OsVTC1-1 RNAi Mutant with Reduction of Ascorbic Acid Synthesis Alters Cell Wall Sugar Composition and Cell Wall-Associated Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. AsA Biosynthesis Gene Expression Analysis by Real-Time Quantitative PCR (qPCR)
2.3. AsA Content Measurement
2.4. Anatomical Observation of Leaves
2.5. Cell Wall Sugar Composition Analysis
2.5.1. Cell Wall Preparation
2.5.2. GC-MS Analysis and Data Analysis
2.6. Transcriptome Analysis
2.7. Proteome Analysis
2.7.1. Protein Extraction
2.7.2. Protein Digestion
2.7.3. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
2.7.4. Protein Identification
2.8. Statistical Analysis
3. Results
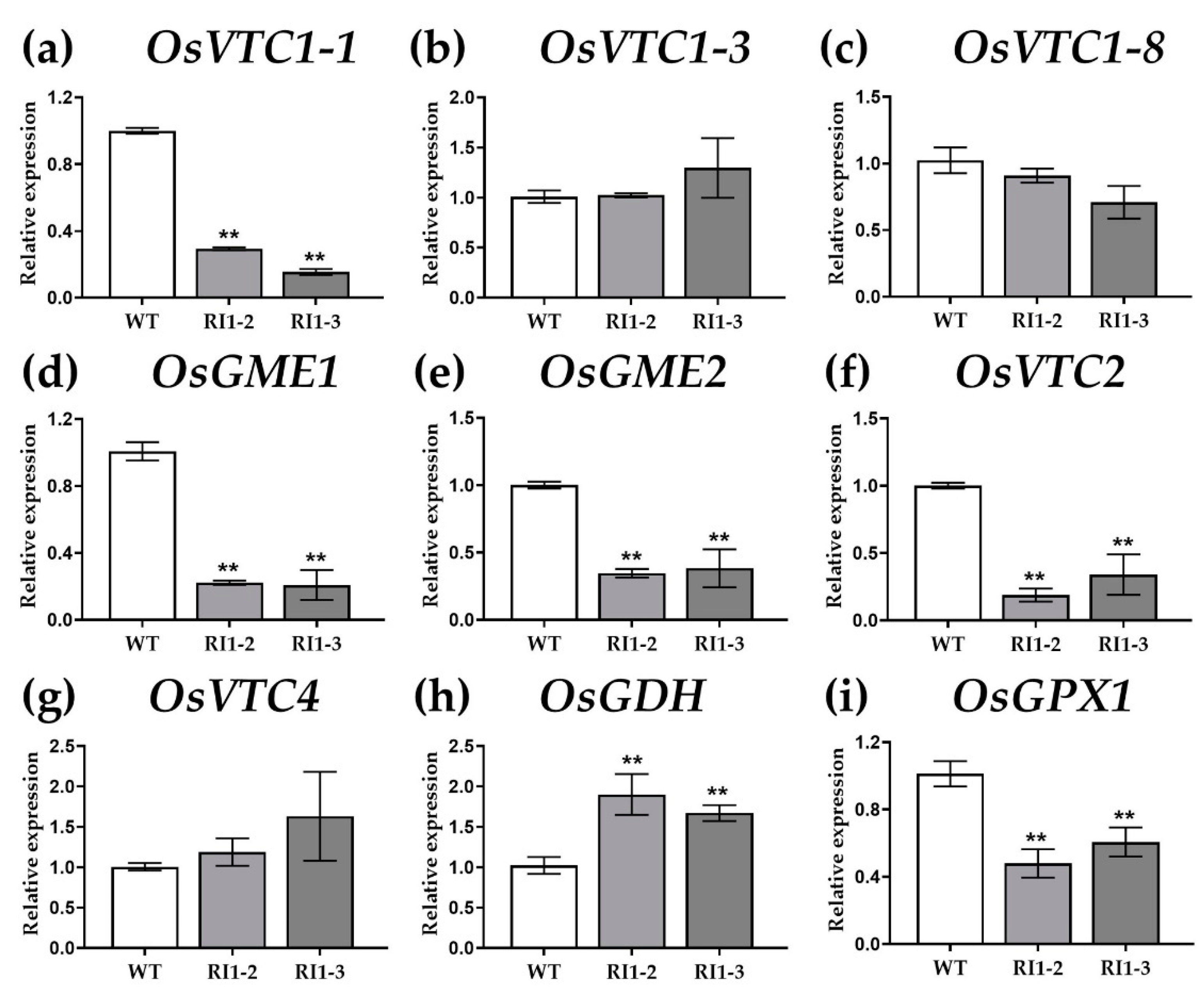
3.1. Gene Expression Analysis by Quantitative Real-Time PCR (qRT-PCR)
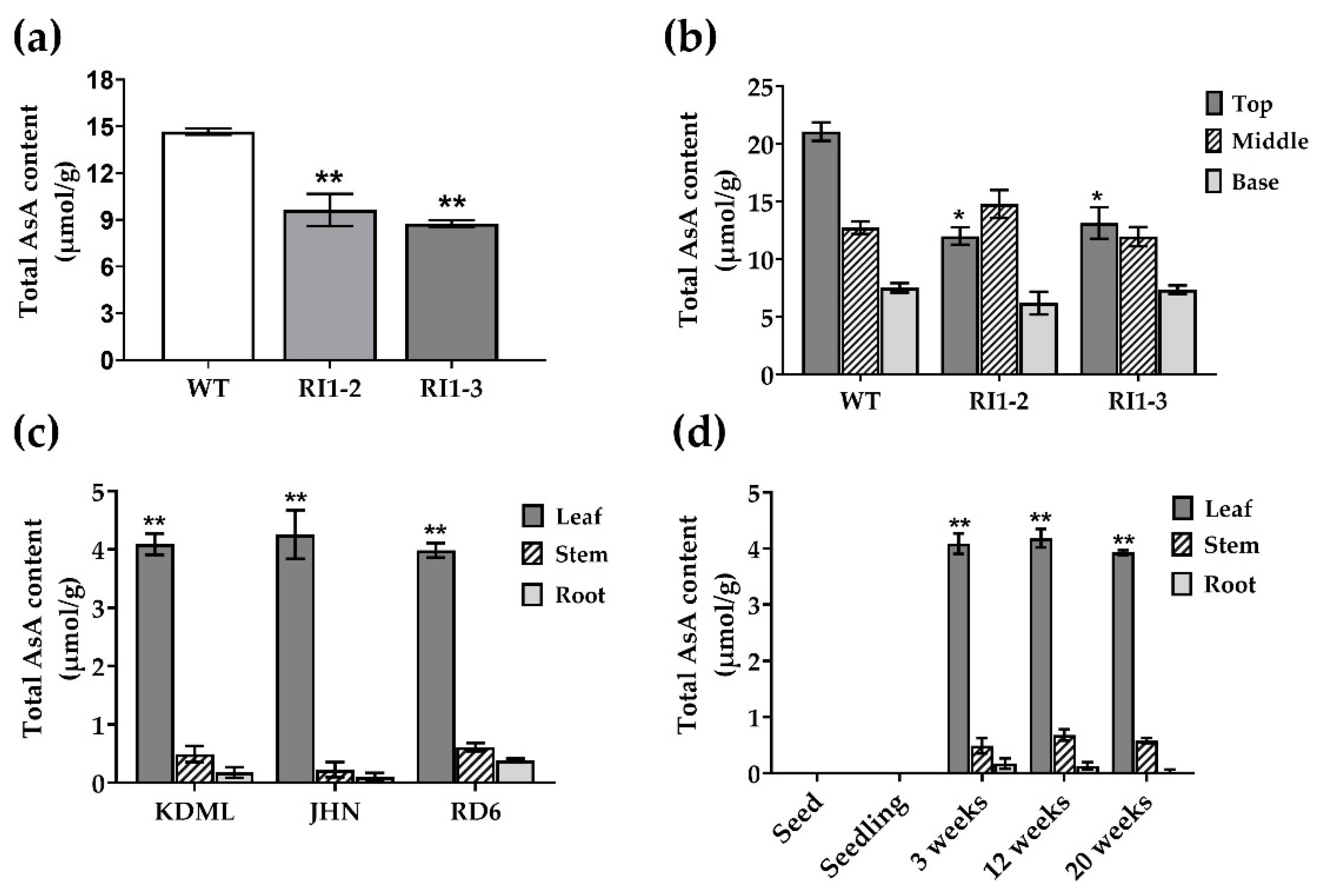
3.2. AsA Measurement
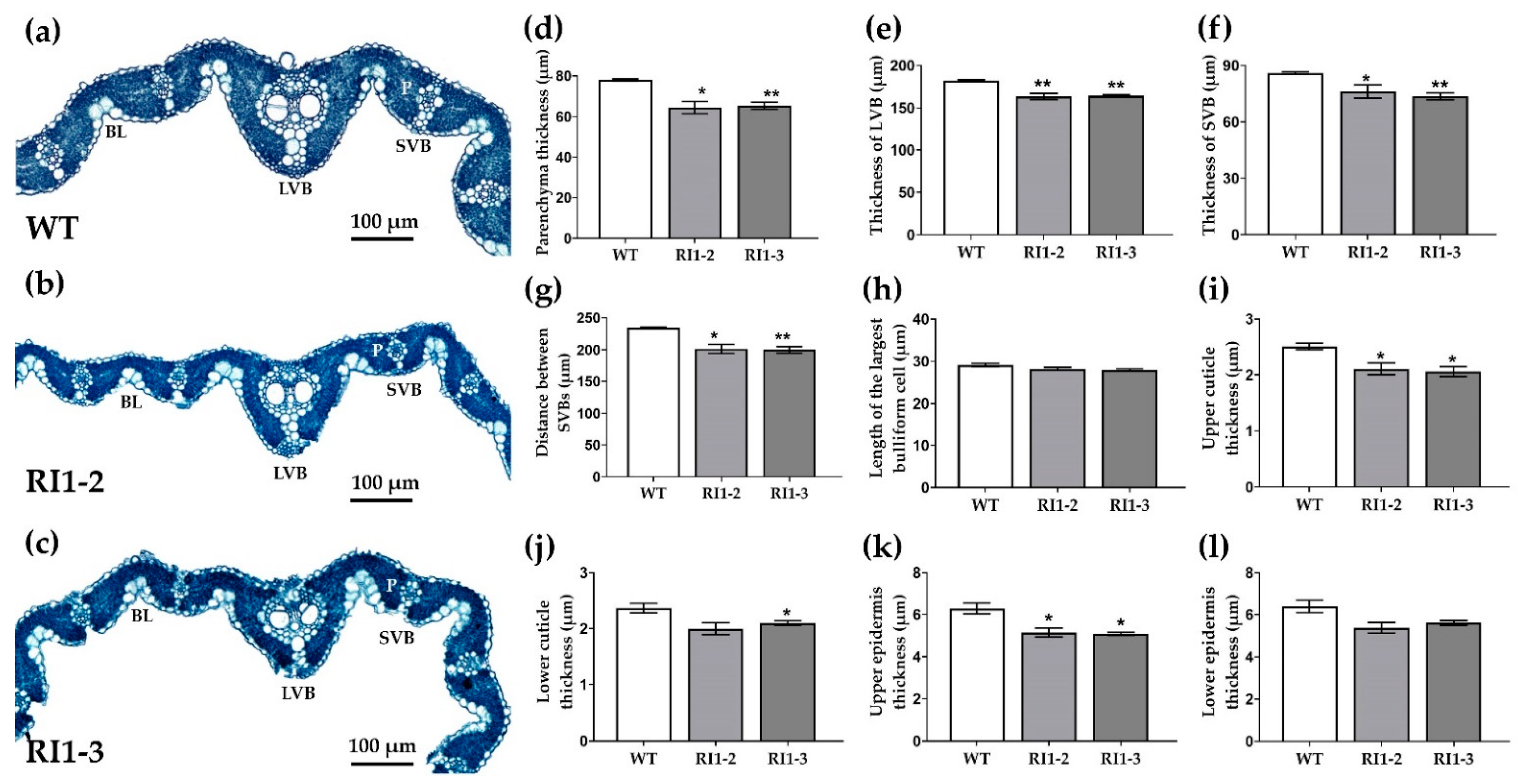
3.3. Leaf Anatomical Comparison between Wild Type and OsVTC1-1 RNAi Lines
3.4. Cell Wall Sugar Composition Analysis
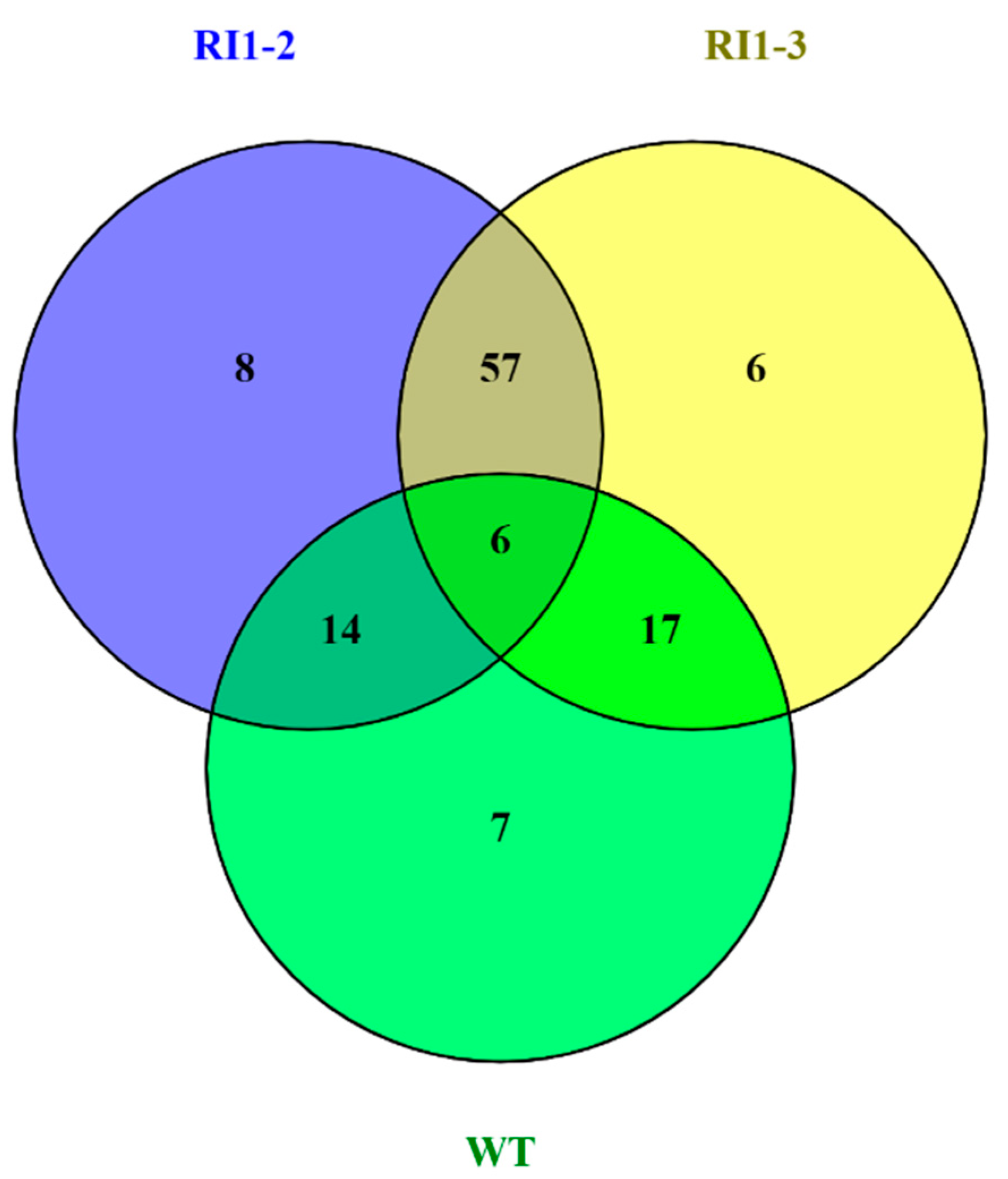
3.5. Transcriptome Analysis
3.6. Proteome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y. Biological Role of Ascorbate in Plants. In Ascorbic Acid in Plants; Springer Briefs in Plant Science; Springer: New York, NY, USA, 2013; pp. 7–33. [Google Scholar]
- Smirnoff, N. The Function and Metabolism of Ascorbic Acid in Plants. Ann. Bot. 1996, 78, 661–669. [Google Scholar] [CrossRef]
- Davey, M.W.; Montagu, M.V.; Inz, D.; Sanmartin, M.; Kanellis, A.; Smirnoff, N.; Benzie, I.J.J.; Strain, J.J.; Favell, D.; Fletcher, J. Plant L-ascorbic acid: Chemistry, function, metabolism, bioavailability and effects of processing. J. Sci. Food Agric. 2000, 80, 825–860. [Google Scholar] [CrossRef]
- Liso, R.; Calabrese, G.; Bitonti, M.B.; Arrigoni, O. Relationship between ascorbic acid and cell division. Exp. Cell Res. 1984, 150, 314–320. [Google Scholar] [CrossRef]
- Smirnoff, N.; Wheeler, G.L. Ascorbic Acid in Plants: Biosynthesis and Function. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 291–314. [Google Scholar] [CrossRef]
- Höller, S.; Hajirezaei, M.-R.; von Wirén, N.; Frei, M. Ascorbate metabolism in rice genotypes differing in zinc efficiency. Planta 2014, 239, 367–379. [Google Scholar] [CrossRef]
- Conklin, P.; Norris, S.; Wheeler, G.; Williams, E.; Smirnoff, N.; Last, R. Genetic evidence for the role of GDP-mannose in plant ascorbic acid (vitamin c) biosynthesis. Proc. Natl. Acad. Sci. USA 1999, 96, 4198–4203. [Google Scholar] [CrossRef]
- Qin, H.; Deng, Z.; Zhang, C.; Wang, Y.; Wang, J.; Liu, H.; Zhang, Z.; Huang, R.; Zhang, Z. Rice GDP-mannose pyrophosphorylase OsVTC1-1 and OsVTC1-3 play different roles in ascorbic acid synthesis. Plant Mol. Biol. 2016, 90, 317–327. [Google Scholar] [CrossRef]
- Pavet, V.; Olmos, E.; Kiddle, G.; Mowla, S.; Kumar, S.; Antoniw, J.; Alvarez, M.; Foyer, C. Ascorbic acid deficiency activates cell death and disease resistance responses in Arabidopsis. Plant Physiol. 2005, 139, 1291–1303. [Google Scholar] [CrossRef]
- Veljovic-Jovanovic, S.D.; Pignocchi, C.; Noctor, G.; Foyer, C.H. Low ascorbic acid in the vtc-1 mutant of Arabidopsis is associated with decreased growth and intracellular redistribution of the antioxidant system. Plant Physiol. 2001, 127, 426–435. [Google Scholar] [CrossRef]
- Qin, H.; Wang, Y.; Wang, J.; Liu, H.; Zhao, H.; Deng, Z.; Zhang, Z.; Huang, R.; Zhang, Z. Knocking down the expression of GMPase gene OsVTC1-1 decreases salt tolerance of rice at seedling and reproductive stages. PLoS ONE 2016, 11, e0168650. [Google Scholar] [CrossRef]
- Qi, T.; Liu, Z.; Fan, M.; Chen, Y.; Tian, H.; Wu, D.; Gao, H.; Ren, C.; Song, S.; Xie, D. GDP-D-mannose epimerase regulates male gametophyte development, plant growth and leaf senescence in Arabidopsis. Sci. Rep. 2017, 7, 10309. [Google Scholar] [CrossRef]
- Ma, L.; Wang, Y.; Liu, W.; Liu, Z. Overexpression of an alfalfa GDP-mannose 3,5-epimerase gene enhances acid, drought and salt tolerance in transgenic Arabidopsis by increasing ascorbate accumulation. Biotechnol. Lett. 2014, 36, 2331–2341. [Google Scholar] [CrossRef]
- Fenech, M.; Amorim-Silva, V.; Esteban del Valle, A.; Arnaud, D.; Ruiz-Lopez, N.; Castillo, A.G.; Smirnoff, N.; Botella, M.A. The role of GDP-l-galactose phosphorylase in the control of ascorbate biosynthesis. Plant Physiol. 2021, 185, 1574–1594. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.Y.; Liu, R.R.; Zhang, C.Q.; Tang, K.X.; Sun, M.F.; Yan, G.H.; Liu, Q.Q. Manipulation of the rice L-galactose pathway: Evaluation of the effects of transgene overexpression on ascorbate accumulation and abiotic stress tolerance. PLoS ONE 2015, 10, e0125870. [Google Scholar] [CrossRef] [PubMed]
- Torabinejad, J.; Donahue, J.; Gunesekera, B.; Allen-Daniels, M.; Gillaspy, G. VTC4 is a bifunctional enzyme that affects myoinositol and ascorbate bosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Fanucchi, M.V. Chapter 11—Development of antioxidant and xenobiotic metabolizing enzyme systems. In The Lung, 2nd ed.; Harding, R., Pinkerton, K.E., Eds.; Academic Press: Boston, MA, USA, 2014; pp. 223–231. [Google Scholar]
- Pehlivan, F. Vitamin C: An antioxidant agent. In Vitamin C; IntechOpen: London, UK, 2017. [Google Scholar]
- Rajput, V.D.; Harish; Singh, R.K.; Verma, K.K.; Sharma, L.; Quiroz-Figueroa, F.R.; Meena, M.; Gour, V.S.; Minkina, T.; Sushkova, S.; et al. Recent developments in enzymatic antioxidant defence mechanism in plants with special reference to abiotic Stress. Biology 2021, 10, 267. [Google Scholar] [CrossRef]
- Held, M.A.; Jiang, N.; Basu, D.; Showalter, A.M.; Faik, A. Plant cell wall polysaccharides: Structure and biosynthesis. In Polysaccharides: Bioactivity and Biotechnology; Ramawat, K.G., Mérillon, J.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 3–54. [Google Scholar]
- Voiniciuc, C.; Pauly, M.; Usadel, B. Monitoring polysaccharide dynamics in the plant cell wall. Plant Physiol. 2018, 176, 2590–2600. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Aispuro, E.; Vargas-Arispuro, I.; Martínez-Téllez, M. Plant cell wall polymers: Function, structure and biological activity of their derivatives. Polymerization 2012, 4, 63–86. [Google Scholar]
- Kumar, M.; Atanassov, I.; Turner, S. Functional analysis of cellulose synthase (CESA) protein class specificity. Plant Physiol. 2017, 173, 970–983. [Google Scholar] [CrossRef]
- Zhang, W.; Qin, W.; Li, H.; Wu, A.M. Biosynthesis and transport of nucleotide sugars for plant hemicellulose. Front. Plant Sci. 2021, 12, 723128. [Google Scholar] [CrossRef]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Annu. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef] [PubMed]
- Harholt, J.; Suttangkakul, A.; Vibe Scheller, H. Biosynthesis of pectin. Plant Physiol. 2010, 153, 384395. [Google Scholar] [CrossRef] [PubMed]
- Yapo, B.M. Pectin rhamnogalacturonan II: On the “small stem with four branches” in the primary cell walls of plants. Int. J. Carbohydr. Chem. 2011, 2011, 964521. [Google Scholar] [CrossRef]
- Yoshida, S.; Forno, D.A.; Cock, J.H.; Gomez, K.A. Laboratory Manual for Physiological Studies of Rice, 3rd ed.; International Rice Research Institutes: Manila, Philippines, 1976; pp. 61–66. [Google Scholar]
- Gregorio, G.; Senadhira, D.; Mendoza, R. Screening Rice for Salinity Tolerance; IRRI Discussion Paper Series; International Rice Research Institute: Los Banos, Philippines, 1997; Volume 22. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Höller, S.; Ueda, Y.; Wu, L.; Wang, Y.; Hajirezaei, M.R.; Ghaffari, M.R.; von Wirén, N.; Frei, M. Ascorbate biosynthesis and its involvement in stress tolerance and plant development in rice (Oryza sativa L.). Plant Mol. Biol. 2015, 88, 545–560. [Google Scholar] [CrossRef]
- Islam, T.; Manna, M.; Kaul, T.; Pandey, S.; Reddy, C.S.; Reddy, M.K. Genome-wide dissection of Arabidopsis and rice for the identification and expression analysis of glutathione peroxidases reveals their stress-specific and overlapping response patterns. Plant Mol. Biol. Report. 2015, 33, 1413–1427. [Google Scholar] [CrossRef]
- Chaipanya, C.; Telebanco-Yanoria, M.J.; Quime, B.; Longya, A.; Korinsak, S.; Korinsak, S.; Toojinda, T.; Vanavichit, A.; Jantasuriyarat, C.; Zhou, B. Dissection of broad-spectrum resistance of the Thai rice variety Jao Hom Nin conferred by two resistance genes against rice blast. Rice 2017, 10, 18. [Google Scholar] [CrossRef]
- Gillespie, K.; Ainsworth, E. Measurement of reduced, oxidized and total ascorbate content in plants. Nat. Protoc. 2007, 2, 871–874. [Google Scholar] [CrossRef]
- Ruzin, S.E. Plant Microtechnique and Microscopy; Oxford University Press: New York, NY, USA, 1999. [Google Scholar]
- Conklin, P.L.; Gatzek, S.; Wheeler, G.L.; Dowdle, J.; Raymond, M.J.; Rolinski, S.; Isupov, M.; Littlechild, J.A.; Smirnoff, N. Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. J. Biol. Chem. 2006, 281, 15662–15670. [Google Scholar] [CrossRef]
- Merchant, N.; Lyons, E.; Goff, S.; Vaughn, M.; Ware, D.; Micklos, D.; Antin, P. The iPlant collaborative: Cyberinfrastructure for enabling data to discovery for the life sciences. PLoS Biol. 2016, 14, e1002342. [Google Scholar] [CrossRef]
- Goff, S.; Vaughn, M.; McKay, S.; Lyons, E.; Stapleton, A.; Gessler, D.; Matasci, N.; Wang, L.; Hanlon, M.; Lenards, A.; et al. The iPlant collaborative: Cyberinfrastructure for plant biology. Front. Plant Sci. 2011, 2, 34. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.; Rosebrough, N.; Farr, A.L.; Randall, R. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. BioTechniques 2003, 34, 374–378. [Google Scholar] [CrossRef]
- Kassambara, A.; Mundt, F. Extract and Visualize the Results of Multivariate Data Analyses [R Package Factoextra Version 1.0.7]. 2020. Available online: https://CRAN.R-project.org/package=factoextra (accessed on 20 May 2022).
- Conklin, P.; Saracco, S.; Norris, S.; Last, R. Identification of ascorbic acid-deficient Arabidopsis thaliana Mutants. Genetics 2000, 154, 847–856. [Google Scholar] [CrossRef]
- Keller, R.; Renz, F.S.; Kossmann, J. Antisense inhibition of the GDP-mannose pyrophosphorylase reduces the ascorbate content in transgenic plants leading to developmental changes during senescence. Plant J. 1999, 19, 131–141. [Google Scholar] [CrossRef]
- Wang, H.-S.; Yu, C.; Zhu, Z.-J.; Yu, X.-C. Overexpression in tobacco of a tomato GMPase gene improves tolerance to both low and high temperature stress by enhancing antioxidation capacity. Plant Cell Rep. 2011, 30, 1029–1040. [Google Scholar] [CrossRef]
- Badejo, A.A.; Tanaka, N.; Esaka, M. Analysis of GDP-d-mannose pyrophosphorylase gene promoter from acerola (Malpighia glabra) and increase in ascorbate content of transgenic tobacco expressing the acerola gene. Plant Cell Physiol. 2008, 49, 126–132. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, Q.C.; Wang, Z.N.; Fan, R.; Li, Y.; Sun, X.F.; Tang, K.X. Engineering ascorbic acid biosynthetic pathway in Arabidopsis leaves by single and double gene transformation. Biol. Plant. 2012, 56, 451–457. [Google Scholar] [CrossRef]
- Cronje, C.; George, G.; Fernie, A.; Bekker, J.; Kossmann, J.; Bauer, R. Manipulation of L-ascorbic acid biosynthesis pathways in Solanum lycopersicum: Elevated GDP-mannose pyrophosphorylase activity enhances L-ascorbate levels in red fruit. Planta 2012, 235, 553–564. [Google Scholar] [CrossRef]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quéméner, B.; Guillon, F.; Bouchet, B.; Faurobert, M.; Gouble, B.; Page, D.; Garcia, V.; et al. GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Lim, B.; Smirnoff, N.; Cobbett, C.; Golz, J. Ascorbate-deficient vtc2 mutants in Arabidopsis do not exhibit decreased growth. Front. Plant Sci. 2016, 7, 1025. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ye, J.; Munir, S.; Yang, T.; Chen, W.; Liu, G.; Zheng, W.; Zhang, Y. Biosynthetic gene pyramiding leads to ascorbate accumulation with enhanced oxidative stress tolerance in tomato. Int. J. Mol. Sci. 2019, 20, 1558. [Google Scholar] [CrossRef] [PubMed]
- Gatzek, S.; Wheeler, G.; Smirnoff, N. Antisense suppression of L-galactose dehydrogenase in Arabidopsis thaliana provides evidence for its role in ascorbate synthesis and reveals light modulated L-galactose synthesis. Plant J. 2002, 30, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef]
- Potters, G.; De Gara, L.; Asard, H.; Horemans, N. Ascorbate and glutathione: Guardians of the cell cycle, partners in crime? Plant Physiol. Biochem. 2002, 40, 537–548. [Google Scholar] [CrossRef]
- Mellidou, I.; Koukounaras, A.; Kostas, S.; Patelou, E.; Kanellis, A.K. Regulation of vitamin C accumulation for improved tomato fruit quality and alleviation of abiotic stress. Genes 2021, 12, 694. [Google Scholar] [CrossRef]
- Zhang, H.; Li, A.; Zhang, Z.; Huang, Z.; Lu, P.; Zhang, D.; Liu, X.; Zhang, Z.-F.; Huang, R. Ethylene response factor TERF1, regulated by ethylene-insensitive3-like factors, functions in reactive oxygen species (ROS) scavenging in tobacco (Nicotiana tabacum L.). Sci. Rep. 2016, 6, 29948. [Google Scholar] [CrossRef]
- Ortiz-Espín, A.M.; Sánchez Guerrero, A.; Sevilla, F.; Jiménez, A. The role of ascorbate in plant growth and development. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 25–45. [Google Scholar]
- Shih, M.C.; Chang, C.M.; Kang, S.M.; Tsai, M.L. Effect of different parts (leaf, stem and stalk) and seasons (summer and winter) on the chemical compositions and antioxidant activity of Moringa oleifera. Int. J. Mol. Sci. 2011, 12, 6077–6088. [Google Scholar] [CrossRef]
- Kka, N.; Rookes, J.; Cahill, D. Quantitation of ascorbic acid in Arabidopsis thaliana reveals distinct differences between organs and growth phases. Plant Growth Regul. 2017, 81, 283–292. [Google Scholar] [CrossRef]
- Arrigoni, O. Ascorbate system in plant development. J. Bioenerg. Biomembr. 1994, 26, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, F.; Paciolla, C.; de Pinto, M.; De Gara, L. A comparative study of glutathione and ascorbate metabolism during germination of Pinus pinea L. seeds. J. Exp. Bot. 2001, 52, 1647–1654. [Google Scholar] [CrossRef] [PubMed]
- Zechmann, B.; Stumpe, M.; Mauch, F. Immunocytochemical determination of the subcellular distribution of ascorbate in plants. Planta 2011, 233, 1–12. [Google Scholar] [CrossRef]
- Antonova, G. The role of ascorbate in growth and development of cells during the formation of annual rings in coniferous trees. In Oxidative Stress in Plants: Causes, Consequences and Tolerance; IK International Publishing House: New Delhi, India, 2011. [Google Scholar]
- Cárcamo, H.J.; Bustos, R.M.; Fernández, F.E.; Bastías, E. Mitigating effect of salicylic acid in the anatomy of the leaf of Zea mays L. lluteño ecotype from the Lluta valley (Arica-Chile) under NaCl stress. Idesia 2012, 30, 55–63. [Google Scholar] [CrossRef][Green Version]
- Schädel, C.; Blöchl, A.; Richter, A.; Hoch, G. Quantification and monosaccharide composition of hemicelluloses from different plant functional types. Plant Physiol. Biochem. 2010, 48, 1–8. [Google Scholar] [CrossRef]
- Lukowitz, W.; Nickle, T.C.; Meinke, D.W.; Last, R.L.; Conklin, P.L.; Somerville, C.R. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA 2001, 98, 2262–2267. [Google Scholar] [CrossRef] [PubMed]
- Gibeaut, D.M.; Carpita, N.C. Biosynthesis of plant cell wall polysaccharides. Faseb J. 1994, 8, 904–915. [Google Scholar] [CrossRef]
- Ciereszko, I. Regulatory roles of sugars in plant growth and development. Acta Soc. Bot. Pol. 2018, 87. [Google Scholar] [CrossRef]
- Pastori, G.M.; Kiddle, G.; Antoniw, J.; Bernard, S.; Veljovic-Jovanovic, S.; Verrier, P.J.; Noctor, G.; Foyer, C.H. Leaf vitamin C contents modulate plant defense transcripts and regulate genes that control development through hormone signaling. Plant Cell 2003, 15, 939–951. [Google Scholar] [CrossRef]
- Newsholme, P.; Stenson, L.; Sulvucci, M.; Sumayao, R.; Krause, M. 1.02—Amino Acid Metabolism. In Comprehensive Biotechnology (Second Edition); Moo-Young, M., Ed.; Academic Press: Burlington, NJ, USA, 2011; pp. 3–14. [Google Scholar]
- Rosenberg, E.; Filer, D.; Zafriti, D.; Kindler, S.H. Aspartokinase activity and the developmental cycle of Myxococcus xanthus. J. Bacteriol. 1973, 115, 29–34. [Google Scholar] [CrossRef]
- Mo, C.; Wan, S.; Xia, Y.; Ren, N.; Zhou, Y.; Jiang, X. Expression patterns and identified protein-protein interactions suggest that cassava CBL-CIPK signal networks function in responses to abiotic stresses. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Wang, P.; Hsu, C.-C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.S.; Macho, A.P.; Tao, W.A.; et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Gray, A.C.; Matthews, B.F. Bifunctional protein in carrot contains both aspartokinase and homoserine dehydrogenase activities. Plant Physiol. 1991, 97, 1323–1328. [Google Scholar] [CrossRef] [PubMed]
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial peptides from plants. Pharmaceuticals 2015, 8, 711–757. [Google Scholar] [CrossRef] [PubMed]
- Pruzinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a rieske-type iron-sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef] [PubMed]
- Webber, A.N.; Packman, L.C.; Gray, J.C. A 10 kDa polypeptide associated with the oxygen-evolving complex of photosystem II has a putative C-terminal non-cleavable thylakoid transfer domain. FEBS Lett. 1989, 242, 435–438. [Google Scholar] [CrossRef]
- McQueen-Mason, S.; Cosgrove, D.J. Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc. Natl. Acad. Sci. USA 1994, 91, 6574–6578. [Google Scholar] [CrossRef]
- Liu, W.; Xu, L.; Lin, H.; Cao, J. Two expansin genes, AtEXPA4 and AtEXPB5, are redundantly required for pollen tube growth and AtEXPA4 is involved in primary root elongation in Arabidopsis thaliana. Genes 2021, 12, 249. [Google Scholar] [CrossRef]
- Arioli, T.; Peng, L.; Betzner, A.S.; Burn, J.; Wittke, W.; Herth, W.; Camilleri, C.; Höfte, H.; Plazinski, J.; Birch, R.; et al. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 1998, 279, 717–720. [Google Scholar] [CrossRef]
- Paredez, A.R.; Somerville, C.R.; Ehrhardt, D.W. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 2006, 312, 1491–1495. [Google Scholar] [CrossRef]
- Taylor, N.G. Cellulose biosynthesis and deposition in higher plants. New Phytol. 2008, 178, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Mishra, L.; Carr, P.; Pilling, M.; Gardner, P.; Mansfield, S.D.; Turner, S. Exploiting cellulose synthase (CESA) class specificity to probe cellulose microfibril biosynthesis. Plant Physiol. 2018, 177, 151–167. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, R.; Feng, S.; Wang, Y.; Wang, Y.; Fan, C.; Ying, L.; Liu, Z.; Schneider, R.; Xia, T.; et al. Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol. J. 2017, 16, 976–988. [Google Scholar] [CrossRef] [PubMed]

| Genes | Primer Sequence (5′-3′) | Product Size (bp) | Reference |
|---|---|---|---|
| OsVTC1-1 | Forward: GTCATGTGAACTAACCCTCC | 229 | [8] |
| Reverse: GAGTTTCTTCTGGTCCTCTTG | |||
| OsVTC1-3 | Forward: CATCTCCAGCAGCATCATC | 239 | [8] |
| Reverse: CATCGTCACCACCATGTAAAC | |||
| OsVTC1-8 | Forward: GATTGTCATGTGAAATAATCC | 144 | [8] |
| Reverse: CTCATTGAGAAGCAGTTATG | |||
| OsGME1 | Forward: AGACTTCCACTGACAGGTTTG | 132 | [15] |
| Reverse: TTCCAATGTTCACTGGCTCAC | |||
| OsGME2 | Forward: GATGCCTATGGCTTGGAAAA | 187 | [31] |
| Reverse: CAAAACGGTCAGTGGAGGTT | |||
| OsVTC2 | Forward: GCAACCATCAACCACCTCCA | 109 | [15] |
| Reverse: CACTATTCATTGTGCCCTCAGC | |||
| OsVTC4 | Forward: GTTGGCCTTGAACATGTGTG | 106 | This study |
| Reverse: CCGAAGAATCAGAGCTCCAG | |||
| OsGDH | Forward: AAGGGGAAAAACATTACAAAG | 146 | [15] |
| Reverse: TTATCAATAGCGGAAGTAGACA | |||
| OsGPX1 | Forward: GCTTACTGCATCACTTTGCC | 76 | [32] |
| Reverse: GCAGTCGCAGGTCTCAATAA | |||
| OsActin | Forward: TCCATCTTGGCATCTCTCA | 337 | [33] |
| Reverse: GTACCCTCATCAGGCATCTG |
| Downregulated DEGs of RI1-2 | |||
| No. | Gene ID | Gene Description | Log2FC |
| 1 | OS07G0677200 | Peroxidase | −5.186 |
| 2 | OS07G0432201 | Similar to thionin-like peptide | −4.666 |
| 3 | OS02G0139500 | Similar to beta-amyrin synthase | −3.203 |
| 4 | OS12G0583300 | Peptidase aspartic, catalytic domain containing protein | −3.163 |
| 5 | OS03G0850400 | Similar to aspartokinase | −3.157 |
| 6 | OS09G0294000 | Similar to bifunctional aspartokinase/homoserine dehydrogenase 2,chloroplast precursor | −2.759 |
| 7 | OS10G0562900 | Non-protein coding transcript | −2.656 |
| 8 | OS01G0791033 | Similar to ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | −2.433 |
| 9 | OS12G0628600 | Similar to thaumatin-like pathogenesis-related protein 3 precursor | −2.352 |
| 10 | OS03G0355300 | Protein of unknown function DUF1618 domain containing protein | −2.136 |
| 11 | OS09G0541600 | Conserved hypothetical protein | −2.089 |
| 12 | OS12G0113500 | Similar to CBL-interacting protein kinase 14 | −2.045 |
| 13 | OS01G0117200 | OsRLCK16 Similar to ARK protein (Fragment) | −1.984 |
| 14 | OS07G0475900 | Similar to kinase family protein | −1.951 |
| 15 | OS06G0581500 | Protein kinase, core domain containing protein | −1.801 |
| 16 | OS12G0141400 | Conserved hypothetical protein | −1.761 |
| 17 | OS12G0428000 | Similar to senescence-associated protein DIN1 | −1.753 |
| 18 | OS03G0805600 | Similar to pheophorbide a oxygenase | −1.514 |
| 19 | OS07G0109700 | Conserved hypothetical protein | −1.421 |
| 20 | OS07G0147500 | Similar to Photosystem II 10 kDa polypeptide, chloroplast precursor | −1.341 |
| 21 | OS01G0762300 | Similar to predicted protein | −1.340 |
| 22 | OS02G0202200 | SPX domain-containing protein | −1.270 |
| 23 | OS08G0117200 | Similar to 40S ribosomal protein S13 (Fragment) | −1.246 |
| 24 | OS10G0465800 | Similar to 60S ribosomal protein L21 | −1.229 |
| 25 | OS01G0949300 | EF-Hand type domain containing protein | −1.113 |
| Downregulated DEGs of RI1-3 | |||
| No. | Gene ID | Gene Description | Log2FC |
| 1 | OS07G0432201 | Similar to thionin-like peptide | −4.554 |
| 2 | OS09G0294000 | Similar to bifunctional aspartokinase/homoserine dehydrogenase 2, chloroplast precursor | −2.996 |
| 3 | OS03G0850400 | Similar to aspartokinase | −2.696 |
| 4 | OS06G0685300 | Similar to predicted protein | −2.668 |
| 5 | OS11G0416900 | ABC transporter-like domain containing protein | −2.440 |
| 6 | OS12G0113500 | Similar to CBL-interacting protein kinase 14 | −2.434 |
| 7 | OS01G0117200 | Similar to ARK protein (Fragment) | −2.182 |
| 8 | OS01G0162200 | Leucine-rich repeat domain containing protein | −2.123 |
| 9 | OS07G0475900 | Similar to kinase family protein | −2.089 |
| 10 | OS08G0100300 | Non-protein coding transcript | −1.966 |
| 11 | OS07G0691200 | Similar to D-alanine--D-alanine ligase B | −1.901 |
| 12 | OS06G0581500 | Protein kinase, core domain containing protein | −1.758 |
| 13 | OS12G0141400 | Conserved hypothetical protein | −1.745 |
| 14 | OS07G0663800 | Similar to oxidoreductase | −1.662 |
| 15 | OS01G0762300 | Similar to predicted protein | −1.584 |
| 16 | OS07G0147500 | Similar to Photosystem II 10 kDa polypeptide, chloroplast precursor | −1.431 |
| 17 | OS10G0396300 | Similar to MOB1 MOB Kinase Activator 1A | −1.357 |
| 18 | OS02G0814400 | Cytochrome c, monohaem domain containing protein | −1.275 |
| 19 | OS03G0805600 | Similar to pheophorbide a oxygenase | −1.244 |
| Protein ID | Protein Name | Peptide Sequence | Log2 Protein Abundance | ||
|---|---|---|---|---|---|
| WT | RI1-2 | RI1-3 | |||
| Q53LQ0 | Protein disulfide isomerase-like 1-1 | FLIGDIEASQGAFQYFGLREDQVPLIIIQDGESKK | 15.67 | 0 | 0 |
| Q0JJZ6 | Anoctamin-like protein | LSAPMGTLGR | 14.95 | 0 | 0 |
| O56834 | Minor outer capsid protein P2 | NLFSLLQKRK | 13.89 | 0 | 0 |
| Q8W1L6 | Peroxisomal fatty acid beta-oxidation multifunctional protein | MNKAMSLLKGALDYSDFK | 13.66 | 0 | 0 |
| Q5QN75 | Mitogen-activated protein kinase | RGKKPHK | 13.59 | 0 | 0 |
| Q84UP7 | Mixed-linked glucan synthase 6 | SHPYMGRAQEEFVNDRR | 12.38 | 0 | 0 |
| C7J0A2 | DNA topoisomerase 3-alpha | ASRYFRMSSEHTMK | 12.03 | 0 | 0 |
| Q9SQX9 | NAC domain containing protein 50 | NASGQAS | 0 | 14.88 | 14.80 |
| Q6ETL8 | Very-long-chain aldehyde decarbonylase | LAAMRLPK | 0 | 13.19 | 13.43 |
| Q0JMH0 | 2-hydroxyacyl-CoA lyase | ARDNVLKMEAQLAK | 0 | 15.58 | 15.05 |
| B9FMJ3 | Kinesin-like protein KIN-13A | LARFQHRLK | 0 | 13.85 | 14.66 |
| Q0IZQ2 | Cysteine-tRNA ligase | QYEKSDEIR | 0 | 12.47 | 14.73 |
| Q9AV50 | Double-stranded RNA-binding protein 6 | ILPLFRPKSNSR | 0 | 16.49 | 14.17 |
| Q6K4V3 | Zinc finger CCCH domain-containing protein 15 | APSSTSK | 0 | 15.22 | 14.92 |
| Q9SP32 | Endoribonuclease dicer homolog 1 | AEENKSKPEER | 0 | 14.70 | 14.53 |
| Q7FAD5 | Synaptonemal complex protein ZEP1 | ARLLYVDSRLECMEQELK | 12.49 | 15.82 | 15.43 |
| Q6ZCZ2 | Brassinosteroid LRR receptor kinase BRL3 | NFARQSVFLAVTLSVLILFSLLIIHYKLWK | 11.70 | 14.56 | 14.02 |
| Q0DV66 | Pheophorbide a oxygenase | AWWQLVPR | 14.21 | 14.26 | 12.17 |
| Q0D5P3 | Formin-like protein 11 | EASKVAPVK | 11.62 | 14.11 | 14.05 |
| Q7XD96 | Endoribonuclease | ASLCLHMSYFK | 12.20 | 13.78 | 14.07 |
| Q67W65 | Transcription initiation factor TFIID subunit 1 | EDELQKAK | 13.86 | 12.72 | 13.59 |
| Q6YUL8 | Kinesin-like protein KIN-4A | ARNIQNKPIVNR | 14.76 | 0 | 11.32 |
| O24230 | Expansin-B2 | MAGASAK | 14.52 | 0 | 14.70 |
| Q6ZJJ0 | Beta-galactosidase 11 | DLHHALR | 13.7 | 13.2 | 0 |
| Q2QNS6 | Cellulose synthase-like protein D4 | LLIAIRLVALGFFLAWRIR | 12.93 | 13.19 | 0 |
| Q5Z6E5 | Cellulose synthase-like protein D5 | ICYIQFPQRFEGIDPSDR | 10.74 | 0 | 13.02 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lamanchai, K.; Salmon, D.L.; Smirnoff, N.; Sutthinon, P.; Roytrakul, S.; Leetanasaksakul, K.; Kittisenachai, S.; Jantasuriyarat, C. OsVTC1-1 RNAi Mutant with Reduction of Ascorbic Acid Synthesis Alters Cell Wall Sugar Composition and Cell Wall-Associated Proteins. Agronomy 2022, 12, 1272. https://doi.org/10.3390/agronomy12061272
Lamanchai K, Salmon DL, Smirnoff N, Sutthinon P, Roytrakul S, Leetanasaksakul K, Kittisenachai S, Jantasuriyarat C. OsVTC1-1 RNAi Mutant with Reduction of Ascorbic Acid Synthesis Alters Cell Wall Sugar Composition and Cell Wall-Associated Proteins. Agronomy. 2022; 12(6):1272. https://doi.org/10.3390/agronomy12061272
Chicago/Turabian StyleLamanchai, Kanyanat, Deborah L. Salmon, Nicholas Smirnoff, Pornsawan Sutthinon, Sittiruk Roytrakul, Kantinan Leetanasaksakul, Suthathip Kittisenachai, and Chatchawan Jantasuriyarat. 2022. "OsVTC1-1 RNAi Mutant with Reduction of Ascorbic Acid Synthesis Alters Cell Wall Sugar Composition and Cell Wall-Associated Proteins" Agronomy 12, no. 6: 1272. https://doi.org/10.3390/agronomy12061272
APA StyleLamanchai, K., Salmon, D. L., Smirnoff, N., Sutthinon, P., Roytrakul, S., Leetanasaksakul, K., Kittisenachai, S., & Jantasuriyarat, C. (2022). OsVTC1-1 RNAi Mutant with Reduction of Ascorbic Acid Synthesis Alters Cell Wall Sugar Composition and Cell Wall-Associated Proteins. Agronomy, 12(6), 1272. https://doi.org/10.3390/agronomy12061272






