.; Kyrpides, N.; Madupu, R.; Markowitz, V.; et al. Toward an Online Repository of Standard Operating Procedures (SOPs) for (Meta)Genomic Annotation. OMICS J. Integr. Biol. 2008, 12, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Korber, B. HIV signature, and sequence variation analysis. In Computational Analysis of HIV Molecular Sequences; Rodrigo, A.G., Learn, G.H., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 2000; Volume 4, pp. 555–572. [Google Scholar]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of Phylogenies after Removing Divergent and Ambiguously Aligned Blocks from Protein Sequence Alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Huson, D.H.; Scornavacca, C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst. Biol. 2012, 61, 1061–1067. [Google Scholar] [CrossRef]
- Bogdanowicz, D.; Giaro, K.; Wróbel, B. TreeCmp: Comparison of Trees in Polynomial Time. Evol. Bioinform. 2012, 8, 475–487. [Google Scholar] [CrossRef]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A Web Server for Inferring, Validating and Visualizing Phylogenetic Trees and Networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed]
- Cholo, M.C.; Boshoff, H.I.; Steel, H.C.; Cockeran, R.; Matlola, N.M.; Downing, K.J.; Mizrahi, V.; Anderson, R. Effects of Clofazimine on Potassium Uptake by a Trk-Deletion Mutant of Mycobacterium tuberculosis. J. Antimicrob. Chemother. 2006, 57, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Zakharyan, E.; Trchounian, A. K+ Influx by Kup in Escherichia coli Is Accompanied by a Decrease in H+ Efflux. FEMS Microbiol. Lett. 2001, 204, 61–64. [Google Scholar] [CrossRef] [PubMed]
- Sayyari, E.; Mirarab, S. Testing for Polytomies in Phylogenetic Species Trees Using Quartet Frequencies. Genes 2018, 9, 132. [Google Scholar] [CrossRef]
- Lin, G.N.; Zhang, C.; Xu, D. Polytomy Identification in Microbial Phylogenetic Reconstruction. BMC Syst. Biol. 2011, 5, S2. [Google Scholar] [CrossRef]
- Maddison, W. Reconstructing character evolution on polytomous cladograms. Cladistics 1989, 5, 365–377. [Google Scholar] [CrossRef]
- Cao, Y.; Jin, X.; Huang, H.; Derebe, M.G.; Levin, E.J.; Kabaleeswaran, V.; Pan, Y.; Punta, M.; Love, J.; Weng, J.; et al. Crystal Structure of a Potassium Ion Transporter, TrkH. Nature 2011, 471, 336–340. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- van Loo, B.; Schober, M.; Valkov, E.; Heberlein, M.; Bornberg-Bauer, E.; Faber, K.; Hyvönen, M.; Hollfelder, F. Structural and Mechanistic Analysis of the Choline Sulfatase from Sinorhizobium meliloti: A Class I Sulfatase Specific for an Alkyl Sulfate Ester. J. Mol. Biol. 2018, 430, 1004–1023. [Google Scholar] [CrossRef]
- Roumiantseva, M.L.; Belova, V.S.; Onishchouk, O.P.; Andronov, E.E.; Kurchak, O.N.; Chizhevskaya, E.P.; Rumyantseva, T.B.; Simarov, B.V. Polymorphism of bet-genes among Sinorhizobium meliloti isolates native to gene centers of alfalfa. Agric. Biol. (Sel’skokhozyaistvennaya Biol.) 2011, 3, 48–54. (In Russian) [Google Scholar]
- Roumiantseva, M.L.; Muntyan, V.S. Root Nodule Bacteria Sinorhizobium Meliloti: Tolerance to Salinity and Bacterial Genetic Determinants. Microbiology 2015, 84, 303–318. [Google Scholar] [CrossRef]
- Baturina, O.A.; Muntyan, V.S.; Afonin, A.M.; Cherkasova, M.E.; Simarov, B.V.; Kabilov, M.R.; Roumiantseva, M.L. Draft Genome Sequence of Sinorhizobium meliloti Strain CXM1-105. Microbiol. Resour. Announc. 2019, 8, e01621-18. [Google Scholar] [CrossRef] [PubMed]
- Baturina, O.A.; Muntyan, V.S.; Cherkasova, M.E.; Saksaganskaya, A.S.; Dzuybenko, N.I.; Kabilov, M.R.; Roumiantseva, M.L. Draft Genome Sequence of Sinorhizobium meliloti Strain AK170. Microbiol. Resour. Announc. 2019, 8, e01571-18. [Google Scholar] [CrossRef] [PubMed]
- Muntyan, V.S.; Baturina, O.A.; Afonin, A.M.; Cherkasova, M.E.; Laktionov, Y.V.; Saksaganskaya, A.S.; Kabilov, M.R.; Roumiantseva, M.L. Draft Genome Sequence of Sinorhizobium meliloti AK555. Microbiol. Resour. Announc. 2019, 8, e01567-18. [Google Scholar] [CrossRef]
- Muntyan, V.S.; Afonin, A.M.; Vladimirova, M.E.; Saksaganskaya, A.S.; Gribchenko, E.S.; Baturina, O.; Roumiantseva, M.L. Complete Genome Sequence of Sinorhizobium meliloti S35m, a Salt-Tolerant Isolate from Alfalfa Rhizosphere in Soil Native to the Caucasus Region. Microbiol. Resour. Announc. 2021, 10, e01417-20. [Google Scholar] [CrossRef]
- Anand, A.; Sharma, A. Use of Proteomics and Transcriptomics to Identify Proteins for Cold Adaptation in Microbes. In Survival Strategies in Cold-adapted Microorganisms; Goel, R., Soni, R., Suyal, D.C., Khan, M., Eds.; Springer Singapore: Singapore, 2022; pp. 285–319. ISBN 9789811626241. [Google Scholar]
- Wood, D.W.; Setubal, J.C.; Kaul, R.; Monks, D.E.; Kitajima, J.P.; Okura, V.K.; Zhou, Y.; Chen, L.; Wood, G.E.; Almeida, N.F.; et al. The Genome of the Natural Genetic Engineer Agrobacterium tumefaciens C58. Science 2001, 294, 2317–2323. [Google Scholar] [CrossRef]
- Galardini, M.; Pini, F.; Bazzicalupo, M.; Biondi, E.G.; Mengoni, A. Replicon-dependent bacterial genome evolution: The case of Sinorhizobium meliloti. Genome Biol. Evol. 2013, 5, 542–558. [Google Scholar] [CrossRef]

| Class | Genera | StrainBioSample | |
|---|---|---|---|
| Species | GenBank | ||
| α-proteobacteria | Sinorhizobium | S. meliloti | Rm1021SAMEA3283068, Rm2011SAMN02603522, USDA1106SAMN07175168, USDA1157SAMN07175169, USDA1021SAMN07175167, B399SAMN06229775, B401SAMN06227501, BL225CSAMN00017103, CCMM B554 (FSM-MA)SAMN06284128, GR4SAMN02603224, KH35cSAMN07175161, KH46SAMN07175162, HM006SAMN07175160, M162SAMN07175163, M270SAMN07175164, Rm41SAMN07175165, RMO17SAMN02952139, RU11/001SAMEA3146337, SM11SAMN02603056, T073SAMN07175166 AK83SAMN00017059 *, AK21SAMN08428886 *, AK555SAMN08826593 *, AK170SAMN10256575 *, S35mSAMN16812329 *, CXM1-105SAMN08826592 * |
| S. medicae | WSM419SAMN02598363 | ||
| Bradyrhizobium | Brad. spp. | GAS369NZ_LT629750.1, ORS 278SAMEA3138227, USDA 6SAMD00060992 and USDA 110SAMN03573437 | |
| Brucella | Bruc. melitensis bv. abortus | 2308SAMEA3138256 | |
| Devosia | D. sp. | A16SAMN04156589 | |
| Chelativorans | Chel. sp. | BNC1SAMN02598260 | |
| Agrobacterium | A. fabrum | C58SAMN02603108 | |
| Rhizobium | Rh. etli | CFN 42SAMN02603106 | |
| Azorhizobium | Azor. caulinodans | ORS 571SAMD00060925 | |
| Gluconacetobacter | Gluc. diazotrophicus | PA1 5SAMN02598444 | |
| Azospirillum | Azo. sp. | B510SAMD00060958 | |
| β-proteobacteria | Cupriavidus | Cupr. taiwanensis | LMG 19424SAMEA3138280 |
| Azoarcus | Azoarc. sp. | BH72SAMEA3138261 | |
| γ-proteobacteria | Klebsiella | K. pneumoniae subsp. pneumoniae | HS11286SAMN02602959 |
| Escherichia | E. coli | K-12SAMN02604091 | |
| Actinobacteria | Frankia | F. spp. | ACN14aSAMEA3138259, EAN1PecSAMN02598325, CcI3SAMN02199398 |
| Corynebacterium | Cor. glutamicum | ATCC 13032SAMD00061105 | |
| Mycolicibacterium | M. smegmatis | MC2 155NZ_LN831039.1 | |
| Mycobacterium | M. spp. | H37RvSAMEA3138326, AF2122/97SAMEA20450668 | |
| Pimelobacter | P. simplex | VKM Ac-2033DSAMN03009415 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muntyan, V.S.; Roumiantseva, M.L. Molecular Phylogenetic Analysis of Salt-Tolerance-Related Genes in Root-Nodule Bacteria Species Sinorhizobium meliloti. Agronomy 2022, 12, 1968. https://doi.org/10.3390/agronomy12081968
Muntyan VS, Roumiantseva ML. Molecular Phylogenetic Analysis of Salt-Tolerance-Related Genes in Root-Nodule Bacteria Species Sinorhizobium meliloti. Agronomy. 2022; 12(8):1968. https://doi.org/10.3390/agronomy12081968
Chicago/Turabian StyleMuntyan, Victoria Spartakovna, and Marina Lvovna Roumiantseva. 2022. "Molecular Phylogenetic Analysis of Salt-Tolerance-Related Genes in Root-Nodule Bacteria Species Sinorhizobium meliloti" Agronomy 12, no. 8: 1968. https://doi.org/10.3390/agronomy12081968
APA StyleMuntyan, V. S., & Roumiantseva, M. L. (2022). Molecular Phylogenetic Analysis of Salt-Tolerance-Related Genes in Root-Nodule Bacteria Species Sinorhizobium meliloti. Agronomy, 12(8), 1968. https://doi.org/10.3390/agronomy12081968






