Genome-Wide Association Study for Resistance to Rhynchosporium in a Diverse Collection of Spring Barley Germplasm
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Field Trials
2.3. Field Phenotyping and Data Preparation
2.4. Genotyping
- More than 8× coverage for at least 50% of the samples (to ensure robust SNP and genotype calls);
- More than 95% of samples represented at SNP locus (for maximum sample representation);
- Minor allele frequency of 1% (to exclude SNPs based on very rare alleles);
- Quality score: less than 1/1000 chance of the SNP having been called by error (for SNP robustness).
2.5. GWAS Using Single-Locus and Multi-Locus Models
3. Results and Discussion
3.1. Genotyping
3.2. Phenotyping
3.3. Genome-Wide Association Studies
3.4. Geographical Distribution of Resistance Alleles
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAO. FAOSTAT Database. 2020. Available online: https://www.fao.org/faostat (accessed on 15 February 2022).
- Avrova, A.; Knogge, W. Rhynchosporium commune: A persistent threat to barley cultivation. Mol. Plant Pathol. 2012, 13, 986–997. [Google Scholar] [PubMed]
- Zhan, J.; Fitt, B.D.L.; Pinnschmidt, H.O.; Oxley, S.J.P.; Newton, A.C. Resistance, epidemiology and sustainable management of Rhynchosporium secalis populations on barley. Plant Pathol. 2008, 57, 1–14. [Google Scholar] [CrossRef]
- Zaffarano, P.L.; McDonald, B.A.; Zala, M.; Linde, C.C. Global Hierarchical Gene Diversity Analysis Suggests the Fertile Crescent Is Not the Center of Origin of the Barley Scald Pathogen Rhynchosporium secalis. Phytopathology 2006, 96, 941–950. [Google Scholar] [CrossRef] [PubMed]
- AHDB. Barley Disease Management Guide; Agriculture and Horticulture Development Board; AHDB: Warwickshire, UK, 2016. [Google Scholar]
- Cooke, L.R.; Locke, T.; Lockley, K.D.; Phillips, A.N.; Sadiq, M.D.S.; Coll, R.; Black, L.; Taggart, P.J.; Mercer, P.C. The effect of fungicide programmes based on epoxiconazole on the control and DMI sensitivity of Rhynchosporium secalis in winter barley. Crop Prot. 2004, 23, 393–406. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Dachbrodt-Saaydeh, S.; Kudsk, P.; Messéan, A. Toward a Reduced Reliance on Conventional Pesticides in European Agriculture. Plant Dis. 2016, 100, 10–24. [Google Scholar] [CrossRef]
- Zaffarano, P.L.; McDonald, B.A.; Linde, C.C. Two new species of Rhynchosporium. Mycologia 2011, 103, 195–202. [Google Scholar] [CrossRef]
- Looseley, M.; Griffe, L.; Buttner, B.; Wright, K.; Bayer, M.; Coulter, M.; Thauvin, J.-N.; Middlefell-Williams, J.; Maluk, M.; Okpo, A.; et al. Characterisation of barley landraces from Syria and Jordan for resistance to rhynchosporium and identification of diagnostic markers for Rrs1Rh4. Theor. Appl. Genet. 2020, 133, 1243–1264. [Google Scholar] [CrossRef]
- Coulter, M.; Buttner, B.; Hofmann, K.; Bayer, M.; Ramsay, L.; Schweizer, G.; Waugh, R.; Looseley, M.; Avrova, A. Characterisation of barley resistance to rhynchosporium on chromosome 6HS. Theor. Appl. Genet. 2019, 132, 1089–1107. [Google Scholar] [CrossRef]
- Hanemann, A.; Schweizer, G.F.; Cossu, R.; Wicker, T.; Röder, M.S. Fine mapping, physical mapping and development of diagnostic markers for the Rrs2 scald resistance gene in barley. Theor. Appl. Genet. 2009, 119, 1507–1522. [Google Scholar] [CrossRef]
- Marzin, S.; Hanemann, A.; Sharma, S.; Hensel, G.; Kumlehn, J.; Schweizer, G.; Röder, M.S. Are pectin esterase inhibitor genes involved in mediating resistance to Rhynchosporium commune in barley? PLoS ONE 2016, 11, e0150485. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Gupta, S.; Zhou, Y.; Wallwork, H.; Zhou, G.; Broughton, S.; Zhang, X.-Q.; Tan, C.; Westcott, S.; et al. Fine mapping QSc.VR4, an effective and stable scald resistance locus in barley (Hordeum vulgare L.), to a 0.38-Mb region enriched with LRR-RLK and GLP genes. Theor. Appl. Genet. 2020, 133, 2307–2321. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ovenden, B.; Milgate, A. Recent insights into barley and Rhynchosporium commune interactions. Mol. Plant Pathol. 2020, 21, 1111–1128. [Google Scholar] [CrossRef] [PubMed]
- Looseley, M.E.; Griffe, L.L.; Buttner, B.; Wright, K.M.; Middlefell-Williams, J.; Bull, H.; Shaw, P.; Macaulay, M.; Booth, A.; Schweizer, G.; et al. Resistance to Rhynchosporium commune in a collection of European spring barley germplasm. Theor. Appl. Genet. 2018, 131, 2513–2528. [Google Scholar] [CrossRef] [PubMed]
- Zhan, J.; Yang, L.; Shang, L.; Newton, A.C. Pathogen Populations Evolve to Greater Race Complexity in Agricultural Systems—Evidence from Analysis of Rhynchosporium secalis Virulence Data. PLoS ONE 2012, 7, e38611. [Google Scholar] [CrossRef][Green Version]
- Milner, S.G.; Jost, M.; Taketa, S.; Mazón, E.R.; Himmelbach, A.; Oppermann, M.; Weise, S.; Knüpffer, H.; Basterrechea, M.; König, P.; et al. Genebank genomics highlights the diversity of a global barley collection. Nat. Genet. 2019, 51, 319–326. [Google Scholar] [CrossRef]
- Alqudah, A.M.; Sallam, A.H.; Baenziger, P.S.; Borner, A. GWAS: Fast-forwarding gene identification and characterization in temperate Cereals: Lessons from Barley—A review. J. Adv. Res. 2020, 22, 119–135. [Google Scholar] [CrossRef] [PubMed]
- Daba, S.D.; Horsley, R.; Brueggeman, R.; Chao, S.; Mohammadi, M. Genome-wide Association Studies and Candidate Gene Identification for Leaf Scald and Net Blotch in Barley (Hordeum vulgare L.). Plant Dis. 2019, 103, 880–889. [Google Scholar] [CrossRef]
- Cope, J.E.; Norton, G.J.; George, T.S.; Newton, A.C. Identifying potential novel resistance to the foliar disease ‘Scald’ (Rhynchosporium commune) in a population of Scottish Bere barley landrace (Hordeum vulgare L.). J. Plant Dis. Prot. 2021, 128, 999–1012. [Google Scholar] [CrossRef]
- Buttner, B.; Draba, V.; Pillen, K.; Schweizer, G.; Maurer, A. Identification of QTLs conferring resistance to scald (Rhynchosporium commune) in the barley nested association mapping population HEB-25. BMC Genom. 2020, 21, 837. [Google Scholar] [CrossRef]
- Hautsalo, J.; Novakazi, F.; Jalli, M.; Goransson, M.; Mannisen, O.; Isolahti, M.; Reitan, L.; Bergersen, S.; Krusell, L.; Damsgard Robertsen, C.; et al. Pyramiding of scald resistance genes in four spring barley MAGIC populations. Theor. Appl. Genet. 2021, 134, 3829–3843. [Google Scholar] [CrossRef]
- Bustos-Korts, D.; Dawson, I.K.; Russell, J.; Tondelli, A.; Guerra, D.; Ferrandi, C.; Nicolazzi, E.L.; Molnar-Lang, M.; Ozkan, H.; Megyeri, M.; et al. Exome sequences and multi-environment field trials elucidate the genetic basis of adaptation in barley. Plant J. 2019, 99, 1172–1191. [Google Scholar] [CrossRef] [PubMed]
- Soboleva, O.N.; Konovalova, G.S.; Radchenko, E.E. Genetic control of scald resistance in barley landraces. Vavilov J. Genet. Breed. 2016, 20, 616–622. [Google Scholar] [CrossRef][Green Version]
- Looseley, M.E.; Newton, A.C.; Atkins, S.D.; Fitt, B.D.; Fraaije, B.A.; Thomas, W.T.B.; Keith, R.; Macaulay, M.; Lynott, J.; Harrap, D. Genetic basis of control of Rhynchosporium secalis infection and symptom expression in barley. Euphytica 2012, 184, 47–56. [Google Scholar] [CrossRef]
- Looseley, M.E.; Keith, R.; Guy, D.; Barral-Baron, G.; Thirugnanasambandam, A.; Harrap, D.; Werner, P.; Newton, A.C. Genetic mapping of resistance to Rhynchosporium commune and characterisation of early infection in a winter barley mapping population. Euphytica 2015, 203, 337–347. [Google Scholar] [CrossRef]
- Zadoks, J.C.; Chang, T.T.; Konzak, C.F. A decimal code for the growth stages of cereals. Weed Res. 1974, 14, 415–421. [Google Scholar]
- Bates, D.; Machler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models Using lme4. J. Stat. Softw. 2015, 7, 48. [Google Scholar] [CrossRef]
- Mosleth, E.F.; Lillehammer, M.; Pellny, T.K.; Wood, A.J.; Riche, A.B.; Hussain, A.; Griffiths, S.; Hawkesford, M.J.; Shewry, P.R. Genetic variation and heritability of grain protein deviation in European wheat genotypes. Field Crops Res. 2020, 255, 107896. [Google Scholar] [CrossRef]
- Mascher, M.; Gundlach, H.; Himmelbach, A.; Beier, S.; Twardziok, S.O.; Wicker, T.; Radchuk, V.; Dockter, C.; Hedley, P.E.; Russell, J.; et al. A chromosome conformation capture ordered sequence of the barley genome. Nature 2017, 544, 427–433. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabrie, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- R Core Team, R. A Language and Environment for Statistical Computing. 2019. Available online: https://www.R-project.org/ (accessed on 22 February 2022).
- Schwender, H.; Fritsch, A. Package Scrime: Analysis of High-Dimensional Categorical Data Such as SNP Data. 2008. Available online: https://cran.r-project.org/web/packages/scrime/ (accessed on 22 March 2022).
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome 2011, 4, 255–258. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.-Y.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-B.; Feng, J.-Y.; Ren, W.-L.; Huang, B.; Zhou, L.; Wen, Y.-J.; Zhang, J.; Dunwell, J.M.; Xu, S.; Zhang, Y.-M. Improving power and accuracy of genome-wide association studies via a multi-locus mixed linear model methodology. Nat. Sci. Rep. 2016, 6, 19444. [Google Scholar] [CrossRef]
- Tamba, C.L.; Zhang, Y.-M. A fast mrMLM algorithm for multi-locus genome-wide association studies. BioRxiv 2018. [Google Scholar] [CrossRef]
- Huang, M.; Liu, X.; Zhou, Y.; Summers, R.M.; Zhang, Z. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience 2019, 8, 154. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Z. GAPIT Version 3: Boosting Power and Accuracy for Genomic Association and Prediction. Genom. Proteom. Bioinform. 2021. [Google Scholar] [CrossRef]
- Tibbs Cortes, L.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, 20077. [Google Scholar] [CrossRef]
- Merrick, L.F.; Burke, A.B.; Zhang, Z.; Carter, A.H. Comparison of Single-Trait and Multi-TraitGenome-Wide Association Models andInclusion of Correlated Traits in the Dissection of the Genetic Architectureof a Complex Trait in a Breeding Program. BioRxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/2021.08.23.457367v2 (accessed on 22 February 2022).
- Mangin, B.; Siberchicot, A.; Nicolas, S.; Doligez, A.; This, P.; Cierco-Ayrolles, C. Novel measures of linkage disequilibrium that correct the bias due to population structure and relatedness. Heredity 2012, 108, 285–291. [Google Scholar] [CrossRef]
- Xu, Y.; Jia, Q.; Zhou, G.; Zhang, X.-Q.; Angessa, T.; Broughton, S.; Yan, G.; Zhang, W.; Li, C. Characterization of the sdw1 semi-dwarf gene in barley. BMC Plant Biol. 2017, 17, 11. [Google Scholar] [CrossRef]
- Zhang, X.; Ovenden, B.; Orchard, B.A.; Zhou, M.; Park, R.F.; Singh, D.; Milgate, A. Bivariate analysis of barley scald resistance with relative maturity reveals a new major QTL on chromosome 3H. Nat. Sci. Rep. 2019, 9, 19258. [Google Scholar] [CrossRef]
- Sayed, H.; Backes, G.; Kayyal, H.; Yahyaoui, A.; Ceccarelli, S.; Grando, S.; Jahoor, A.; Baum, M. New molecular markers linked to qualitative and quantitative powdery mildew and scald resistance genes in barley for dry areas. Euphytica 2004, 135, 225–228. [Google Scholar] [CrossRef]
- von Korff, M.; Wang, H.; Léon, J.; Pillen, K. AB-QTL analysis in spring barley. I. Detection of resistance genes against powdery mildew, leaf rust and scald introgressed from wild barley. Theor. Appl. Genet. 2005, 111, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Wagner, C.; Schweizer, G.; Kramer, M.; Dehmer-Badani, A.G.; Ordon, F.; Friedt, W. The complex quantitative barley–Rhynchosporium secalis interaction: Newly identified QTL may represent already known resistance genes. Theor. Appl. Genet. 2008, 118, 113–122. [Google Scholar] [CrossRef]
- Genger, R.K.; Nesbitt, K.; Brown, A.H.D.; Abbott, D.C.; Burdon, J.J. A novel barley scald resistance gene: Genetic mapping of the Rrs15 scald resistance gene derived from wild barley, Hordeum vulgare ssp. spontaneum. Plant Breed. 2005, 124, 137–141. [Google Scholar] [CrossRef]
- Zantinge, J.; Xue, S.; Holtz, M.; Xi, K.; Juskiw, P. The identification of multiple SNP markers for scald resistance in spring barley through restriction-site associated sequencing. Euphytica 2019, 215, 8. [Google Scholar] [CrossRef]
- Stotz, H.; Mitrousia, G.; de Wit, P.; Fitt, B.D.L. Effector-triggered defence against apoplastic fungal pathogens. Trends Plant Sci. 2014, 19, 491–500. [Google Scholar] [CrossRef]
- Saintenac, C.; Lee, W.S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat receptor kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368–374. [Google Scholar] [CrossRef]
- Liebrand, T.W.H.; van den Berg, G.C.M.; Zhang, Z.; Smit, P.; Cordewener, J.H.G.; America, A.H.P.; Sklenar, J.; Jones, A.M.E.; Tameling, W.I.L.; Robatzek, S.; et al. Receptor like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef]
- Larkan, N.J.; Lydiate, D.J.; Parkin, I.A.P.; Nelson, M.N.; Epp, D.J.; Cowling, W.A.; Rimmer, S.R.; Borhan, M.H. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New. Phytol. 2013, 197, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Saintenac, C.; Cambon, F.; Aouini, L.; Verstappen, E.; Ghaffary, S.M.T.; Poucet, T.; Marande, W.; Berges, H.; Xu, S.; Jaouannet, M.; et al. A wheat cysteine-rich receptor-like kinase confers broad-spectrum resistance against Septoria tritici blotch. Nat. Commun. 2021, 12, 433. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhou, S.-Y.; Zhao, W.-S.; Su, S.-C.; Peng, Y.-L. A novel wall-associated receptor-like protein kinase gene, OsWAK1, plays important roles in rice blast disease resistance. Plant Mol. Biol. 2009, 69, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, G.; Baumlein, H.; Mock, H.-P.; Himmelbach, A.; Schweizer, P. The multigene family encoding germin-like proteins of barley. Regulation and function in Basal host resistance. Plant Physiol. 2006, 142, 181–192. [Google Scholar] [CrossRef]
- Christensen, A.B.; Thordal-Christensen, H.; Zimmermann, G.; Gjetting, T.; Lyngkjær, M.F.; Dudler, R.; Schweizer, P. The Germinlike Protein GLP4 Exhibits Superoxide Dismutase Activity and Is an Important Component of Quantitative Resistance in Wheat and Barley. MPMI 2007, 17, 109–117. [Google Scholar] [CrossRef]
- Manosalva, P.M.; Davidson, R.M.; Liu, B.; Zhu, X.; Hulbert, S.H.; Leung, H.; Leach, J.E. A Germin-Like Protein Gene Family Functions as a Complex Quantitative Trait Locus Conferring Broad-Spectrum Disease Resistance in Rice. Plant Physiol. 2009, 149, 286–296. [Google Scholar] [CrossRef]

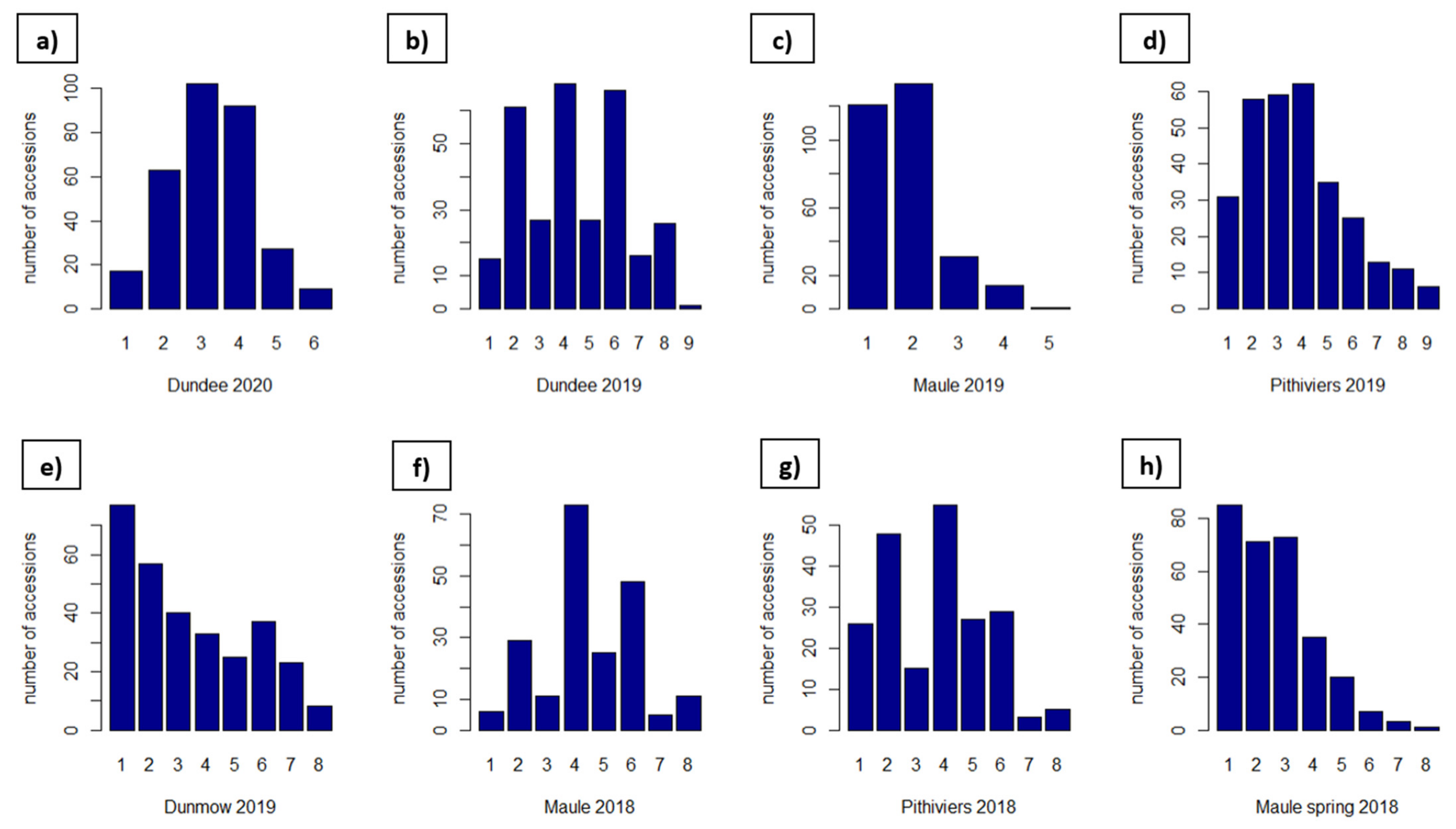
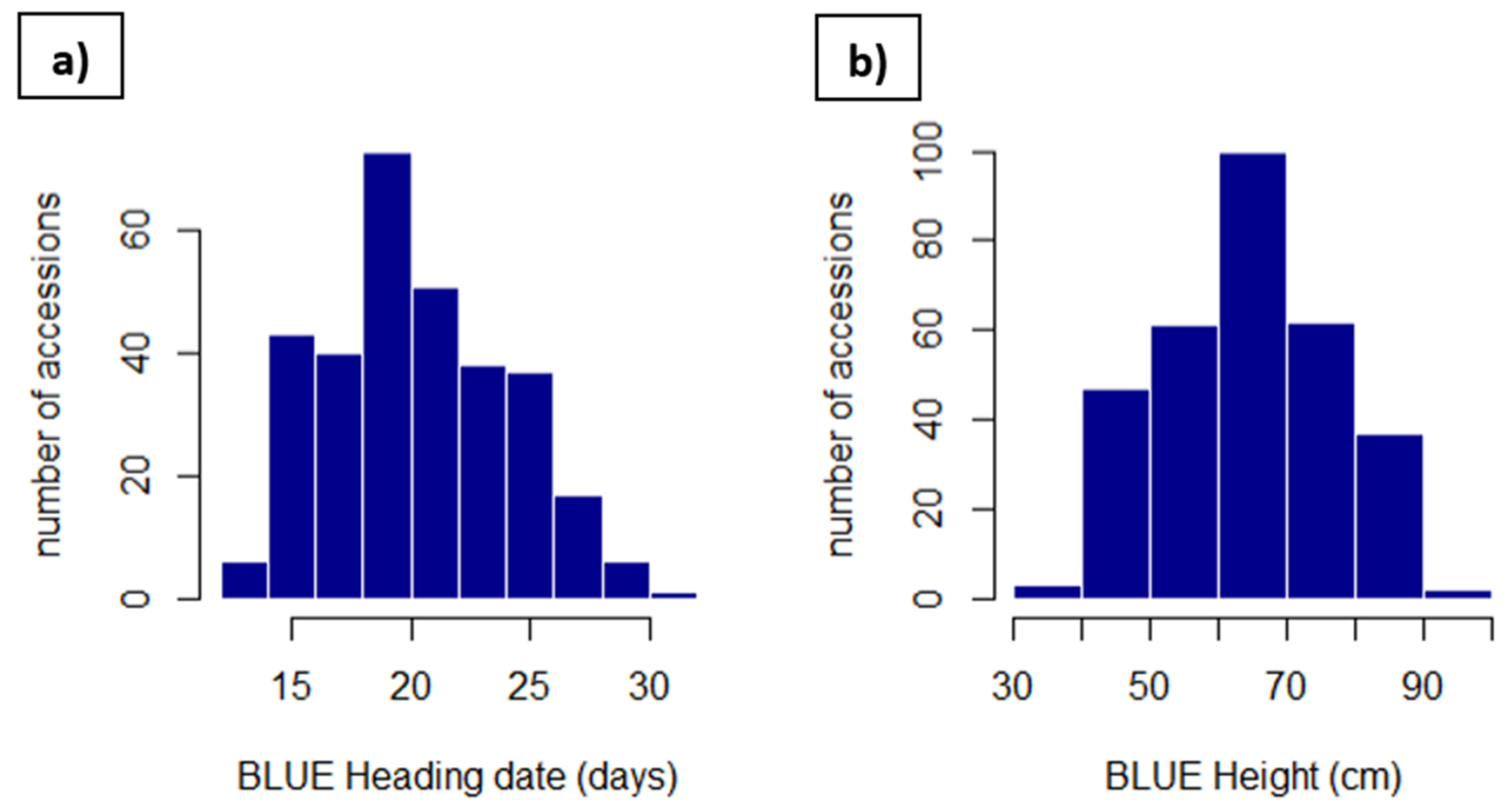
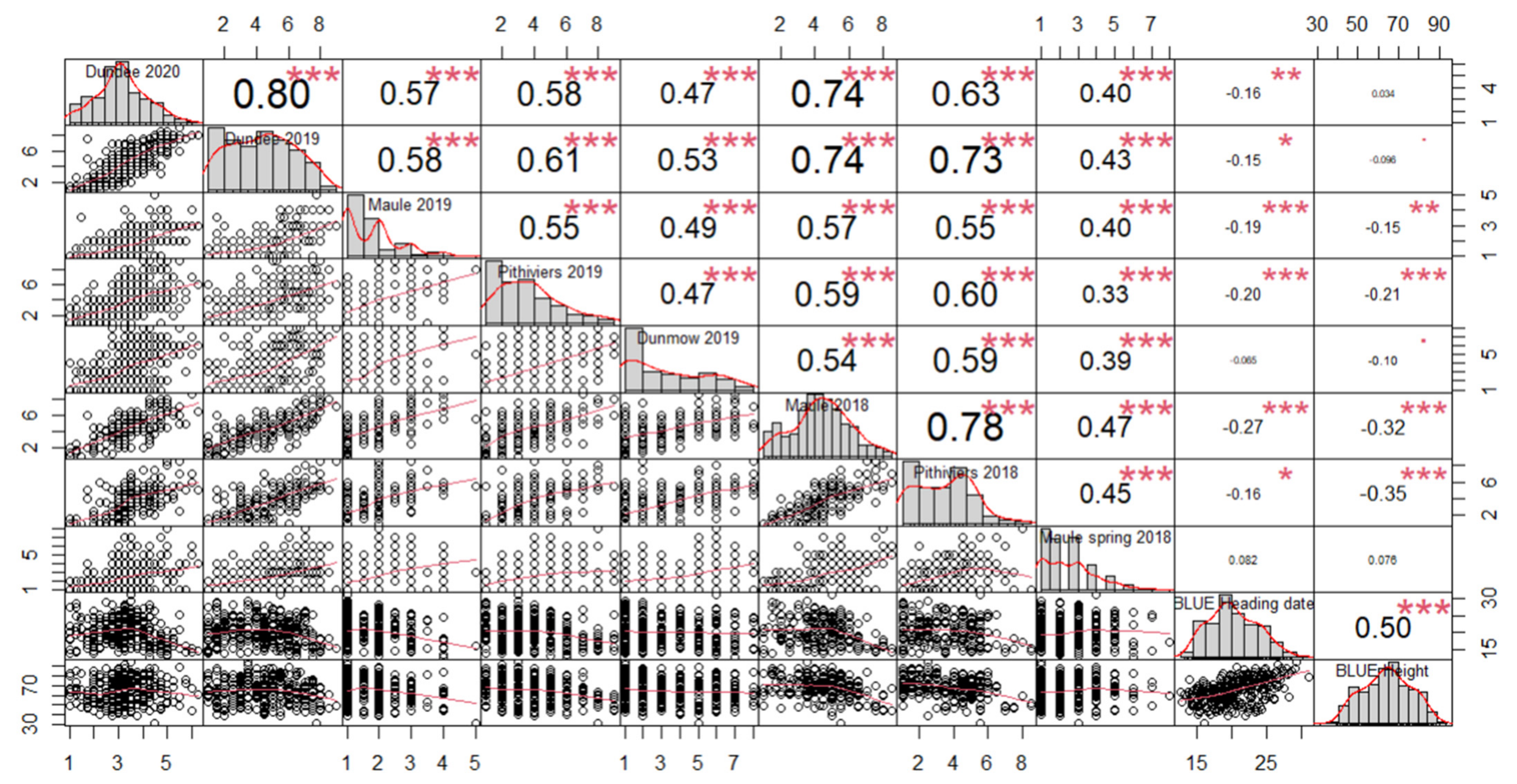
| Site | Soil Type | GPS Coordinates | Number of Field Trials | Years and Sowing Season (A = Autumn, S = Spring) |
|---|---|---|---|---|
| Dundee (Scotland) | Sandy loam | 56.45 N, −3.07 E | 2 | 2019 (A), 2020 (A) |
| Dunmow (England) | Loam | 51.88 N, 0.41 E | 1 | 2019 (A) |
| Maule (France) | Clay loam | 48.92 N, 1.82 E | 3 | 2018 (A), 2018 (S), 2019 (A) |
| Pithiviers (France) | Sandy loam | 48.12 N, 2.14 E | 2 | 2018 (A), 2019 (A) |
| Score | Description |
|---|---|
| 1 | No infection observed |
| 2 | 1% infection on lower leaves |
| 3 | 5% infection on lower leaves |
| 4 | 25% infection on lower leaves |
| 5 | 50% infection on lower leaves |
| 6 | Leaves appear 1/2 infected 1/2 green |
| 7 | Leaves appear more infected than green |
| 8 | Very little green leaf tissue left |
| 9 | Leaves dead no green leaf left |
| QTL Name | Chr a | Physical Interval b | Top SNP c | Max −log10P d | RAF e | Nb Genes f | Dataset g | Methods h | References i |
|---|---|---|---|---|---|---|---|---|---|
| QTL-3H-1 | chr3H | 673759655–674277867 | chr3H_674155754 | 6.08 | 0.84 | 16 | Dundee19; Pithiviers18 | BLINK; FASTmrMLM | QRh.S42-3H.a [49], QTLRS6 and QTLRS7b [48] |
| QTL-5H-1 | chr5H | 5560028–5561932 | chr5H_5561636 | 5.01 | 0.85 | 1 | Dundee19; Maule19 | BLINK; FASTmrMLM | reported in the current study only |
| QTL-5H-2 | chr5H | 394034355–423047705 | chr5H_400584464 | 5.52 | 0.91 | 178 | Dunmow19; Dundee19 | BLINK; EMMAX | reported in the current study only |
| QTL-7H-1 | chr7H | 5389278–5394956 | chr7H_5389278 | 5.31 | 0.22 | 2 | Maule18; Dundee20; Dundee19 | EMMAX; FASTmrMLM | Rrs2 [11], Qsc_7H_1 [22] |
| QTL-7H-2 | chr7H | 68708954–68768630 | chr7H_68766673 | 7.28 | 0.25 | 3 | Dundee19; Pithiviers19 | BLINK; FASTmrMLM | QTLSR-7H-2017 [47] |
| QTL-7H-3 | chr7H | 113870715–193866189 | chr7H_151131820 | 5.59 | 0.85 | 512 | Dundee19; Dundee20 | BLINK; FASTmrMLM | QTLVixenRrs7H271 [50], Qsc-7H.3 [19] |
| QTL-7H-4 | chr7H | 638442686–638453794 | chr7H_638446781 | 6.18 | 0.64 | 1 | Dundee19; Maule19 | BLINK; FASTmrMLM | qS271_7 [10], Rrs15 [51], Qsc_7H_2 [22] |
| QTL Most Significant SNP | Asia | Ethiopia | Europe | Middle East | North Africa | North America | South America | Total |
|---|---|---|---|---|---|---|---|---|
| chr3H_674155754 | 0.88 | 1.00 | 0.85 | 0.77 | 0.83 | 0.90 | 1.00 | 0.84 |
| chr5H_5561636 | 0.50 | 1.00 | 0.98 | 0.91 | 0.72 | 1.00 | 1.00 | 0.85 |
| chr5H_400584464 | 0.77 | 1.00 | 0.90 | 0.96 | 0.97 | 1.00 | 0.88 | 0.91 |
| chr7H_5389278 | 0.17 | 0.00 | 0.06 | 0.45 | 0.07 | 0.30 | 0.13 | 0.22 |
| chr7H_68766673 | 0.15 | 0.83 | 0.03 | 0.39 | 0.14 | 0.30 | 0.38 | 0.25 |
| chr7H_151131820 | 0.48 | 0.94 | 0.93 | 0.93 | 0.79 | 1.00 | 1.00 | 0.85 |
| chr7H_638446781 | 0.33 | 0.83 | 0.41 | 0.88 | 0.90 | 0.60 | 0.75 | 0.64 |
| Average frequency | 0.47 | 0.80 | 0.59 | 0.76 | 0.63 | 0.73 | 0.73 | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thauvin, J.-N.; Russell, J.; Vequaud, D.; Looseley, M.; Bayer, M.; Le Roux, P.-M.; Pin, P.; Waugh, R.; Avrova, A. Genome-Wide Association Study for Resistance to Rhynchosporium in a Diverse Collection of Spring Barley Germplasm. Agronomy 2022, 12, 782. https://doi.org/10.3390/agronomy12040782
Thauvin J-N, Russell J, Vequaud D, Looseley M, Bayer M, Le Roux P-M, Pin P, Waugh R, Avrova A. Genome-Wide Association Study for Resistance to Rhynchosporium in a Diverse Collection of Spring Barley Germplasm. Agronomy. 2022; 12(4):782. https://doi.org/10.3390/agronomy12040782
Chicago/Turabian StyleThauvin, Jean-Noël, Joanne Russell, Dominique Vequaud, Mark Looseley, Micha Bayer, Pierre-Marie Le Roux, Pierre Pin, Robbie Waugh, and Anna Avrova. 2022. "Genome-Wide Association Study for Resistance to Rhynchosporium in a Diverse Collection of Spring Barley Germplasm" Agronomy 12, no. 4: 782. https://doi.org/10.3390/agronomy12040782
APA StyleThauvin, J.-N., Russell, J., Vequaud, D., Looseley, M., Bayer, M., Le Roux, P.-M., Pin, P., Waugh, R., & Avrova, A. (2022). Genome-Wide Association Study for Resistance to Rhynchosporium in a Diverse Collection of Spring Barley Germplasm. Agronomy, 12(4), 782. https://doi.org/10.3390/agronomy12040782






