Abstract
All land plants seal their above ground body parts with a lipid-rich hydrophobic barrier called the cuticle to protect themselves from dehydration and other terrestrial threats. Mutational studies in several model species have identified multiple loci regulating cuticular metabolism and development. Of particular importance are the eceriferum (cer) mutants characterized by a loss of cuticular wax. Some barley cer mutants, including cer-x, show defects in the distinctive β-diketone-enriched wax bloom on reproductive stage leaf sheaths, stems, and spikes. We exploited extensive allelic populations, near-isogenic lines, and powerful genotyping platforms to identify variation in the HvWAX INDUCER1 (HvWIN1) gene, encoding a SHINE transcription factor, as underlying cer-x. Comparing the cer-x allelic glossy sheath4.l Bowman Near Isogenic Line BW407 to cv. Bowman revealed an increased cuticular permeability in tissues showing reduced accumulation of β-diketones and altered cuticular metabolic gene expression in BW407. Analyses across the barley pangenome and hundreds of exome-capture datasets revealed high sequence conservation of HvWIN1 and two non-synonymous variants exclusive to the cultivated germplasm. Taken together, we suggest that variation in HvWIN1 controls multiple cuticular features in barley.
1. Introduction
Land plants have a waxy, reflective cuticle to protect them from dangers of terrestrial life, including desiccation, pathogen attack, and UV damage [1,2]. The cuticle forms on epidermal plant tissues exposed to the atmosphere, and consists of a cutin matrix embedded with polysaccharides and intracuticular waxes, which is covered by an epicuticular wax layer [1,3,4]. Epicuticular waxes are mostly composed of acyl aliphatics arranged as films or crystals, which can impart a glossy or glaucous appearance, respectively [1]. Biochemical and genetic studies have shown that cuticular waxes derive from the elongation of C16 or C18 fatty acids (FAs) into very-long-chain fatty acids (VLCFAs, >20 carbons) by Fatty Acid Elongase complexes, including β-ketoacyl-CoA synthases (KCSs), in the endoplasmic reticulum. These VLCFAs are either recruited into the decarbonylation pathway, making odd-numbered alkanes, secondary alcohols, and ketones, or into the alcohol forming pathway, producing even-numbered primary alcohols and wax esters [1,5,6]. While most land plant cuticles share these compounds, exact cuticle composition and structure vary depending on tissue, species, developmental stage, and environmental conditions, which likely reflect adaptive responses to diverse niches and growth habits [7,8,9,10,11,12]. For instance, grasses, including our cereal staples, show cuticular properties that may help these plants cope with drought-prone environments and influence grain yields [7,13,14,15,16,17]. In particular, the distinctive blue-green glaucous wax bloom produced on exposed leaf sheaths, stem internodes, and inflorescences during the reproductive phase of many graminoid crops, including barley and wheat, may provide protection from pests, UV damage, and water loss [14,15,18,19,20,21]. While wild barley and wheat species are often glossy, many modern cultivars are glaucous, suggesting an association between cultivation and the glaucous trait [22,23].
To learn more about the development and genetic control of cereal cuticles, we studied cuticular mutants in barley. Many of these are glossy or eceriferum (cer) mutants, identified in mutagenesis screens for visibly reduced glaucousness and subsequently resolved into more than 75 independent allelism groups [24,25]. To date, researchers have identified the genes underlying several cuticular mutants in barley, including Hv3-KETOACYL-COA SYNTHASE 1 (HvKCS1) and Hv3-KETOACYL-COA SYNTHASE 6 (HvKCS6) [26,27]. In addition, cloning of the CER-CQU metabolic gene cluster revealed the genes necessary for the elongation of C16 and shorter precursors to form the C29-33 β-diketone and hydroxy-β-diketone crystalline tubes that comprise the main component of the barley and wheat epicuticular wax bloom [28]. The cluster encodes three enzymes—a diketone synthase (DKS), a lipase/carboxyl transferase, and a P450 hydroxylase—which underlie the barley Cer-C, Cer-Q, and Cer-U loci, respectively, and whose orthologs are also present within the W1 metabolic gene cluster responsible for β-diketone biosynthesis in wheat [22,29,30,31]. However, we know little about the upstream regulation of the expression and activity of these components [28].
Several transcription factors are known to control cuticular wax in plants. Of particular interest are the SHINE1/WAX INDUCER1 (SHN1/WIN1) clade of the APETALA2 (AP2) domain superfamily, hereafter referred to as SHINE (SHN) transcription factors, a group named after its founding member, the Arabidopsis SHN1/WIN1 transcription factor shown to control the expression of genes involved in cuticular wax and cutin biosynthesis, as well as the formation of surface features on reproductive organs [32,33,34,35,36]. Homologous genes in rice, cotton, and wheat are also associated with cuticular wax production, cuticle permeability, and responses to pathogens and abiotic stress [37,38,39]. In barley, a gene called NUDUM (NUD) encodes a putative SHN transcription factor whose deletion leads to a loss of cuticular modifications on the barley grain pericarp essential for hull adherence but is not associated with altered cuticular features on other tissues [40]. In contrast, the barley glossy sheath4 (gsh4, synonym cer-x) mutants are characterized by a loss of surface wax on leaf sheaths, spikes, and stems [41], which previous analyses showed was associated with dramatic reductions in β-diketone wax classes and alkan-2-ol containing esters on spikes [42]. We hypothesized that mutations in the CER-X gene underlie the surface wax phenotypes in gsh4/cer-x mutants, and our study aimed to identify and characterise the hypothesized causal variation in this gene. To do this, we mobilized advanced genotyping technologies and exploited germplasm resources at NordGen and USDA-ARS. We discovered that multiple alleles show the predicted deleterious polymorphisms in a single gene encoding an SHN transcription factor, distinct from NUD. (2016) named this gene HvWIN1 and showed that reducing HvWIN1 gene expression via virus-induced gene silencing led to reduced cuticular FA and increased susceptibility to fungal infection [43]. Consistent with a role in the cuticular barrier, we found that putative impaired function of HvWIN1 in the gsh4.l allele was associated with increased cuticular permeability in tissues showing reduced surface wax and changes in gene expression, including CER-C, CER-Q, and CER-U. We also show that while HvWIN1 is largely conserved among barley genotypes, two non-synonymous variants specifically occur in cultivated barleys. Our data suggest that variation in HvWIN1 is associated with changes in surface wax and metabolic gene expression. Based on homology with SHN transcription factors, we speculate that HvWIN1 likely plays a regulatory role in controlling gene expression important for cuticle integrity and wax bloom formation.
2. Materials and Methods
2.1. Plant Material and Growth Conditions
Barley (Hordeum vulgare) cultivar (cv.) Bowman and the Bowman Near Isogenic Lines (BWNILs) were obtained from the James Hutton Institute [44] for two available cer-x mutants, cer-x.60 (BW126) and gsh4.l (BW407), to characterise the Cer-X locus (Table 1), as well as for the nud1.a mutant (BW638). Twenty-five original cer-x alleles and their backgrounds, cultivars (cv.) Bonus, Nordal, Carlsberg II, Foma, and Kristina, were used for allele resequencing to confirm CER-X gene identity. These were obtained from NordGen, Alnarp, Sweden (www.nordgen.org 11 August 2020), except for gsh4.l and its background cv. Gateway, which were obtained from the National Plant Germplasm System (NPGS) at the U.S. Department of Agriculture-Agricultural Research Service (USDA-ARS; www.ars.usda.gov 11 August 2020). All germplasm is detailed in Table S1 in Supplementary Materials. Visual scoring of wax coverage for each line is reported in Table S2. BW407 was selected for major experiments due to the original mutant, gsh4.l, displaying a more severe phenotype than that of cer-x.60 (Table S2 in Supplementary Materials). Plants were grown in general-purpose cereal compost (peat/wood fibre 70%/30%, magnesium limestone, Osmocote® start 12 + 14 + 24, Osmocote® standard 16 + 9 + 12, H2Gro® wetting agent, perlite, and sand) under long-day glasshouse conditions (16 h light 18 °C/8 h dark 14 °C), with supplemental light as required during short seasonal photoperiods to reach a light intensity of 150 Wm2. Plants for visual wax scoring were grown in 5-inch square pots, while all other plants were grown in trays of 3.5-inch pots. Genotypes were randomly arranged on the glasshouse bench.

Table 1.
Major germplasm used within this study.
2.2. Cuticle Integrity Measurements
Cuticle integrity was assessed using chlorophyll leaching and Toluidine Blue (TB) staining assays in BW407 compared to Bowman and BW638 compared to Bowman. For chlorophyll leaching assays, 10 cm fresh tissue sections were isolated from the second fully expanded leaf blade (growth stage 12, GS12; [45]), flag leaf blade (GS55), and leaf sheath (GS55), with three (leaf sheath) or four (leaf blade) biological replicates per genotype. Fresh weight was recorded for each tissue sample before immediately immersing samples in 80% ethanol and agitating them at 60 rpm in the dark at room temperature. Following tissue immersion, the absorbance of the ethanol solution was measured with a Varioskan Lux using Skanit microplate reader software (Thermo Fisher Scientific, Waltham, MA, USA) every hour for 6 h. For each sample, absorbance was recorded at 664 and 647 nm as maximum absorbance of chlorophyll A and B, respectively; at 720 nm to subtract the background signal and at 900 and 975 nm to correct for path length. Absorbance values were normalised to a 1 cm pathlength using the formulae: path length = (A977 − A900)/0.036, Aχ (1 cm) = (Aχ − A720)/path length (where Aχ is the absorbance to be corrected) [46,47]. Total chlorophyll concentration was then calculated using the formula: total chlorophyll (µg/mL) = 7.93 (A664) + 19.53 (A647) [48]. A sample of 80% ethanol was used as blank. Data were normalised to fresh weight. Area under the curve was calculated for each genotype, and unpaired Student’s t-tests were used to check for statistical significance between genotypes separately for each tissue, comparing BW407 versus Bowman and BW638 versus Bowman. Data were plotted using the ggplot2 R function [49]. Epidermal staining with TB was used to examine cuticle permeability in flag leaf blade, flag leaf sheath, and spike. Samples were immersed in 0.05% (w/v) aqueous TB for 5 h (spike) or 24 h (leaf blade and leaf sheath), rinsed with water, and photographed.
2.3. Genotyping and Candidate Gene Sequencing
BW407 and BW126, along with Bowman, Bonus, and Gateway, were genotyped using the barley 50K iSelect SNP chip [50]. Markers were ordered based on barley cv. Morex genome assembly (Morex V3 [51]) and plotted to visualise their positions along the barley chromosomes using the ggplot2 R function [49]. We attempted to amplify HvWIN1 (HORVU6Hr1G038120) from 27 cer-x alleles, their background cv’s, and BWNILs using 15 µL reactions of 0.075 µL GoTaq DNA Polymerase (Promega, Madison, WI, USA), 3.0 µL 5X Colorless GoTaq Reaction Buffer (Promega, Madison, WI, USA), 0.3 µL dNTPs, 0.3 µL of each primer at 10 µM, 9.025 µL DNase free water and 2 µL genomic DNA for each sample using the following PCR cycling conditions: 95 °C for 2 min; 40 cycles: 95 °C for 60 s, 60 °C for 60 s and 72 °C for 60 s; 72 °C for 2 min. The amplicons were cleaned with ExoSAP (Applied Biosystems, Bedford, MA, USA) and sequenced at the Genome Technology sequencing facilities of the James Hutton Institute. Primers used for PCR and sequencing are listed in Table S3. The HvWIN1 gene model was plotted using the ggplot2 R function [49].
2.4. Gene Expression
Hull tissues at five days post-anthesis (DPA) were harvested from BW407 and Bowman, snap-frozen, and ground to a fine powder in liquid nitrogen. Total RNA was isolated from 0.1 g of each sample using the Qiagen RNeasy Plant Mini Kit, and cDNA was synthesised using ProtoScript II First Strand cDNA Synthesis Kit (New England Biolabs, Ipswich, MA, USA). SYBR Green Power Up (Thermo Fisher Scientific, Waltham, MA, USA) was used to measure transcript levels of HvCER-C, HvCER-Q, HvCER-U, HvCER1, HvCER1.2, HvWAX ESTER SYNTHASE/DIACYLGLYCEROL ACYLTRANSFERASE 1 (HvWSD1), HvLONG-CHAIN ACYL-COA SYNTHETASE 2 (HvLACS2), HvKCS1, and HvKCS6 in four biological replicates. Primers for HvCER-C, HvCER-Q, and HvCER-U were taken from [22], primers for HvCER1, HvKCS6, and HvWSD1 were taken from [52], primers for HvKCS1 were taken from [26], and primers for HvLACS2 were taken from [43]. The qRT-PCR was normalised as in [53] with HvACT7 as an endogenous control. Primer sets are listed in Table S3. Data were plotted using the ggplot2 R function [49]. We retrieved HvWIN1 expression data from the Barley Expression Database (EoRNA, ref. [54]) using the corresponding barley gene reference transcript model (BART1_0-p44305; BaRTV1.0, ref. [55]) to profile HvWIN1 expression across 16 different tissues in Morex.
2.5. Wax Quantification
Flag leaf blades, flag leaf sheaths, and spikes at GS55 were harvested from BW407 and Bowman and immediately stored at −80 °C. Surface waxes were extracted from one spike, one flag leaf blade, and one 10 cm segment of leaf sheath for four biological replicates by dipping the sample for 1 min in 20 mL dichloromethane containing 10 µg methyl-nonadecanoate as the internal standard and dried under a vacuum evaporator. Extracts were derivatised by resuspending them in 200 µL (spikes and leaf sheaths) or 100 µL (leaf blades) N-O-bis-trimethylsilyltrifluoroacetamide (BSTFA) and incubating at 140 °C for 1 h. Wax components were identified using Gas Chromatography-Mass Spectrometry (GC-MS) using a Trace DSQTM II Series Quadrupole system (Thermo Electron Corporation, Hemel Hempstead, UK), fitted with a CTC CombiPAL autosampler (CTC Analytics, Zwingen, Switzerland) as previously described [56] with the following modifications: the programmable temperature vaporising (PTV) injector operated in split mode (40:1 ratio), and solvent delay for mass spectrum acquisition was 2.8 min. Data were acquired and analysed using Xcalibur™ (version 2.0.7, Thermo Fisher Scientific, Waltham, MA, USA): specific ions, characteristic of each compound, were selected and used for compound detection and quantification in a processing method. Processed data were manually checked and corrected where necessary. Compounds found in less than three (out of four) replicates were assigned as “traces”. Data were plotted using the ggplot2 R function [49].
2.6. Sequence Retrieval and Phylogeny
We identified SHN proteins across plants by conducting a BLASTP search with the HvWIN1 protein sequence as the reference sequence against the following databases: Physcomitrium patens, Selaginella moellendorffii, Marchantia polymorpha, Amborella trichopoda, Arabidopsis thaliana, Solanum lycopersicum, Solanum tuberosum, Brassica rapa, Oryza sativa Japonica, Zea mays, Sorghum bicolor, Brachypodium distachyon, Triticum aestivum, and Hordeum vulgare sequences were obtained from EnsemblPlants (https://plants.ensembl.org/ [57] 20 September 2021), Pinus sylvestris and Picea abies from PLAZA Gymnosperms (https://bioinformatics.psb.ugent.be/plaza/versions/gymno-plaza/ 22 September 2021), and Azolla filiculoides and Salvinia cucullata from FernBase (https://www.fernbase.org/ 23 September 2021). Additionally, the SlSHN2 (Solyc12g009490.1.1) protein sequence was obtained from Sol Genomics (https://solgenomics.net/ [58] 20 September 2021) and Traes_6DS_E6A0BE6CD protein sequence was obtained from Phytozome [59] (https://phytozome-next.jgi.doe.gov/ 20 September 2021). Sequences with apparent assembly errors were manually corrected by genomic sequence comparison against other putative SHINEs. Orthologues were selected based on the presence of three conserved motifs (AP2 domain, middle motif, and c-terminal motif) and further explored using MEME motif discovery (http://meme-suite.org/tools/meme 26 January 2022). Site distribution was set to one occurrence per sequence (OOPS) for a maximum of six motifs per sequence with motif widths of 5–70 amino acids [60].
Full protein sequences were aligned in Molecular Evolutionary Genetics Analysis (MEGA) 11 [61] using MUSCLE. The evolutionary history was inferred using the Maximum Likelihood method and JTT matrix-based model [62] and tested using 300 bootstrap replications. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with a superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.7322)). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 43 amino acid sequences. All positions containing gaps and missing data were eliminated (complete deletion option), resulting in 149 positions in the final dataset [61].
2.7. Haplotype Analysis
SNP data of HvWIN1 were retrieved from published exome-capture datasets of H. spontaneum and H. vulgare lines [63,64]. The dataset collected included 3 kb upstream and 3 kb downstream of the coding region and was filtered to retain sites with ≥98% of samples homozygous. Accessions with missing data points or heterozygosity at these sites were excluded. The resulting dataset, including 456 accessions, was used to build two HvWIN1 haplotype networks, one containing only exonic SNPs and another containing all SNP sites in the dataset, including those in the 3 kb upstream and downstream sequences. Median-Joining haplotype network construction was performed using PopArt (popart.otago.ac.nz [65]). Exonic site haplotypes were plotted on a world map using the rworldmap R package (v1.3-6; [66]). Where available, latitude and longitude of sampling or based on sub-national production mid-point as in [63,64] were used. If unknown, the latitude and longitude of the capital city of the country of origin were used. This information was not available for nine lines which were therefore excluded from this analysis. In addition, HvWIN1 sequences from 19 barley accessions were retrieved from published sequencing data [67]. Full-length genomic sequences, including 3 kb upstream and 3 kb downstream, were aligned using ClustalW in MEGA-X (version 10.1.8, ref. [61]), and the alignment was used to identify SNPs and define haplotypes.
3. Results
3.1. CER-X Controls Cuticular Integrity and Epicuticular Wax Composition
To investigate the mechanisms underlying cuticle formation in barley, we examined the Bowman Near Isogenic Line [44] BW407 which contains the glossy sheath4.l (gsh4.l) allele originally identified as a radiation-induced mutant in Gateway (Table S1 in Supplementary Materials) and allelic to cer-x [41]. Compared to the glaucous appearance of Gateway, the gsh4.l mutant displays a striking glossy appearance on spikes, leaf sheaths, and exposed nodes and internodes, characteristic of a loss of crystalline epicuticular waxes [24,68,69]. We confirmed that BW407 exhibits glossy phenotypes similar to the original gsh4.l allele and in contrast with the glaucous recurrent parent Bowman (Figure 1A–C, Table S2 in Supplementary Materials).
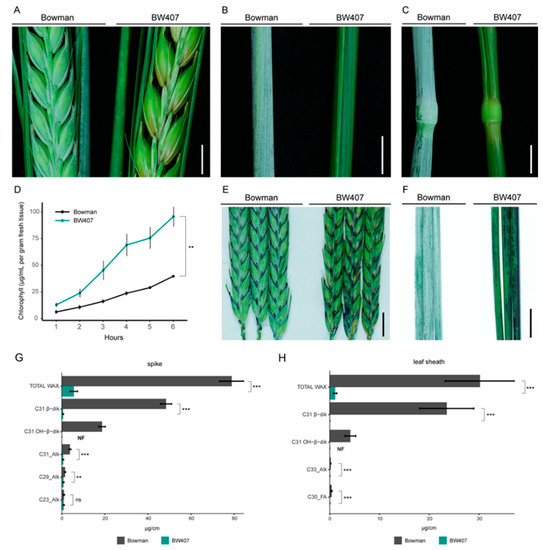
Figure 1.
The CER-X gene regulates specific cuticle structural and chemical properties in barley. (A–C) Wax crystal deposition on spikes, flag leaf sheaths, and stems (nodes and internodes) of Bowman and BW407. Scale bars, 5 mm. (D) Chlorophyll leaching of Bowman and BW407 flag leaf sheaths. Bars represent standard error (N = 3). p = 0.00145 (unpaired Student’s t-test). (E,F) Toluidine blue staining of spikes and flag leaf sheaths of Bowman and BW407. Scale bars, 10 mm. (G,H) Quantification of surface wax components extracted from (G) spikes and (H) flag leaf sheaths at GS55. NF = not found. Alk = Alkane, FA = Fatty Acid; β-dik = β-diketones; OH-β-dik = hydroxy-β-diketones. * p < 0.05, ** p < 0.01, *** p < 0.001 (two-tailed Student’s t-test).
We next examined whether the gsh4.l allele influences cuticular properties in tissues showing the glossy phenotype (flag leaf sheaths and spikes) as well as those without a visible phenotype (fully expanded leaf blades from the second leaf and flag leaf). We used chlorophyll leaching as a proxy for cuticle permeability, with an increased permeability associated with cuticular defects [32]. We first measured how readily chlorophyll leaches from BW407 versus Bowman tissues following immersion in 80% ethanol. Both flag leaf and second leaf blades showed no differences in chlorophyll leaching between genotypes, but chlorophyll leached more quickly from flag leaf sheaths in BW407 compared to Bowman (p = 0.00145; Figure 1D and Figure S1A,B), suggesting increased cuticular permeability in BW407. Next, we tested cuticle integrity by immersing tissues in a toluidine blue (TB) solution, which penetrates and stains tissues with discontinuous or defective cuticles [70]. Both spikes and leaf sheaths in BW407 compared to Bowman were more permeable to TB when immersed into a 0.05% TB solution for 5 h and 24 h, respectively, while no significant difference was observed in the leaf blades after 24 h (Figure 1E,F and Figure S1C). Based on these data, we suggest that BW407 has weakened cuticle integrity on leaf sheaths and spikes compared to Bowman, but not in leaves. Thus, we conclude that the CER-X gene is important for cuticle integrity in tissues that develop a wax bloom.
We next assessed whether decreases in the β-diketone aliphatics which make up the epicuticular wax tubes [69] could explain the loss of glaucous wax in BW407. We used gas chromatography-mass spectrometry (GC–MS) to perform partial compositional analysis on flag leaf blade, flag leaf sheath, and spike wax extracts from BW407 and Bowman. As reported previously [71], barley leaf blade wax is predominantly composed of C26 alcohols, which we found made up 69% and 71% of total leaf blade wax in Bowman and BW407, respectively. Neither total wax accumulation nor levels of identified single components differed between BW407 and Bowman leaf blades (Figure S1D; Table S4). In contrast, BW407 leaf sheaths and spikes showed 96% and 93% reductions in total wax, respectively, compared to Bowman (Figure 1G,H; Table 2). As expected, C31 β-diketones (β-diketones and hydroxy-β-diketones) contribute 92% and 85% of the total wax from Bowman flag leaf sheaths and spikes, respectively; BW407 showed reductions in most wax components compared to Bowman, with β-diketones showing the greatest drop and accumulating to only 0.044% and 0.86% Bowman levels in BW407 flag leaf sheaths and spikes, respectively (Figure 1G,H; Table 2 and Table S4; p < 0.001, t-test), consistent with the previous report of trace amounts of β-diketone wax classes in cer-x.60 compared to its wild-type parent Bonus [42]. We also detected lower levels of C30 FAs in leaf sheaths and reductions in several long-chain alkanes in spikes (Table 2 and Table S4). Interestingly, while C18 FAs were not robustly detected in Bowman leaf sheaths, higher levels were detectable in BW407 leaf sheaths. Altogether, our evidence supports that loss of β-diketones leads to the loss of glaucousness in BW407 leaf sheaths and spikes, the same tissues showing compromised cuticular integrity.

Table 2.
Relative abundance of major wax components on flag leaf sheaths and spikes a.
3.2. CER-X Is HvWIN1
We exploited existing germplasm resources and applied high throughput genotyping platforms to identify the gene underlying the Cer-X locus. We used the barley 50k iSelect SNP chip [50] to genotype two BWNILs, the aforementioned BW407 (gsh4.l, generated in Gateway) [68], and BW126, which contains the cer-x.60 allele originally generated in Bonus [24]. We identified Gateway alleles across chromosome 6H in BW407, consistent with previous mapping data [72], but did not distinguish clear introgression borders. However, two Bonus introgressions across chromosome 6H in BW126 overlapped with the BW407 region containing Gateway alleles (Figure 2A; Table S5). The first introgression spans a region between 452–457 Mb and contains the rob1 locus previously shown to be closely linked to the gsh4.l locus in BW407 [73]. This introgression also includes the gene HORVU6Hr1G038120 (HORVU.MOREX.r3.6HG0578240.1 from Morex V3 [51]), which encodes a predicted protein containing characteristic SHN domains—an AP2/ERF DNA binding domain, a “middle motif” which is unique to SHN family proteins, and a “c-terminal motif”—identifying this gene as encoding a barley homolog of the SHN1/WIN1 transcription factor controlling wax and cutin biosynthesis in Arabidopsis [32,36]. This gene was previously named HvWIN1 by Kumar et al. (2016), who associated HvWIN1 expression with cuticular FA levels and resistance to fungal infection [43]. We speculated that loss of function alleles in HvWIN1 may underlie the glossy mutant phenotypes associated with the Cer-X locus.
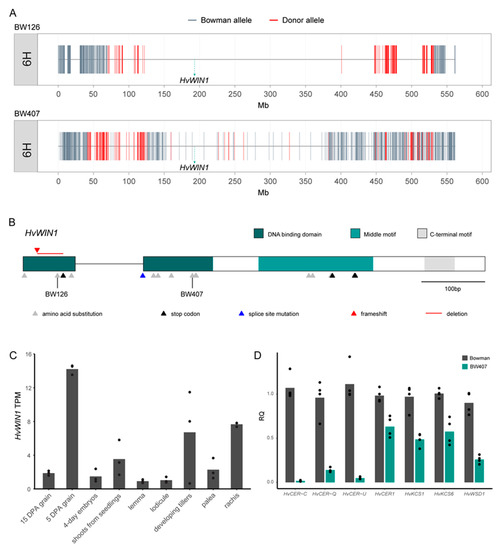
Figure 2.
HvWIN1 underlies the Cer-X locus in barley. (A) Barley 50k iSelect SNP chip [50] plotted over chromosome 6H of BW126 and BW407. (B) HvWIN1 gene model (HORVU6Hr1G038120). Variants in independent cer-x alleles are represented as triangles. Triangles are coloured based on mutation type. (C) Expression of HvWIN1 in Morex tissues from the Barley Expression Database (EoRNA [54]). Data expressed in transcripts per million (TPM). Bars represent mean expression. N = 3. (D) Expression of HvWIN1 putative downstream targets in Bowman and BW407 hull tissues at five days post anthesis (DPA). Expressed as relative quantity (RQ). N = 3.
Direct sequencing of HvWIN1 in BW407 and BW126 identified point mutations in each line which cause single amino acid substitutions in the predicted AP2 domain of HvWIN1: M54R in BW407 and W19R in BW126 (Figure 2B; Table S2). These point mutations were confirmed in their donor mutants, gsh4.l and cer-x.60, compared to their parent cultivars (Table S2). Resequencing HvWIN1 in 23 other independent cer-x alleles, which all exhibit loss or reduction in surface waxes, identified four alleles that failed to amplify a HvWIN1 product, suggesting a potential deletion, while the remaining 19 alleles all contained mutations in HvWIN1 compared to their parent cultivars (Figure 2B; Table S2). One of these, cer-x.407, contained a 42 bp deletion in the first exon, which led to an early stop codon, while the other 18 were single point mutations (Table S2). Of the cer-x alleles containing point mutations, including gsh4.l and cer-x.60, 18 were unique: 13 caused single amino acid substitutions, four caused premature stop codons, and one interrupted an intron splicing site (Figure 2B; Table S2). All these mutations impacted either the AP2 domain or the SHN-specific middle motif in the predicted protein, suggesting these regions are crucial to HvWIN1 function. We then used the Barley Expression Database (EoRNA, [54] to examine HvWIN1 expression across different tissues in cv. Morex. Transcripts were abundant in developing tissues, including grain, tillers, and shoots from seedlings, as well as inflorescence tissues such as the rachis (Figure 2C and Figure S2A), supporting HvWIN1’s role in barley cuticle development. From these data, we conclude that CER-X is HvWIN1.
3.3. HvWIN1 Influences Gene Expression Associated with Cuticle Development
Since HvWIN1 encodes a putative transcription factor, we speculated that loss of β-diketones and decreased cuticular integrity in mutants reflected differences in HvWIN1-dependent gene expression. To explore this hypothesis, we compared the expression levels of selected candidate genes in Bowman and BW407 based on their demonstrated or predicted roles in wax synthesis and cuticle formation in barley. Consistent with β-diketone and hydroxy-β-diketone deficiency, BW407 showed strikingly reduced HvCER-C, HvCER-Q, and HvCER-U expression levels (Figure 2D). The expression of key genes controlling cuticular lipid biosynthesis was also reduced in BW407, including HvKCS1 and HvKCS6 (Figure 2D) which encode condensing enzymes producing VLCFAs. These substrates are necessary for various wax and cutin biosynthetic pathways, supporting our observation that most wax components are reduced in BW407 and potentially explaining the increase in BW407 flag leaf sheaths of C18 FAs, essential precursors in FA elongation for the ubiquitous aliphatic classes but not β-diketones (Table S4). In addition, BW407 had decreased expression of HvWAX ESTER SYNTHASE/DIACYLGLYCEROL ACYLTRANSFERASE 1 (HvWSD1), a homolog of the Arabidopsis wax synthase AtWSD1 involved in ester biosynthesis [74], and two homologs of alkane synthase AtCER1 [75], HvCER1 and HvCER1.2 (Figure 2D and Figure S2B), consistent with the reduction of long-chain wax esters and long-chain alkanes in BW407 (Figure 1G; Table S4). We also detected reductions in the barley orthologue of LONG-CHAIN ACYL-COA SYNTHETASE 2 (LACS2; Figure S2B) involved in generation of long-chain acyl pools important for cutin biosynthesis and cuticle permeability in Arabidopsis [76,77]. Based on these results, we suggest that HvWIN1 may participate in a regulatory network to control the expression of genes important for cuticular integrity and the wax bloom in barley.
3.4. HvWIN1 Is Part of a Highly Conserved Gene Family
The extent of glaucousness varies across grasses [23,28,78,79,80,81], with wild Triticeae species generally producing alcohol-rich wax at the reproductive stage [82], suggesting that the β-diketone rich wax characteristic of cultivated wheat and barley may have been selected during domestication. We conducted haplotype analyses to explore whether variation in HvWIN1 could contribute to differences in reproductive wax between cultivated and wild germplasm. We retrieved HvWIN1 sequence data from whole-exome HvWIN1 sequences from 456 wild (H. spontaneum) and cultivated (H. vulgare) barley accessions [63,64]. We identified only six variations in the coding region, all in the second exon of HvWIN1, suggesting that HvWIN1 is highly conserved. Of these variants, two caused non-synonymous changes (D136N and T113I) in the SHN-specific middle motif, with the T113I variant changing a highly conserved amino acid in SHN1/WIN1 orthologs across species. The variants were arranged in seven haplotypes (HAP; Figure 3A; Tables S6 and S7). HAP1 and HAP2 contain over 90% of the germplasm (417 out of 456 accessions) and are the only two haplotypes represented across cultivars, landraces, and wild barleys. HAP3, with 19 lines, was shared between landraces and wild barley only. Interestingly, the two non-synonymous variants, T113I and D136N, were only found in HAP4 (13 sequences) and HAP5 (5 sequences), respectively, haplotypes made up of cultivars and landraces only. Finally, HAP6 and HAP7 were found in one landrace and one wild barley, respectively. The geographical distribution of HvWIN1 haplotypes did not identify strong associations between a specific variant and its origin; however, all HAP5 lines and eight out of 13 HAP4 lines were collected from central, southern, and eastern Asian countries, including Afghanistan, Pakistan, Tajikistan, Nepal, India, and China, with a strong presence in regions of the Himalayan plateau (Figure S3B, Table S7). Analysis of the 3 kb upstream and 3 kb downstream of the HvWIN1 coding sequence identified an additional 15 and 11 variants, respectively, which formed 25 haplotypes, of which six included 90% of the lines (412 out of 456, Tables S7 and S8; Figure S3). We also compared HvWIN1 genomic sequences across the barley pangenome, 19 barley accessions representative of global barley diversity [67]. We found little variation in the coding region, with only two-second exon SNP sites, both present in the previous dataset, including the T113I variant in two landraces: HvZDM01467 (also called Du-Li Huang or Dulihuang), one of the founders of the Chinese breeding program [67], and HvHOR7552, a landrace from Pakistan (Tables S7 and S9). Analysis of the 3 kb upstream and 3 kb downstream of the HvWIN1 coding sequence in the pangenome lines identified an additional 15 and 10 SNPs, respectively, and no big structural changes (such as large deletions or introgressions) in the regions putatively containing regulatory domains (Table S9). Collectively, these data suggest that HvWIN1 is broadly conserved in barley, although two minor haplotypes exclusive to landraces and cultivated barley show changes to conserved amino acids.
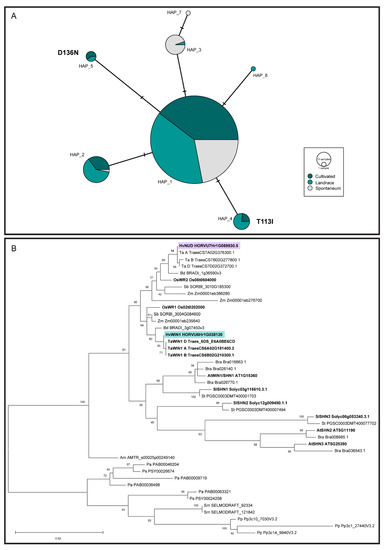
Figure 3.
Sequence analyses of HvWIN1 and SHN transcription factors. (A) Median-joining network for HvWIN1 haplotypes. SNPs were identified comparing HvWIN1 exonic regions of 456 barley lines. Node size is relative to haplotype frequency. Bars between two nodes indicate the number of nucleotides within the sequence that differ between haplotypes. Amino acid changes are indicated in bold. (B) Phylogenetic relationship of SHN transcription factors across representative land plant species. Evolutionary analysis was inferred by using the Maximum Likelihood method and JTT matrix-based model in MEGA 11 [61,62] The tree with the highest log likelihood (−3217.61) is shown. The percentage of trees in which the associated taxa clustered together in 300 bootstrap replications is shown next to the branches. Hv, Hordeum vulgare; Ta, Triticum aestivum; Bd, Brachypodium distachyon; Os, Oryza sativa; Sb, Sorghum bicolor; Zm, Zea mays; Bra, Brassica rapa; At, Arabidopsis thaliana; Sl, Solanum lycopersicum; St, Solanum tuberosum; Am, Amborella trichopoda; Pa, Picea abies; Ps, Pinus sylvestris; Sm, Selaginella moellendorffii; Pp, Physcomitrium patens. HvNUD is highlighted in purple, HvWIN1 is highlighted in green. Gene models in bold indicate functionally characterised genes.
We expanded our analyses to examine SHN transcription factors across representative species in the green plant lineage. Consistent with previous work [83], SHN proteins appear land plant-specific and likely emerged during plant adaptation to drier terrestrial environments, with SHN homologs detected in the bryophyte Physcomitrium patens but not in other bryophytes or algal species (Figure 3B and Figure S4). SHN homologs were highly conserved in all other land plants examined, with intriguing exceptions in water ferns. The closest homologs in the water ferns, Azolla filiculoides and Salvinia cucullata, contained partial SHN motifs but low conservation of gene sequence and structure (Figure S4), though we note that the lack of clear SHN homologs in water ferns could also represent assembly errors. SHN homologs form distinct subclades within both the dicot and monocot groups, suggesting that SHN expansion and diversification events occurred separately in dicot and monocot lineages (Figure 3B). Three SHN transcription factors (SHN1/WIN1, SHN2, and SHN3) act redundantly to pattern surface features in Arabidopsis floral organs [33], while the barley SHN ortholog NUD seems to have undergone neofunctionalisation. In barley, the deletion of the NUD gene underlies ‘naked’ varieties where hulls shed freely due to a loss of a lipid-rich cementing layer on the grain pericarp, which normally adheres to the hull [40]. Comparing Bowman and the BWNIL introgressed with the nud1.a deletion (BW638) showed no change in leaf blade or leaf sheath cuticle permeability (Figure S5) and no visual wax phenotype (Table S2), suggesting that, unlike HvWIN1, NUD does not regulate these traits.
4. Discussion
The original gsh4.l and cer-x alleles were characterized by loss of surface wax coating on spikes, leaf sheaths, and stems, which we confirmed reflect a loss of β-diketones, VLC alkanes, and VLCFAs in the BW407 gsh4.l introgression line. We also found that cuticular permeability increased in BW407. However, while total wax was reduced in BW407, changes in composition rather than total wax load may play more important roles in explaining increased cuticular permeability in glossy BW407 tissues. Firstly, across multiple species and tissues, total wax content is not correlated to cuticle permeability [84,85]. Secondly, β-diketone-deficient glossy iw1Iw2 wheat lines also show increased chlorophyll leaching compared to glaucous β-diketone rich lines, despite equivalent total wax in both groups [14]. Altogether, our data is consistent with the role of β-diketones and alkanes in cuticular permeability in barley, which may underlie responses to drought and improved yield performance in glaucous varieties under arid conditions [13,14,16,86]
Our results show that variation in HvWIN1 influences cuticular properties in both vegetative and reproductive tissues, consistent with previous work manipulating SHN1/WIN1 activity that showed roles in cuticular lipid metabolism as well as resistance to stress [32,33,34,35,36,37,38,39]. In Arabidopsis, the overexpression of AtWIN1/SHN1 significantly increased wax and cutin accumulation and improved drought tolerance [32,35,36], while silencing of a tomato SHINE gene SlSHINE3 (SlSHN3) caused reduced accumulation of fruit cuticular lipids, including cutin, and increased susceptibility to fungal infection and drought [87]. Similarly, transgenic knock-down of wheat TaWIN1 reduced cuticular waxes and cutin, associated with increased susceptibility to fungal infection [88]. Overexpression of rice SHN orthologs, the wax synthesis regulatory gene 1 (OsWR1) and wax synthesis regulatory gene 2 (OsWR2), each increased drought tolerance and promoted wax synthesis; cutin levels also increased in the OsWR2 overexpression line, although these were not examined for OsWR1 [37,89]. Barley spikes rub-inoculated with HvWIN1 silencing constructs showed decreases in free FAs important for cutin biosynthesis and increased susceptibility to fungal infection, which the authors suggested reflects a role for HvWIN1 in reinforcing the cuticle [43]. Our data show that variation in HvWIN1 reduces cuticular waxes and alters cuticular integrity, but whether HvWIN1 function also influences cutin levels in barley remains a pressing question. Moreover, we are curious whether non-synonymous cultivar-specific variation in HvWIN1 has any influence on cuticular properties which could have been selected during cultivation, given the association between SHN function and resistance to drought and infection.
Consistent with our work showing HvWIN1-responsive changes in cuticular metabolic gene expression, transgenic manipulation of SHN-encoding gene expression in other plants also suggested that SHN transcription factors regulate the expression of genes involved in both wax and cutin biosynthetic pathways [36]. Silencing of SlSHN3 in tomato decreased expression of SlCYP86A69, a gene necessary for cutin accumulation in the tomato fruit cuticle [87], while OsWR1 overexpression and RNA interference lines in rice showed increased and decreased expression of OsCER1, OsKCS1, and OsCYP86A7 homologs, respectively [37]. OsWR2 overexpression similarly promoted the expression of OsCER1, OsKCS1, OsLACS1, and OsCYP86A7 homologs [89]. Silencing HvWIN1 in barley was also correlated with reduced expression of HvCYP86A2, HvCYP89A2, and HvLACS2 genes [43]. Overexpressing AtWIN1/SHN1 rapidly and directly induced the expression of cutin biosynthetic genes such as AtCYP86A7, AtCYP86A4, and AtLACS2 [35], while wax biosynthetic genes such as AtKCS1 and AtCER1 are induced more slowly, suggesting that they operate further downstream [35]. Finally, heterologous expression of a durum wheat homologue in tobacco, called TdWIN1, showed that the protein localized to the nucleus and could activate reporter constructs, consistent with transcription factor activity [38]. These data all point to a role for SHN proteins in regulating the transcription of genes important for surface characteristics.
We speculate that HvWIN1 may also control different gene targets depending on developmental stage and tissue. In barley, cutin and wax deposition reduced cuticular permeability in expanding leaves, but further decreases in cuticular permeability reflected additional wax deposition in cells at their final length [90], suggesting carefully coordinated metabolic gene expression linked to differentiation. Environmental signals may be involved as deposition of epicuticular waxes appears linked to exposure to the atmosphere rather than age per se [71]. Moreover, barley and wheat leaf wax changes from predominantly primary alcohols and alkanes in seedlings to β-diketones in reproductive stage tissues such as sheaths, stems, and spikes [14,31,91,92]. Learning more about HvWIN1 control of metabolic gene expression during development will be important to determine how HvWIN1 may control individual events and/or responses to environmental signals during vegetative and reproductive development to regulate cuticular integrity and cuticular waxes in specific tissues.
To the best of our knowledge, the only other SHN-encoding gene studied in barley is the NUD gene shown to control cuticular lipids on the grain pericarp but not on other surfaces [40]. A recent study showed that heterologous overexpression of an SHN coding sequence amplified from barley, which these authors called HvSHN1, increased tolerance to stress and altered cuticle permeability in tobacco [93]. We suspect that this HvSHN1 is NUD since the protein interaction network displayed in [93] names HvSHN1 as NUD, and the protein sequence reported in their previous manuscript [94] differs from NUD by only four amino acids, while HvWIN1 differs from NUD by 63 amino acids; our searches of the barley genome do not detect a third SHN orthologue in barley. This suggests that NUD can influence the cuticular properties of other tissues when overexpressed. We speculate that the different roles of HvWIN1 and NUD may reflect their different expression profiles, with HvWIN1 expressed in most tissues (Figure 2C) and NUD being expressed in grain but not the hull or leaf tissues [40]. It will be interesting to explore the origin of HvWIN1 and NUD tissue expression differences.
In summary, we discovered that variation in HvWIN1 underlies alleles at the Cer-X locus, which we conclude are responsible for changes in cuticular integrity and cuticular waxes in barley. Our work was accelerated by a synergistic combination of access to invaluable germplasm resources and the development of advanced genotyping platforms. Unlike most other genes so far identified, which control cuticular waxes in barley, HvWIN1 encodes a transcription factor. Learning more about the regulatory network controlling cuticular features and how this may differ between cultivars and wild species may become increasingly important to develop more resilient cereal varieties better equipped to respond to environmental challenges.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/agronomy12051088/s1: Figure S1. The Cer-X locus does not affect cuticular and wax properties in barley leaf blades. Figure S2. Expression of HvWIN1 in barley tissues from Barley Expression Database (EoRNA). Figure S3. Sequence variation of HvWIN1 in barley. Figure S4. Motif analysis of SHINE family transcription factors. Figure S5. The NUD locus does not affect leaf blade and leaf sheath cuticle properties in barley. Supplemental Tables.xlsx: Table S1. Barley germplasm. Table S2. cer-x allele resequencing and visual scoring of wax coverage. Table S3. Primer sequences. Table S4. Relative abundance of wax components on flag leaf blades, flag leaf sheaths and spikes. Table S5. Genotyping data of Bowman, BW407, BW126, Gateway and Bonus using barley 50k iSelect SNP chip. Table S6. Exonic haplotypes of HvWIN1 discovered from 456 wild (H. spontaneum) and cultivated (H. vulgare) barley accessions. Table S7. Lines used for HvWIN1 haplotype analysis Table S8. Up and downstream and whole genomic haplotypes of HvWIN1 discovered from 456 wild (H. spontaneum) and cultivated (H. vulgare) barley accessions. Table S9. Haplotypes of HvWIN1 in the barley pangenome.
Author Contributions
Conceptualization, S.M.M., C.C. and T.M.; methodology, S.M.M., C.C. and T.M.; formal analysis, T.M. and C.C.; investigation, T.M., C.C., L.L. and M.E.; writing—original draft preparation, S.M.M. and T.M.; writing—review and editing, S.M.M., C.C. and T.M.; supervision, S.M.M. and C.C.; project administration, S.M.M.; funding acquisition, S.M.M. and C.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Biological and Biotechnological Research Council grant number BB/R010315/1 to S.M.M., C.C. and M.E. were supported by BB/R010315/1. M.E. was also supported by the University of Dundee and the Council for Academics at Risk (Cara). L.L. was supported by the China Scholarship Council and the University of Dundee. T.M. was supported by a Carnegie-Cant-Morgan PhD Scholarship and the University of Dundee.
Data Availability Statement
Large datasets were not generated; however, R scripts were developed and will be deposited upon manuscript publication.
Acknowledgments
We are deeply grateful to the NordGen, National Plant Germplasm System at the U.S. Department of Agriculture-Agricultural Research Service and the James Hutton Institute for germplasm. We also acknowledge guidance from Alexandre Foito in generation of GC-MS data and Joanne Russell in haplotype analysis.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Yeats, T.H.; Rose, J.K.C. The Formation and Function of Plant Cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffree, C.E. The Fine Structure of the Plant Cuticle. Ann. Plant Rev. 2006, 23, 11–125. [Google Scholar]
- Shepherd, T.; Wynne Griffiths, D. The Effects of Stress on Plant Cuticular Waxes. New Phytol. 2006, 171, 469–499. [Google Scholar] [CrossRef] [PubMed]
- Guzmán, P.; Fernández, V.; García, M.L.; Khayet, M.; Fernández, A.; Gil, L. Localization of Polysaccharides in Isolated and Intact Cuticles of Eucalypt, Poplar and Pear Leaves by Enzyme-Gold Labelling. Plant Physiol. Biochem. 2014, 76, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuels, L.; DeBono, A.; Lam, P.; Wen, M.; Jetter, R.; Kunst, L. Use of Arabidopsis Eceriferum Mutants to Explore Plant Cuticle Biosynthesis. J. Vis. Exp. 2008, 16, e709. [Google Scholar] [CrossRef]
- Kolattukudy, P.E. (Ed.) Kolattukudy PE Biochemistry of Plant Waxes. In Chemistry and Biochemistry of Natural Waxes; Elsevier: Amsterdam, The Netherlands, 1976; pp. 289–347. [Google Scholar]
- Xue, D.; Zhang, X.; Lu, X.; Chen, G.; Chen, Z.-H. Molecular and Evolutionary Mechanisms of Cuticular Wax for Plant Drought Tolerance. Front. Plant Sci. 2017, 8, 621. [Google Scholar] [CrossRef]
- Edwards, D.; Kenrick, P. The Early Evolution of Land Plants, from Fossils to Genomics: A Commentary on Lang (1937) “On the Plant-Remains from the Downtonian of England and Wales”. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140343. [Google Scholar] [CrossRef] [Green Version]
- Domínguez, E.; Heredia-Guerrero, J.A.; Heredia, A. The Plant Cuticle: Old Challenges, New Perspectives. J. Exp. Bot. 2017, 68, 5251–5255. [Google Scholar] [CrossRef]
- Gosney, B.J.; Potts, B.M.; O’Reilly-Wapstra, J.M.; Vaillancourt, R.E.; Fitzgerald, H.; Davies, N.W.; Freeman, J.S. Genetic Control of Cuticular Wax Compounds in Eucalyptus Globulus. New Phytol. 2016, 209, 202–215. [Google Scholar] [CrossRef]
- Jetter, R.; Kunst, L.; Samuels, A.L. Composition of Plant Cuticular Waxes. In Annual Plant Reviews Volume 23: Biology of the Plant Cuticle; Wiley: Hoboken, NJ, USA, 2006; pp. 145–181. [Google Scholar]
- Buschhaus, C.; Jetter, R. Composition Differences between Epicuticular and Intracuticular Wax Substructures: How Do Plants Seal Their Epidermal Surfaces? J. Exp. Bot. 2011, 62, 841–853. [Google Scholar] [CrossRef] [Green Version]
- Bi, H.; Kovalchuk, N.; Langridge, P.; Tricker, P.J.; Lopato, S.; Borisjuk, N. The Impact of Drought on Wheat Leaf Cuticle Properties. BMC Plant Biol. 2017, 17, 85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wang, W.; Li, W. Genetic Interactions Underlying the Biosynthesis and Inhibition of β-Diketones in Wheat and Their Impact on Glaucousness and Cuticle Permeability. PLoS ONE 2013, 8, e54129. [Google Scholar]
- Laskoś, K.; Czyczyło-Mysza, I.M.; Dziurka, M.; Noga, A.; Góralska, M.; Bartyzel, J.; Myśków, B. Correlation between Leaf Epicuticular Wax Composition and Structure, Physio-Biochemical Traits and Drought Resistance in Glaucous and Non-Glaucous near-Isogenic Lines of Rye. Plant J. 2021, 108, 93–119. [Google Scholar] [CrossRef]
- Guo, J.; Xu, W.; Yu, X.; Shen, H.; Li, H.; Cheng, D.; Liu, A.; Liu, J.; Liu, C.; Zhao, S.; et al. Cuticular Wax Accumulation Is Associated with Drought Tolerance in Wheat Near-Isogenic Lines. Front. Plant Sci. 2016, 7, 1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dodd, R.S.; Poveda, M.M. Environmental Gradients and Population Divergence Contribute to Variation in Cuticular Wax Composition in Juniperus Communis. Biochem. Syst. Ecol. 2003, 31, 1257–1270. [Google Scholar] [CrossRef]
- Tulloch, A.P.; Hoffman, L.L. Epicuticular Wax of Panicum Virgatum. Phytochemistry 1980, 19, 837–839. [Google Scholar] [CrossRef]
- Richards, R.A.; Rawson, H.M.; Johnson, D.A. Glaucousness in Wheat: Its Development and Effect on Water-Use Efficiency, Gas Exchange and Photosynthetic Tissue Temperatures. Funct. Plant Biol. 1986, 13, 465–473. [Google Scholar] [CrossRef]
- González, A.; Ayerbe, L. Effect of Terminal Water Stress on Leaf Epicuticular Wax Load, Residual Transpiration and Grain Yield in Barley. Euphytica 2010, 172, 341–349. [Google Scholar] [CrossRef]
- Febrero, A.; Fernández, S.; Molina-Cano, J.L.; Araus, J.L. Yield, Carbon Isotope Discrimination, Canopy Reflectance and Cuticular Conductance of Barley Isolines of Differing Glaucousness. J. Exp. Bot. 1998, 49, 1575–1581. [Google Scholar] [CrossRef]
- Hen-Avivi, S.; Savin, O.; Racovita, R.C.; Lee, W.-S.; Adamski, N.M.; Malitsky, S.; Almekias-Siegl, E.; Levy, M.; Vautrin, S.; Bergès, H.; et al. A Metabolic Gene Cluster in the Wheat W1 and the Barley Cer-Cqu Loci Determines β-Diketone Biosynthesis and Glaucousness. Plant Cell 2016, 28, 1440–1460. [Google Scholar] [CrossRef] [Green Version]
- Nice, L.M.; Steffenson, B.J.; Brown-Guedira, G.L.; Akhunov, E.D.; Liu, C.; Kono, T.J.Y.; Morrell, P.L.; Blake, T.K.; Horsley, R.D.; Smith, K.P.; et al. Development and Genetic Characterization of an Advanced Backcross-Nested Association Mapping (AB-NAM) Population of Wild × Cultivated Barley. Genetics 2016, 203, 1453–1467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundqvist, U.; Wettstein, D. Induction of Eceriferum Mutants in Barley by Ionizing Radiations and Chemical Mutagens. Hereditas 1962, 48, 342–362. [Google Scholar] [CrossRef]
- Lundqvist, U.; Lundqvist, A. Mutagen Specificity in Barley for 1580 Eceriferum Mutants Localized to 79 Loci. Hereditas 1988, 108, 1–12. [Google Scholar] [CrossRef]
- Li, C.; Haslam, T.M.; Kr�ger, A.; Schneider, L.M.; Mishina, K.; Samuels, L.; Yang, H.; Kunst, L.; Schaffrath, U.; Nawrath, C.; et al. The β-Ketoacyl-CoA Synthase HvKCS1, Encoded by Cer-Zh, Plays a Key Role in Synthesis of Barley Leaf Wax and Germination of Barley Powdery Mildew. Plant Cell Physiol. 2018, 59, 811–827. [Google Scholar] [CrossRef]
- Weidenbach, D.; Jansen, M.; Franke, R.B.; Hensel, G.; Weissgerber, W.; Ulferts, S.; Jansen, I.; Schreiber, L.; Korzun, V.; Pontzen, R. Evolutionary Conserved Function of Barley and Arabidopsis 3-KETOACYL-CoA SYNTHASES in Providing Wax Signals for Germination of Powdery Mildew Fungi. Plant Physiol. 2014, 166, 1621–1633. [Google Scholar] [CrossRef] [Green Version]
- von Wettstein-Knowles, P. The Polyketide Components of Waxes and the Cer-Cqu Gene Cluster Encoding a Novel Polyketide Synthase, the β-Diketone Synthase, DKS. Plants 2017, 6, 28. [Google Scholar] [CrossRef] [Green Version]
- Simpson, D.; von Wettstein-Knowles, P. Structure of Epicuticular Waxes on Spikes and Leaf Sheaths of Barley as Revealed by a Direct Platinum Replica Technique. Carlsberg Res. Commun. 1980, 45, 465–481. [Google Scholar] [CrossRef] [Green Version]
- Schneider, L.M.; Adamski, N.M.; Christensen, C.E.; Stuart, D.B.; Vautrin, S.; Hansson, M.; Uauy, C.; von Wettstein-Knowles, P. The Cer-Cqu Gene Cluster Determines Three Key Players in a β-Diketone Synthase Polyketide Pathway Synthesizing Aliphatics in Epicuticular Waxes. J. Exp. Bot. 2016, 67, 2715–2730. [Google Scholar] [CrossRef] [Green Version]
- von Wettstein-Knowles, P. Genetic Control of β-Diketone and Hydroxy-β-Diketone Synthesis in Epicuticular Waxes of Barley. Planta 1972, 106, 113–130. [Google Scholar] [CrossRef]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE Clade of AP2 Domain Transcription Factors Activates Wax Biosynthesis, Alters Cuticle Properties, and Confers Drought Tolerance When Overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.X.; Malitsky, S.; De Oliveira, S.; Branigan, C.; Franke, R.B.; Schreiber, L.; Aharoni, A. SHINE Transcription Factors Act Redundantly to Pattern the Archetypal Surface of Arabidopsis Flower Organs. PLoS Genet. 2011, 7, e1001388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshima, Y.; Shikata, M.; Koyama, T.; Ohtsubo, N.; Mitsuda, N.; Ohme-Takagi, M. MIXTA-Like Transcription Factors and WAX INDUCER1/SHINE1 Coordinately Regulate Cuticle Development in Arabidopsis and Torenia Fournieri. Plant Cell 2013, 25, 1609–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Höfte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The Transcription Factor WIN1/SHN1 Regulates Cutin Biosynthesis in Arabidopsis Thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broun, P.; Poindexter, P.; Osborne, E.; Jiang, C.-Z.; Riechmann, J.L. WIN1, a Transcriptional Activator of Epidermal Wax Accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 4706–4711. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An Ethylene Response Factor OsWR1 Responsive to Drought Stress Transcriptionally Activates Wax Synthesis Related Genes and Increases Wax Production in Rice. Plant Mol. Biol. 2012, 78, 275–288. [Google Scholar] [CrossRef]
- Djemal, R.; Khoudi, H. Isolation and Molecular Characterization of a Novel WIN1/SHN1 Ethylene-Responsive Transcription Factor TdSHN1 from Durum Wheat (Triticum Turgidum. L. Subsp. Durum). Protoplasma 2015, 252, 1461–1473. [Google Scholar] [CrossRef]
- Li, X.; Liu, N.; Sun, Y.; Wang, P.; Ge, X.; Pei, Y.; Liu, D.; Ma, X.; Li, F.; Hou, Y. The Cotton GhWIN2 Gene Activates the Cuticle Biosynthesis Pathway and Influences the Salicylic and Jasmonic Acid Biosynthesis Pathways. BMC Plant Biol. 2019, 19, 379. [Google Scholar] [CrossRef]
- Taketa, S.; Amano, S.; Tsujino, Y.; Sato, T.; Saisho, D.; Kakeda, K.; Nomura, M.; Suzuki, T.; Matsumoto, T.; Sato, K.; et al. Barley Grain with Adhering Hulls Is Controlled by an ERF Family Transcription Factor Gene Regulating a Lipid Biosynthesis Pathway. Proc. Natl. Acad. Sci. USA 2008, 105, 4062–4067. [Google Scholar] [CrossRef] [Green Version]
- Lundqvist, U.; Franckowiak, J.D. BGS 354, Glossy Sheath4, Gsh4. Barley Genet. Newsl. 2011, 41, 146–148. [Google Scholar]
- von Wettstein-Knowles, P. Biosynthetic Relationships between β-Diketones and Esterified Alkan-2-Ols Deduced from Epicuticular Wax of Barley Mutants. Mol. Gen. Genet. MGG 1976, 144, 43–48. [Google Scholar] [CrossRef]
- Kumar, A.; Yogendra, K.N.; Karre, S.; Kushalappa, A.C.; Dion, Y.; Choo, T.M. WAX INDUCER1 (HvWIN1) Transcription Factor Regulates Free Fatty Acid Biosynthetic Genes to Reinforce Cuticle to Resist Fusarium Head Blight in Barley Spikelets. J. Exp. Bot. 2016, 67, 4127–4139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druka, A.; Franckowiak, J.; Lundqvist, U.; Bonar, N.; Alexander, J.; Houston, K.; Radovic, S.; Shahinnia, F.; Vendramin, V.; Morgante, M.; et al. Genetic Dissection of Barley Morphology and Development. Plant Physiol. 2011, 155, 617–627. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A Decimal Code for the Growth Stages of Cereals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Warren, C.R. Rapid Measurement of Chlorophylls with a Microplate Reader. J. Plant Nutr. 2008, 31, 1321–1332. [Google Scholar] [CrossRef]
- Lampinen, J.; Raitio, M.; Perälä, A.; Oranen, H.; Harinen, R.-R. Microplate Based Pathlength Correction Method for Photometric DNA Quantification Assay. Thermo Fisher Scientific. 2012. Available online: https://docslib.org/doc/8198812/microplate-based-pathlength-correction-method-for-photometric (accessed on 7 March 2022).
- Lolle, S.J.; Berlyn, G.P.; Engstrom, E.M.; Krolikowski, K.A.; Reiter, W.-D.; Pruitt, R.E. Developmental Regulation of Cell Interactions in the Arabidopsis Fiddlehead-1 Mutant: A Role for the Epidermal Cell Wall and Cuticle. Dev. Biol. 1997, 189, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Bayer, M.M.; Rapazote-Flores, P.; Ganal, M.; Hedley, P.E.; Macaulay, M.; Plieske, J.; Ramsay, L.; Russell, J.; Shaw, P.D.; Thomas, W.; et al. Development and Evaluation of a Barley 50k ISelect SNP Array. Front. Plant Sci. 2017, 8, 1792. [Google Scholar] [CrossRef] [Green Version]
- Mascher, M.; Wicker, T.; Jenkins, J.; Plott, C.; Lux, T.; Koh, C.S.; Ens, J.; Gundlach, H.; Boston, L.B.; Tulpová, Z.; et al. Long-Read Sequence Assembly: A Technical Evaluation in Barley. Plant Cell 2021, 33, 1888–1906. [Google Scholar] [CrossRef]
- Duan, R.; Xiong, H.; Wang, A.; Chen, G. Molecular Mechanisms Underlying Hull-Caryopsis Adhesion/Separation Revealed by Comparative Transcriptomic Analysis of Covered/Naked Barley (Hordeum vulgare L.). Int. J. Mol. Sci. 2015, 16, 14181–14193. [Google Scholar] [CrossRef] [Green Version]
- Shoesmith, J.; Solomon, C.; Yang, X.; Wilkinson, L.; Sheldrick, S.; van Eijden, E.; Couwenberg, S.; Pugh, L.; Eskan, M.; Stephens, J.; et al. APETALA2 Functions as a Temporal Factor Together with BLADE-ON-PETIOLE2 and MADS29 to Control Flower and Grain Development in Barley. Development 2021, 148, dev194894. [Google Scholar] [CrossRef]
- Milne, L.; Bayer, M.; Rapazote-Flores, P.; Mayer, C.-D.; Waugh, R.; Simpson, C.G. EORNA, a Barley Gene and Transcript Abundance Database. Sci. Data 2021, 8, 90. [Google Scholar] [CrossRef]
- Rapazote-Flores, P.; Bayer, M.; Milne, L.; Mayer, C.-D.; Fuller, J.; Guo, W.; Hedley, P.E.; Morris, J.; Halpin, C.; Kam, J.; et al. BaRTv1.0: An Improved Barley Reference Transcript Dataset to Determine Accurate Changes in the Barley Transcriptome Using RNA-Seq. BMC Genom. 2019, 20, 968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brennan, M.; Shepherd, T.; Mitchell, S.; Topp, C.F.E.; Hoad, S.P. Husk to Caryopsis Adhesion in Barley Is Influenced by Pre- and Post-Anthesis Temperatures through Changes in a Cuticular Cementing Layer on the Caryopsis. BMC Plant Biol. 2017, 17, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolser, D.; Staines, D.M.; Pritchard, E.; Kersey, P. Ensembl Plants: Integrating Tools for Visualizing, Mining, and Analyzing Plant Genomics Data BT—Plant Bioinformatics: Methods and Protocols; Edwards, D., Ed.; Springer: New York, NY, USA, 2016; pp. 115–140. ISBN 978-1-4939-3167-5. [Google Scholar]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—from Genotype to Phenotype to Breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A Comparative Platform for Green Plant Genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a Mixture Model by Expectation Maximization to Discover Motifs in Biopolymers. In Proceedings of the 2nd International Conference on Intelligent Systems for Molecular Biology, Stanford, CA, USA, 14–17 August 1994; ISCB: Leesburg, VA, USA, 1994; Volume 2, pp. 28–36. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The Rapid Generation of Mutation Data Matrices from Protein Sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Russell, J.; Mascher, M.; Dawson, I.K.; Kyriakidis, S.; Calixto, C.; Freund, F.; Bayer, M.; Milne, I.; Marshall-Griffiths, T.; Heinen, S.; et al. Exome Sequencing of Geographically Diverse Barley Landraces and Wild Relatives Gives Insights into Environmental Adaptation. Nat. Genet. 2016, 48, 1024–1030. [Google Scholar] [CrossRef] [Green Version]
- Bustos-Korts, D.; Dawson, I.K.; Russell, J.; Tondelli, A.; Guerra, D.; Ferrandi, C.; Strozzi, F.; Nicolazzi, E.L.; Molnar-Lang, M.; Ozkan, H.; et al. Exome Sequences and Multi-Environment Field Trials Elucidate the Genetic Basis of Adaptation in Barley. Plant J. 2019, 99, 1172–1191. [Google Scholar] [CrossRef] [Green Version]
- Leigh, J.W.; Bryant, D. Popart: Full-Feature Software for Haplotype Network Construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- South, A. Rworldmap: A New R Package for Mapping Global Data. R J. 2011, 3, 35. [Google Scholar] [CrossRef] [Green Version]
- Jayakodi, M.; Padmarasu, S.; Haberer, G.; Bonthala, V.S.; Gundlach, H.; Monat, C.; Lux, T.; Kamal, N.; Lang, D.; Himmelbach, A.; et al. The Barley Pan-Genome Reveals the Hidden Legacy of Mutation Breeding. Nature 2020, 588, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Rasmusson, D.C.; Lambert, J.W. Inheritance of the Glossy-Sheath Character in Barley, Hordeum vulgare L. 1. Crop Sci. 1965, 5, 251–253. [Google Scholar] [CrossRef]
- Mikkelsen, J.D. Structure and Biosynthesis of β-Diketones in Barley Spike Epicuticular Wax. Carlsberg Res. Commun. 1979, 44, 133–147. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, T.; Tanaka, H.; Machida, C.; Watanabe, M.; Machida, Y. A New Method for Rapid Visualization of Defects in Leaf Cuticle Reveals Five Intrinsic Patterns of Surface Defects in Arabidopsis. Plant J. 2004, 37, 139–146. [Google Scholar] [CrossRef]
- von Wettstein-Knowles, P. The molecular phenotypes of the eceriferum mutants. In Barley Genetics II; Nilan, R.A., Ed.; Washington State University Press: Pullman, WA, USA, 1971; pp. 146–193. [Google Scholar]
- Shahla, A.; Tsuchiya, T. Genetic Analysis in Six Telotrisomic Lines in Barley (Hordeum vulgare L.). J. Hered. 1990, 81, 127–130. [Google Scholar] [CrossRef]
- Lundqvist, U.; Franckowiak, J.D. BGS 351; Glossy Sheath 1. Barley Genet. Newsl. 1997, 26, 294–295. [Google Scholar]
- Li, F.; Wu, X.; Lam, P.; Bird, D.; Zheng, H.; Samuels, L.; Jetter, R.; Kunst, L. Identification of the Wax Ester Synthase/Acyl-Coenzyme A:Diacylglycerol Acyltransferase WSD1 Required for Stem Wax Ester Biosynthesis in Arabidopsis. Plant Physiol. 2008, 148, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Bourdenx, B.; Bernard, A.; Domergue, F.; Pascal, S.; Léger, A.; Roby, D.; Pervent, M.; Vile, D.; Haslam, R.P.; Napier, J.A.; et al. Overexpression of Arabidopsis ECERIFERUM1 Promotes Wax Very-Long-Chain Alkane Biosynthesis and Influences Plant Response to Biotic and Abiotic Stresses. Plant Physiol. 2011, 156, 29–45. [Google Scholar] [CrossRef] [Green Version]
- Bessire, M.; Chassot, C.; Jacquat, A.; Humphry, M.; Borel, S.; Petétot, J.M.; Métraux, J.; Nawrath, C. A Permeable Cuticle in Arabidopsis Leads to a Strong Resistance to Botrytis Cinerea. EMBO J. 2007, 26, 2158–2168. [Google Scholar] [CrossRef]
- Schnurr, J.; Shockey, J.; Browse, J. The Acyl-CoA Synthetase Encoded by LACS2 Is Essential for Normal Cuticle Development in Arabidopsis. Plant Cell 2004, 16, 629–642. [Google Scholar] [CrossRef] [Green Version]
- De la Fuente Cantó, C.; Russell, J.; Hackett, C.A.; Booth, A.; Dancey, S.; George, T.S.; Waugh, R. Genetic Dissection of Quantitative and Qualitative Traits Using a Minimum Set of Barley Recombinant Chromosome Substitution Lines. BMC Plant Biol. 2018, 18, 340. [Google Scholar] [CrossRef] [PubMed]
- Uddin, M.N.; Marshall, D.R. Variation in Epicuticular Wax Content in Wheat. Euphytica 1988, 38, 3–9. [Google Scholar] [CrossRef]
- Qi, P.; Pendergast, T.H.; Johnson, A.; Bahri, B.A.; Choi, S.; Missaoui, A.; Devos, K.M. Quantitative Trait Locus Mapping Combined with Variant and Transcriptome Analyses Identifies a Cluster of Gene Candidates Underlying the Variation in Leaf Wax between Upland and Lowland Switchgrass Ecotypes. Theor. Appl. Genet. 2021, 134, 1957–1975. [Google Scholar] [CrossRef] [PubMed]
- Würschum, T.; Langer, S.M.; Longin, C.F.H.; Tucker, M.R.; Leiser, W.L. Refining the Genetic Architecture of Flag Leaf Glaucousness in Wheat. Theor. Appl. Genet. 2020, 133, 981–991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tulloch, A.P.; Baum, B.R.; Hoffman, L.L. A Survey of Epicuticular Waxes among Genera of Triticeae. 2. Chemistry. Can. J. Bot. 1980, 58, 2602–2615. [Google Scholar] [CrossRef]
- Kong, L.; Liu, Y.; Zhi, P.; Wang, X.; Xu, B.; Gong, Z.; Chang, C. Origins and Evolution of Cuticle Biosynthetic Machinery in Land Plants. Plant Physiol. 2020, 184, 1998–2010. [Google Scholar] [CrossRef]
- Jetter, R.; Riederer, M. Localization of the Transpiration Barrier in the Epi-and Intracuticular Waxes of Eight Plant Species: Water Transport Resistances Are Associated with Fatty Acyl Rather than Alicyclic Components. Plant Physiol. 2016, 170, 921–934. [Google Scholar] [CrossRef] [Green Version]
- Seufert, P.; Staiger, S.; Arand, K.; Bueno, A.; Burghardt, M.; Riederer, M. Building a Barrier: The Influence of Different Wax Fractions on the Water Transpiration Barrier of Leaf Cuticles. Front. Plant Sci. 2022, 12, 3170. [Google Scholar] [CrossRef]
- Merah, O.; Deléens, E.; Souyris, I.; Monneveux, P. Effect of Glaucousness on Carbon Isotope Discrimination and Grain Yield in Durum Wheat. J. Agron. Crop Sci. 2000, 185, 259–265. [Google Scholar] [CrossRef]
- Shi, J.X.; Adato, A.; Alkan, N.; He, Y.; Lashbrooke, J.; Matas, A.J.; Meir, S.; Malitsky, S.; Isaacson, T.; Prusky, D.; et al. The Tomato SlSHINE3 Transcription Factor Regulates Fruit Cuticle Formation and Epidermal Patterning. New Phytol. 2013, 197, 468–480. [Google Scholar] [CrossRef]
- Kong, L.; Chang, C. Suppression of Wheat TaCDK8/TaWIN1 Interaction Negatively Affects Germination of Blumeria Graminis f.Sp. Tritici by Interfering with Very-Long-Chain Aldehyde Biosynthesis. Plant Mol. Biol. 2018, 96, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jenks, M.A.; Liu, J.; Liu, A.; Zhang, X.; Xiang, J.; Zou, J.; Peng, Y.; Chen, X. Overexpression of Transcription Factor OsWR2 Regulates Wax and Cutin Biosynthesis in Rice and Enhances Its Tolerance to Water Deficit. Plant Mol. Biol. Rep. 2014, 32, 719–731. [Google Scholar] [CrossRef]
- Richardson, A.; Wojciechowski, T.; Franke, R.; Schreiber, L.; Kerstiens, G.; Jarvis, M.; Fricke, W. Cuticular Permeance in Relation to Wax and Cutin Development along the Growing Barley (Hordeum vulgare) Leaf. Planta 2007, 225, 1471–1481. [Google Scholar] [CrossRef] [PubMed]
- Tulloch, A.P. Composition of Leaf Surface Waxes of Triticum Species: Variation with Age and Tissue. Phytochemistry 1973, 12, 2225–2232. [Google Scholar] [CrossRef]
- Kosma, D.K.; Jenks, M.A. Eco-Physiological and Molecular-Genetic Determinants of Plant Cuticle Function in Drought and Salt Stress Tolerance BT—Advances in Molecular Breeding Toward Drought and Salt Tolerant Crops; Jenks, M.A., Hasegawa, P.M., Jain, S.M., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 91–120. ISBN 978-1-4020-5578-2. [Google Scholar]
- Djemal, R.; Khoudi, H. The Barley SHN1-Type Transcription Factor HvSHN1 Imparts Heat, Drought and Salt Tolerances in Transgenic Tobacco. Plant Physiol. Biochem. 2021, 164, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Djemal, R.; Mila, I.; Bouzayen, M.; Pirrello, J.; Khoudi, H. Molecular Cloning and Characterization of Novel WIN1/SHN1 Ethylene Responsive Transcription Factor HvSHN1 in Barley (Hordeum vulgare L.). J. Plant Physiol. 2018, 228, 39–46. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).