Abstract
Mung bean is vulnerable to bruchids (Callosobruchus spp.), resulting in low quality and losses worldwide. Developing resistant cultivars is the most effective, economical, and eco-friendly way to protect mung bean seeds from the damage of bruchids. Previously, we identified two gene loci that are tightly linked with bruchid resistance, which makes new cultivar development possible using molecular assisted selection (MAS). In the present study, marker-assisted backcross (MABC) breeding was employed to introgress the bruchid resistance gene VrPGIP2 locus from the donor parent V2802 into a popular cultivar, Kamphaeng Saen 1. Markers VrBR-SSR013 and DMB-SSR158 were used for foreground selection, and phenotypic selection was used for background selection. Three advanced inbred lines (R67-11, R67-22, and R92-15) carrying the resistance gene VrPGIP2 were developed from the BC3F6 population. Further analysis showed that the line R67-22 performed best; it was highly resistant to bruchids and showed excellent agronomic traits in the field. Therefore, R67-22 could be further evaluated in large-scale trials for release as a variety.
1. Introduction
Mung bean (Vigna radiata (L.) Wilczek), also known as green gram, is a key legume crop member of the family Leguminosae, subfamily Papilionaceae, and genus Vigna. Mung bean originated on the Indian subcontinent and is adapted to tropical and subtropical regions [1]. About 90% of global mung bean production is in Asia, where India is the largest producer, followed by China. Mung bean has also spread to other countries of Africa, South America, and Australia [1,2]. Mung bean occupies a unique position in agricultural systems due to its multiple benefits, including high nutritional value, shorter life span, wider adaptability, soil-ameliorative properties (nitrogen fixing capacity), high biomass production, and multifarious uses in numerous food preparations [1,2,3,4]. Usually, mung bean is cultivated as a monoculture, as part of a crop rotation, or in intercropping with diverse cereals [5,6]. Therefore, mung bean plays a crucial role in agricultural sustainability, the adjustment of planting structures, and agricultural disaster recovery. However, the yield potentiality and the stability of grain quality of mung bean are restricted by abiotic and biotic constraints [7,8,9].
Bruchids (Callosobruchus spp.), also known as seed weevils, are detrimental storage pests in mung bean and other legume crops. About 20 species, belonging to six genera, of bruchids infest different grain legumes [10]. Two species, Callosobruchus maculatus F. (cowpea weevil) and Callosobruchus chinensis L. (azuki bean weevil), both belonging to order Coleoptera and family Bruchidae, cause the most damage worldwide in mung bean crops [11,12]. The initial infestation occurs in the field, which causes minor damage. After harvesting, a secondary infestation will happen during storage, which is more damaging, usually resulting in the compete loss of a seed lot within a few months [13]. Conventional approaches, including cultural, physical, biological, phytochemical, and chemical methods, do not effectively control the occurrence of bruchids and may also induce issues concerning food safety and environmental hazards [14]. Thus, breeding resistant cultivars is the most effective, economical, and safe way to solve the problems caused by this pest, which is also a major goal in mung bean breeding programs. Several sources of bruchid resistance have been identified both in wild and cultivated mung bean. TC1966 was the first reported wild mung bean (V. radiata var. Sublobata (Roxb.) Verdc.) possessing high resistance to bruchids (C. chinensis, C. maculatus, Callosobruchus phaseoli (Gyllenhal), and Zabrotes subfasciatus Boheman) [15], and another two sublobata accessions, ACC23 and ACC41, from Australia also show resistance to C. chinensis, C. maculatus, and C. phaseoli [11]. However, the cultivars derived from wild mung bean may have food safety risks [16], as well as linkage drag of undesirable traits, such as pod dehiscence [17]. Four cultivated mung bean accessions, including V1128, V2709, V2802, and V2817, have been reported to show moderate to high resistance to C. chinensis and C. maculatus [12,18], which was controlled by a single dominant gene (Br locus) with modifiers [19].
Although mung bean germplasms possessing resistance to bruchids were identified a long time ago [11,12,15,18], not many improved cultivars with bruchid resistance have been developed. The progress in the development of bruchid-resistant cultivar(s) is slow due to the fact that the seed resistance evaluation takes at least 50–60 days. Therefore, selection time in a single generation is relatively long, at about 130–140 days (including seed production). Additionally, bruchid evaluation can be expensive and tedious, since breeders have to maintain bruchid cultures all the time and have growth chamber(s) or insect room for the bruchids. Marker-assisted selection (MAS) is one of the most promising tools for breeding, using molecular markers to identify and select genes or genomic regions of interest without regular phenotyping [20]. Using MAS can overcome the difficulties in breeding for bruchid resistance and accelerate the development of bruchid-resistant mung bean. Several years ago, a sequence-tagged site (STS)-based marker, STSbr1, linked with the bruchid resistance gene in ACC41, showed polymorphism among an Indian sublobata accession (Sub2) and 12 other mung bean cultivars [21], while the simple sequence repeat (SSR) marker DMB-SSR158 (DMB158 in the original article) was found to be co-segregated perfectly with the Br locus in V2802 [22]. Subsequently, Chotechung et al. [23] showed that VrPGIP2 is the candidate gene at the Br locus in V2802, which explained 93.34–93.84% of the total variation in the proportion of damaged seeds caused by C. chinensis and C. maculatus, and V2802, TC1966, ACC41, V1128, and V2817 shared the same VrPGIP2 gene. They also showed that two tightly linked SSR markers, VrBR-SSR013 and DMB-SSR158, with a physical distance of 38 Kb, covered VrPGIP2. Subsequently, VrPGIP1 and VrPGIP2, two tightly linked genes associated with the Br locus, were reported to confer bruchid resistance in V2709 [24], while VrPGIP1 is associated with the Br locus in ACC41 [25]. The function of these two genes in bruchid resistance has been verified in our previous study [26].
Marker-assisted backcross (MABC) breeding is a MAS method that is popularly used in breeding programs to incorporate gene(s) into an elite variety [27]. In this study, we utilized MABC breeding to introgress the bruchid resistance gene VrPGIP2 locus from V2802 into the popular cultivar Kamphaeng Saen 1 (hereafter called KPS1). We obtained an advanced line, R67-22, with high bruchid resistance and similar agronomic traits to KPS1, which could be further evaluated in large-scale trials for release as a variety.
2. Materials and Methods
This project was conducted at the Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences (JAAS), Nanjing City, Jiangsu Province, China, from 2016 to 2021. The experimental field was located in the Luhe District, Nanjing (latitude 32.34222° N and longitude 118.8413° W).
2.1. Parental Materials
The recurrent parent KPS1 is a commercial cultivar from Thailand, which was selected from VC1973A, an elite breeding line developed by the Asian Vegetable Research and Development Center (AVRDC; current name World Vegetable Center). KPS1 is a high-yield and large-seeded cultivar, with 6.13 g per 100 seeds, but is highly susceptible to bruchids [12]. VC1973A is also the parental line of ‘Zhonglv 1’, the first nationwide popular mung bean cultivar of China, which has revolutionized mung bean production in China due to its excellent characteristics, including compact plant type, lodging resistance, early maturity, and high yield [28].
The donor parent V2802 (PI 25461) is a landrace from the Philippines with moderate to high resistance to C. chinensis and C. maculatus [12,18]. It is a medium-seeded cultivar (4.45 g per 100 seeds) with a dull green seed coat luster [19]. In China, the agricultural traits of V2802, including the trailing growth habit, late maturity (response to a long photoperiod), and seed size, are not suitable for commercial production.
The phenotype and seed characteristics of KPS1 and V2802 are shown in Supplementary Figure S1.
2.2. Marker-Assisted Backcross (MABC) Breeding Strategy for Bruchid Resistance
The breeding strategy in this study is depicted in Figure 1. Hybridization between KPS1 (female) and V2802 (male) plants was conducted at the Luhe experimental field during the period from August to October 2016. The F1 plants were identified by hypocotyl color (purple), and the true hybrid plants were backcrossed with KPS1 for three generations until BC3F1, which took about two years (from May 2017 to July 2018). About 20 individuals showing heterozygous markers and with similar phenotypes to KPS1 were selected to conduct backcrossing until the BC3F1 generation. Then, self-pollination was performed to obtain a BC3F2 population in 2018 (from August to October). The single seed descent method was used to obtain a stable BC3F6 generation during the period from 2019 to 2021.
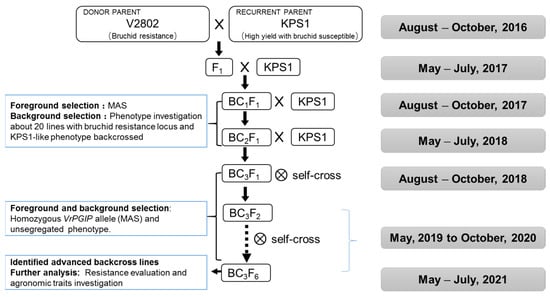
Figure 1.
Introgression of the VrPGIP2 gene, conferring bruchid resistance, from V2802 into KPS1 using the MABC breeding strategy.
For foreground selection, markers VrBR-SSR013 and DMB-SSR158, which co-segregated perfectly with the Br locus in V2802 [23], were used (Supplementary Table S1) in each generation from BC1F1 until BC3F6. For background selection, phenotypic selection was carried out. Because the difference in morphological characteristics between KPS1 and V2802 was large (V2802 showed a trailing growth habit and late maturity), phenotypic selection was effective in this study.
2.3. DNA Extraction and Polymerase Chain Reaction (PCR) Amplification
Genomic DNA was extracted from young leaves of the mung bean plant using a modified CTAB method [29]. PCR reactions were performed using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China), following the manufacturer’s instructions, with 10 µL PCR mixture. The amplified products were electrophoresed using 8% non-denaturing polyacrylamide gels in 0.5 × TBE buffer, and the bands were visualized by silver staining [30].
2.4. RNA Extraction and cDNA Synthesis
Total RNA was extracted using an RNA prep Pure plant kit (Tiangen, Beijing, China) from the young leaves of mung bean plants, and cDNA was synthesized using HiScript II Q RT SuperMix reagent Kit (Vazyme, Nanjing, China), following the manufacturer instructions. The coding sequences of VrPGIP2 (LOC106760237) were amplified using Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China) and inserted into the pClone007 Vector (Tsingke, Beijing, China) for sequencing.
2.5. Evaluation of Bruchid Resistance
Seeds from the parents (KPS1and V2802) and the advanced lines (R67-11, R67-22, and R92-15) developed in this study (see Results) were used for the bruchid resistance test and replicated three times following the previously described method [19]. Fifty seeds from each plant were placed in a small round box (10 cm in diameter and 3 cm in height). Then, 30 (15 males and 15 females) newly emerged (1–3 days old) C. maculatus adults were introduced into the box for egg oviposition for 7 days and then removed. The experiment was performed at 28 °C, 70% RH. Twenty days after bruchid introduction, the number of emerging adult bruchids was counted, and the number of damaged seeds was counted after 60 days.
Based on the percentage of damaged seeds, the resistance to bruchids was classified into five classes, as follows [31]:
- (1)
- Highly resistant (HR): 0% to 10%.
- (2)
- Resistant (R): 10% to 35%.
- (3)
- Moderately resistant (MR): 35% to 65%.
- (4)
- Susceptible (S): 65% to 90%.
- (5)
- Highly susceptible: 90% to 100%.
2.6. Investigation of Agronomic Traits
The agronomic traits [31], including plant height (cm), main stem node number, branch number, seed number per pod, 100-seed weight (g), and seed yield per plant (g), were recorded in the field plot experiment. Twelve individual plants of each variety (V2802, KPS1, R67-11, R67-22 and R92-15) were analyzed. Statistical analysis was performed with GraphPad and SPSS 24.0 software.
2.7. Genetic Background Analysis
The whole genomes of R67-22, V2802, and KPS1 underwent re-sequencing and paired-end sequencing via the Illumina HiSeq 2500 platform (Illumina Technologies, BIOZERON, Shanghai, China) to generate reads in accordance with a previous study [30]. After filtering out low-quality reads, 118 SNPs (Supplementary Table S2) were selected for genetic background characterization of R67-22.
3. Results
3.1. Development of Bruchid-Resistant Lines through MABC Breeding
The breeding scheme for improving bruchid resistance in KPS1 through MABC is shown in Figure 1. We obtained 20 true F1 hybrid plants of the cross KPS1 × V2808, which could be easily distinguished by the purple color of the hypocotyl, a dominant trait derived from V2802. Then, all the F1 plants were backcrossed with KPS1. From BC1F1 to BC3F1 generations, about 100 lines were planted in each generation, and MAS was conducted to identify and select 20 backcross progenies carrying the Br locus from V2802 for backcrossing. Starting with the BC3F1 generation, the single-seed descent method was used to develop the BC3F6 generation. MAS and phenotypic selection were carried out until we obtained stable lines.
The polymorphism of the markers VrBR-SSR013 and DMB-SSR158 was reconfirmed in the BC3F6 population (Supplementary Figure S2). Eventually, three advanced lines (R67-11, R67-22, and R92-15) were selected from the BC3F6 population. These lines had similar phenotypes to KPS1 and carried the Br locus from V2802, detected by both VrBR-SSR013 and DMB-SSR158. Further, to detect whether the VrPGIP2 gene (LOC106760237) from V2802 was indeed introgressed into R67-11, R67-22, and R92-15, the coding sequences (CDS) of the VrPGIP2 gene from these lines were amplified and sequenced. Sequence alignment showed that VrPGIP2 of the R67-11, R67-22, and R92-15 lines were all identical to that of V2802 (Figure 2). Thus, the introgression was successful, and these three potential lines were selected for further analysis.

Figure 2.
Alignment of the coding sequence of VrPGIP2 from KPS1, V2802, R67-11, R67-22, and R92-15 (portion containing differential bases). The numbers labeled under the bases indicate the location of SNPs from the start codon.
3.2. R67-11, R67-22, and R92-15 Lines All Showed Resistance to Bruchid
To further examine the bruchid resistance of mung bean seeds from R67-11, R67-22, and R92-15, mature seeds from these lines, KPS1 and V2802 were tested for resistance against C. maculatus. The developmental period of bruchids, the number of adults that emerged, and the percentage of damaged seeds were statistically analyzed, respectively, and the resistance level was assessed (Table 1). The results showed that both R67-11 and R67-22 were highly resistant to C. maculatus, while R92-15 was resistant (Table 1 and Figure 3). As for the parents, the seeds from V2802 appeared moderately resistant (MR), and the seeds from KPS1 were almost completely damaged (HS) by C. maculatus (Table 1 and Figure 3), which was consistent with the results of previous studies [12,23]. Resistant mung bean seeds can restrict the growth and development of C. maculatus, as the developmental period of C. maculatus larva was prolonged significantly on the seeds of the R67-11, R67-22, and R92-15 lines (about 40–44 days) compared to KPS1 (about 25 days) and V2802 (about 32 days). Meanwhile, the number of adults that emerged was significantly decreased in the seeds of the R67-11, R67-22, and R92-15 lines (Table 1). It is worth noting that the advanced backcross lines expressed higher resistance to C. maculatus than their donor parent V2802.

Table 1.
C. maculatus resistance in advanced backcross lines and their parents.

Figure 3.
The resistance to C. maculatus of the seeds from R67-11, R67-22, and R92-15 compared with KPS1 and V2802.
3.3. The Agronomic Traits of Bruchid-Resistant Lines
To identify whether the introgression of bruchid resistance locus Br from V2802 would affect the agronomic traits of R67-11, R67-22, and R92-15, we investigated six main traits concerning yield, including plant height, main stem node number, branch number, seed number per pod, 100-seed weight (seed size), and seed yield per plant. As expected, the agronomic traits of the three advanced backcross lines were far superior to the donor parent V2802 (Table 2). However, R67-11 and R92-15 were still inferior to KPS1 for yield-related traits. The seed size and seed yield per plant of R67-11 and R92-15 were significantly less than those of KPS1. Nonetheless, R67-22 performed on par with KPS1 (Table 2 and Figure 4). Six agronomic traits of R67-22 were not significantly different to those of KPS1, albeit that it showed a slightly higher branch number and main stem node number, as well as a slightly lower seed yield per plant. Thus, R67-22 appeared to be a potential improved line for release as a variety.

Table 2.
Comparison of six agronomic traits between parental lines and advanced backcross lines.

Figure 4.
The plant phenotype and seed traits contrasted between R67-22 and KPS1. The comparison included the plant morphology of KPS1 (A), R67-22 (B), ripe pods (C), and mature seeds (D).
3.4. The Genetic Background of R67-22 Showed Highly Similarity to KPS1
In this study, background selection was performed through phenotypic selection. The genetic background of the line R67-22 was assessed by whole-genome resequencing (10× depth). In total, 118 SNPs between KPS1 and V2802 (Supplementary Table S2) were identified and used for background recovery analysis. As shown in Figure 5, the Br locus containing VrPGIP genes on chromosome 5 from V2802 was introgressed into the genome of R67-22. The sequences of chromosomes 1, 2, 3, 7, and 9 of R67-22 were almost identical to the KPS1 genome. On the other chromosomes of R67-22, there were only a few fragments derived from V2802, and this genomic residue was reasonable in a BC3 progeny. In addition, R67-22 was homozygous at all 118 SNPs (Supplementary Table S2), indicating the high genome stability of this line.
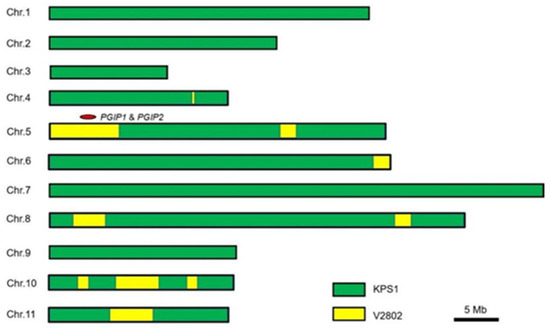
Figure 5.
Genetic background of R67-22 determined by whole-genome resequencing.
4. Discussion
To the best of our knowledge, improved cultivars with bruchid resistance are only available in China and Korea [28,32]. These cultivars were developed by conventional breeding methods. However, because bruchids evolve rapidly and may overcome resistance conferred by a single resistance gene in existing cultivars [33], more resistant cultivars with different resistance genes must be developed. Although bruchid resistance was the first trait subjected to gene mapping in the mung bean, developing bruchid-resistant mung bean cultivars through marker-assisted breeding (MAB) is still in its infancy due to the unavailability of closely linked markers [13,19]. Nonetheless, recent progress in gene mapping of bruchid resistance [23,24,25,26] has made the improvement of popular mung bean cultivars’ bruchid resistance possible through MABC. In this study, we successfully developed a bruchid-resistant mung bean using the VrPGIP2 gene from V2802 via MABC. MABC is regarded as one of the most efficient approaches to crop breeding [27] and has been applied widely in major crops such as rice [34,35], wheat [36], maize [37], and soybean [38]. In fact, the use of MAB in mung bean has been very limited. Only recently, Papan et al. [39] reported MAB in mung bean for the first time. They developed mung bean lines with resistance to both Cercospora leaf spot and powdery mildew diseases through MABC.
In this study, MAB was performed only by foreground selection of VrPGIP2, although MAB for background selection can increase background genome recovery and reduce the number of backcross generations in MABC [20]. Background selection was carried out by phenotypic selection because marker polymorphism (3%) between V2802 and KPS1 is very low [22], and thus background selection by MAS is unsuitable. Three bruchid-resistant lines, R67-11, R67-22, and R92-15, with high yields and good agronomic traits, were developed, and R67-22 showed potential as a new cultivar (Table 2 and Figure 4). Whole-genome resequencing confirmed that the VrPGIP2 gene from V2802 was successfully introgressed into KPS1 and revealed only 52 SNPs between KPS1 and R67-22, indicating a high percentage of the genome of KPS1 recovery in R67-22 (Supplementary Table S2 and Figure 5). Interestingly, R67-11 and R67-22 showed higher resistance than the donor parent V2802 (Table 1). This suggested that expression of the VrPGIP2 gene and/or modifying gene(s) in the KPS1 background genome was stronger than in V2802, and that some unknown factor(s) from KPS1 may promote resistance to bruchids. Additional studies should be conducted to investigate this transgressive segregation of bruchid resistance.
In conclusion, we successfully developed new bruchid-resistant lines by introgressing the Br locus (VrPGIPs) from V2802 into a popular cultivar, KPS1, using the MABC breeding strategy. The markers VrBR-SSR013 and DMB-SSR158 provided precision selection of the Br locus and saved 2–3 years in selection and backcrossing for resistance. Finally, we obtained one advanced line (R67-22) from the BC3F6 population. The bruchid resistance of this line was stronger than that of V2802, while its agronomic traits were on par with KPS1. Thus, the R67-22 line shows promise for release as a variety, which will be further evaluated in large-scale trials. MABC, using the markers VrBR-SSR013 and DMB-SSR158, is therefore a highly efficient approach for the breeding of bruchid-resistant mung bean cultivars.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12061271/s1, Supplementary Figure S1. The phenotype (A and B) and seed characteristics (C) of KPS1 and V2802. Supplementary Figure S2. Reconfirmation of the polymorphism of VrBR-SSR013 and DMB-SSR158 in genotyping. Supplementary Table S1. Primers used for marker-assisted selection of the VrPGIP2 gene conferring bruchid resistance in V2802. Supplementary Table S2. Information on 118 SNPs from the genetic background analysis.
Author Contributions
Conceptualization, X.Y. and X.C.; methodology, P.S., J.C. and Y.L.; software, J.L.; validation, R.W. and C.X.; formal analysis, Q.Y. and C.X.; investigation, Q.Z., J.C., Y.L. and P.S.; writing—original draft preparation, R.W.; writing—review and editing, Y.L. and P.S.; visualization, Q.Z. and J.L.; supervision, X.Y.; project administration, X.C. and X.Y.; funding acquisition, R.W., X.C., P.S. and X.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (2019YFD1001300, 2019YFD1001301), the Natural Science Foundation of Jiangsu Province (BK20190257), China Agriculture Research System of MOF and MARA-Food Legumes (CARS-08-G15), the Jiangsu Seed Industry Revitalization Project (JBGS (2021)004), and the Office of National Higher Education Science Research and Innovation Policy Council, Thailand, via the Program Management Unit for Human Resources and Institutional Development, Research and Innovation (B16F640185).
Data Availability Statement
Supporting information is available from the Wiley Online Library or from the authors.
Acknowledgments
We are thankful to Heping Gu for carrying out the field experiment.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tomooka, N.; Vaughan, D.A.; Moss, H.; Maxted, N. The Asian Vigna: Genus Vigna Subgenus Ceratotropis Genetic Resources; Springer: Dordrecht, The Netherlands, 2002. [Google Scholar]
- Nair, R.; Schreinemachers, P. Global Status and Economic Importance of Mungbean. In The Mungbean Genome; Nair, R.M., Schafleitner, R., Lee, S.-H., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–8. [Google Scholar]
- Somta, P.; Srinives, P. Genome research in mungbean (Vigna radiata (L.) Wilczek) and blackgram (V. mungo (L.) Hepper). ScienceAsia 2007, 33, 69–74. [Google Scholar] [CrossRef]
- Lambrides, C.J.; Godwin, I.D. Mungbean. In Pulses, Sugar and Tuber Crops. Genome Mapping & Molecular Breeding in Plants; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 3, pp. 69–90. [Google Scholar]
- Wang, L.X.; Wang, S.H.; Luo, G.L.; Zhang, J.T.; Chen, Y.H.; Chen, H.L.; Cheng, X.Z. Evaluation of the production potential of mung bean cultivar “Zhonglv 5”. Agronomy 2022, 12, 707. [Google Scholar] [CrossRef]
- Mandal, B.K.; Dhara, M.C.; Mandal, B.B.; Das, S.K.; Nandy, R. Rice, mungbean, soybean, peanut, ricebean, and blackgram yields under different intercropping systems. Agron. J. 1990, 82, 1063–1066. [Google Scholar] [CrossRef]
- Laosatit, K.; Somta, P.; Xin, C.; Srinives, P. Approaches to Biotic Stresses. In The Mungbean Genome; Nair, R.M., Schafleitner, R., Lee, S.-H., Eds.; Springer: Cham, Switzerland, 2020; pp. 133–167. [Google Scholar]
- Batzer, J.C.; Singh, A.; Rairdin, A.; Chiteri, K.; Mueller, D.S. Mungbean: A preview of disease management challenges for an alternative U.S. cash crop. J. Integr. Pest. Manag. 2022, 13, 4. [Google Scholar] [CrossRef]
- Basu, P.S.; Pratap, A.; Gupta, S.; Sharma, K.; Tomar, R.; Singh, N.P. Physiological traits for shortening crop duration and improving productivity of greengram (Vigna radiata L. Wilczek) under high temperature. Front. Plant Sci. 2019, 10, 1508. [Google Scholar] [CrossRef] [Green Version]
- Southgate, B.J. Biology of the Bruchidae. Annu. Rev. Entomol. 1979, 24, 449–473. [Google Scholar] [CrossRef]
- Lambrides, C.J.; Imrie, B.C. Susceptibility of mungbean varieties to the bruchid species Callosobruchus maculatus (F.), C.phaseoli (Gyll.), C.chinensis (L.), and Acanthoscelides obtectus (Say.) (Coleoptera: Chrysomelidae). Aust. J. Agric. Res. 2000, 51, 85–89. [Google Scholar] [CrossRef]
- Somta, C.; Somta, P.; Tomooka, N.; Ooi, P.a.C.; Vaughan, D.A.; Srinives, P. Characterization of new sources of mungbean (Vigna radiata (L.) Wilczek) resistance to bruchids, Callosobruchus spp. (Coleoptera: Bruchidae). J. Stored Prod. Res. 2008, 44, 316–321. [Google Scholar] [CrossRef]
- Somta, P. Genetics and breeding of resistance to bruchids (Callosobruchus spp.) in Vigna Crops: A review. NU. Int. J. Sci. 2007, 4, 1–17. [Google Scholar]
- Mishra, S.K.; Macedo, M.L.R.; Panda, S.K.; Panigrahi, J. Bruchid pest management in pulses: Past practices, present status and use of modern breeding tools for development of resistant varieties. Ann. Appl. Biol. 2018, 172, 4–19. [Google Scholar] [CrossRef]
- Fujii, K.; Miyazaki, S. Infestation resistance of wild legumes (Vigna Sublobata) to azuki bean weevil, Callosobruchus Chinensis (L.) (Coleoptera, Bruchidae) and its relationship with cytogenetic classification. Appl. Entomol. Zool. 1987, 22, 229–230. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Ishimotob, M.; Yamanakaa, N.; Miyazaki, S.; Hiramatsu, M.; Nakajima, Y.; Hirano, K.; Krtamura, K.; Miyazaki, S. Effects of bruchid-resistant mungbean meal on growth and blood-biochemical values in mice. JIRCAS J. 1996, 3, 23–31. [Google Scholar]
- Watanasit, A.; Pichitporn, S. Improvement of mung-bean for resistance to bruchids. In Mungbean Germ-Plasm: Collection, Evaluation and Utilization Forbreeding Program; Srinives, P., Kitbamroong, C., Miyazaki, S., Eds.; Japan International Research Center for Agricultural Sciences: Tsukuba, Japan, 1996; pp. 67–71. [Google Scholar]
- Talekar, N.S.; Lin, C.P. Characterization of Callosobruchus Chinensis (Coleoptera, Bruchidae) resistance in mungbean. J. Econ. Entomol. 1992, 85, 1150–1153. [Google Scholar] [CrossRef]
- Somta, P.; Ammaranan, C.; Ooi, P.a.C.; Srinives, P. Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata, L. Wilczek). Euphytica 2007, 155, 47–55. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Ghosh, S.; Chatterjee, M.; Das, P.; Lahari, T.; Maji, A.; Mondal, N.; Pradhan, K.K.; Bhattacharyya, S. Molecular markers linked with bruchid resistance in Vigna radiata var. sublobata and their validation. J. Plant Biochem. Biotechnol. 2011, 20, 155–160. [Google Scholar] [CrossRef]
- Chotechung, S.; Chankaew, S.; Srinives, P.; Somta, P. Identification of DNA markers associated with bruchid resistance in mungbean. Khon Khan Agric. J. 2011, 39, 221–226, (In Thai with English Abstract). [Google Scholar]
- Chotechung, S.; Somta, P.; Chen, J.; Yimram, T.; Chen, X.; Srinives, P. A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: Bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 2016, 129, 1673–1683. [Google Scholar] [CrossRef]
- Kaewwongwal, A.; Chen, J.B.; Somta, P.; Kongjaimun, A.; Yimram, T.; Chen, X.; Srinives, P. Novel alleles of two tightly linked genes encoding polygalacturonase-inhibiting proteins (VrPGIP1 and VrPGIP2) associated with the Br. locus that confer bruchid (Callosobruchus spp.) resistance to mungbean (Vigna radiata) accession V2709. Front. Plant Sci. 2017, 8, 1692. [Google Scholar] [CrossRef]
- Kaewwongwal, A.; Liu, C.Y.; Somta, P.; Chen, J.B.; Tian, J.; Yuan, X.X.; Chen, X. A second VrPGIP1 allele is associated with bruchid resistance (Callosobruchus spp.) in wild mungbean (Vigna radiata var. sublobata) accession ACC41. Mol. Genet. Genom. 2020, 295, 275–286. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Yan, Q.; Yuan, X.X.; Lin, Y.; Chen, J.B.; Wu, R.R.; Xue, C.C.; Zhu, Y.L.; Chen, X. Two polygalacturonase-inhibiting proteins (VrPGIP) of Vigna radiata confer resistance to bruchids (Callosobruchus spp.). J. Plant Physiol. 2021, 258–259, 153376. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Melchinger, A.E. Selection theory for marker-assisted backcrossing. Genetics 2005, 170, 909–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, J.; Cheng, X.Z.; Fan, B.J.; Wang, L.X.; Liu, J.J.; Liu, C.Y.; Wang, S.H.; Cao, Z.M.; Chen, H.L.; Wang, Y.; et al. Current situation and development trend of mung bean varieties in China. Crops 2021, 6, 15–21. (In Chinese) [Google Scholar]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular-weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.; Laosatit, K.; Chen, J.B.; Yuan, X.X.; Wu, R.R.; Amkul, K.; Chen, X.; Somta, P. Mapping and functional characterization of Stigma Exposed 1, a DUF1005 gene controlling petal and stigma cells in mungbean (Vigna radiata). Front. Plant Sci. 2020, 11, 575922. [Google Scholar] [CrossRef]
- Cheng, X.Z.; Wang, S.H.; Wang, L.X. Descriptors and Data Standard for Mungbean [Vigna radiata (L.) Wilczek]; China Agriculture Press: Beijing, China, 2006. (In Chinese) [Google Scholar]
- Lee, Y.H.; Moon, J.K.; Park, K.Y.; Ku, J.H.; Chung, M.N. A new mungbean cultivar with bruchid resistance, ‘Jangannogdu’. Korean J. Breed. 2000, 32, 296–297. [Google Scholar]
- Fox, C.W.; Bush, M.L.; Messina, F.J. Biotypes of the seed beetle Callosobruchus maculatus have differing effects on the germination and growth of their legume hosts. Agric. For. Entomol. 2010, 12, 353–362. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Nayak, D.K.; Pandit, E.; Behera, L.; Anandan, A.; Mukherjee, A.K.; Lenka, S.; Barik, D.P. Incorporation of bacterial blight resistance genes into lowland rice cultivar through marker-assisted backcross breeding. Phytopathology 2016, 106, 710–718. [Google Scholar] [CrossRef] [Green Version]
- Hasan, M.M.; Rafii, M.Y.; Ismail, M.R.; Mahmood, M.; Rahim, H.A.; Alam, M.A.; Ashkani, S.; Malek, M.A.; Latif, M.A. Marker-assisted backcrossing: A useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015, 29, 237–254. [Google Scholar] [CrossRef] [Green Version]
- Mallick, N.; Sharma, J.B.; Tomar, R.S.; Sivasamy, M.; Prabhu, K.V. Marker-assisted backcross breeding to combine multiple rust resistance in wheat. Plant Breed. 2015, 134, 172–177. [Google Scholar] [CrossRef]
- Yang, L.Q.; Wang, W.; Yang, W.P.; Wang, M.C. Marker-assisted selection for pyramiding the waxy and opaque-16 genes in maize using cross and backcross schemes. Mol. Breed. 2013, 31, 767–775. [Google Scholar] [CrossRef]
- Maranna, S.; Verma, K.; Talukdar, A.; Lal, S.K.; Kumar, A.; Mukherjee, K. Introgression of null allele of Kunitz trypsin inhibitor through marker-assisted backcross breeding in soybean (Glycine max L. Merr.). BMC Genet. 2016, 17, 106. [Google Scholar] [CrossRef] [Green Version]
- Papan, P.; Chueakhunthod, W.; Jinagool, W.; Tharapreuksapong, A.; Masari, A.; Kaewkasi, C.; Ngampongsai, S.; Girdthai, T.; Tantasawat, P.A. Improvement of Cercospora leaf spot and powdery mildew resistance of mungbean variety KING through marker-assisted selection. J. Agric. Sci. 2022, 159, 676–687. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).