Genetic Bases of Flow- and Sink-Related Traits in Rice Revealed by Genome-Wide Association Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Phenotypic Investigation
2.2. Genotypic Data
2.3. Genome-Wide Association Study (GWAS)
2.4. Candidate Gene Analysis
3. Results
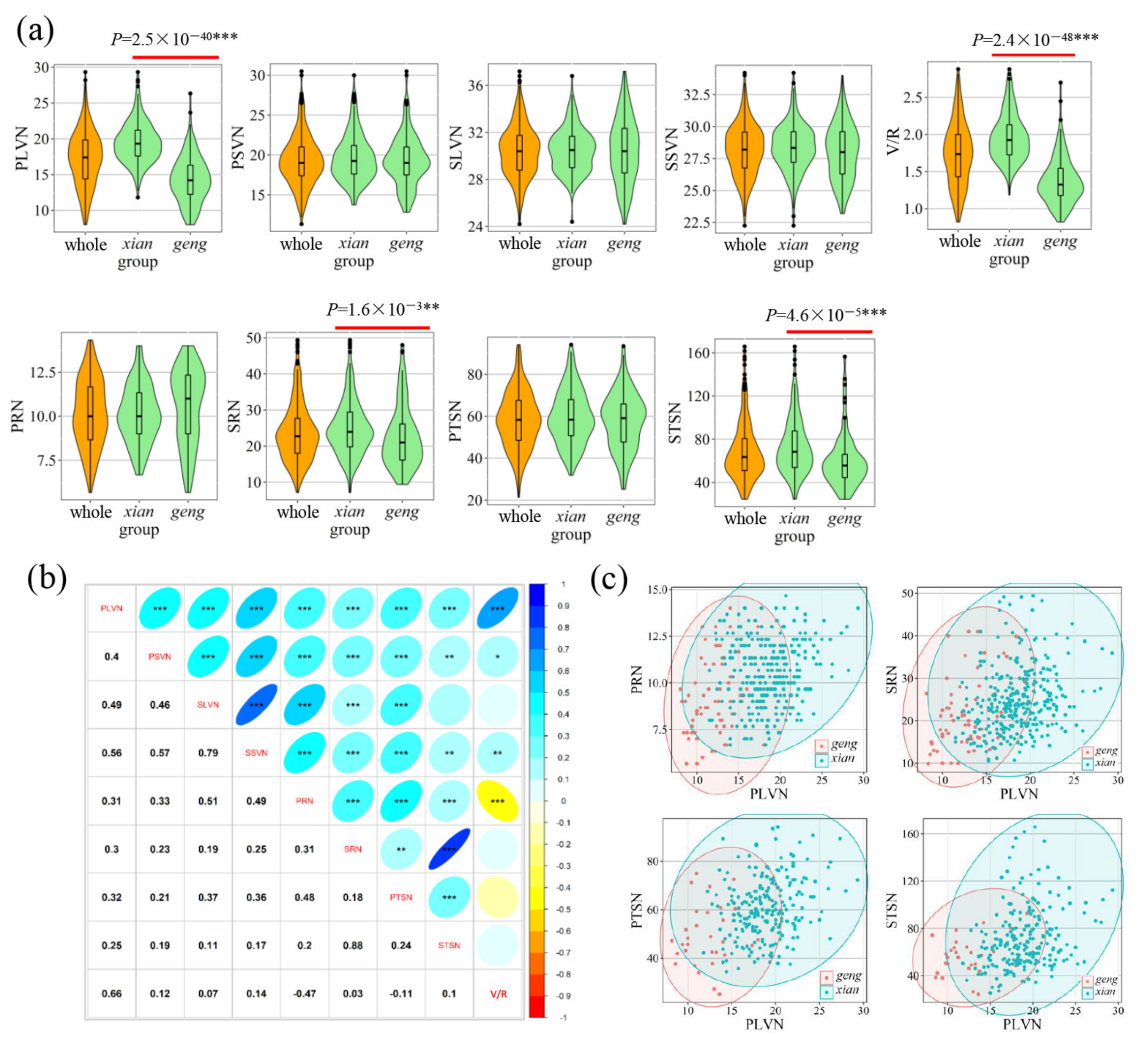
3.1. Phenotypic Variation and Correlation
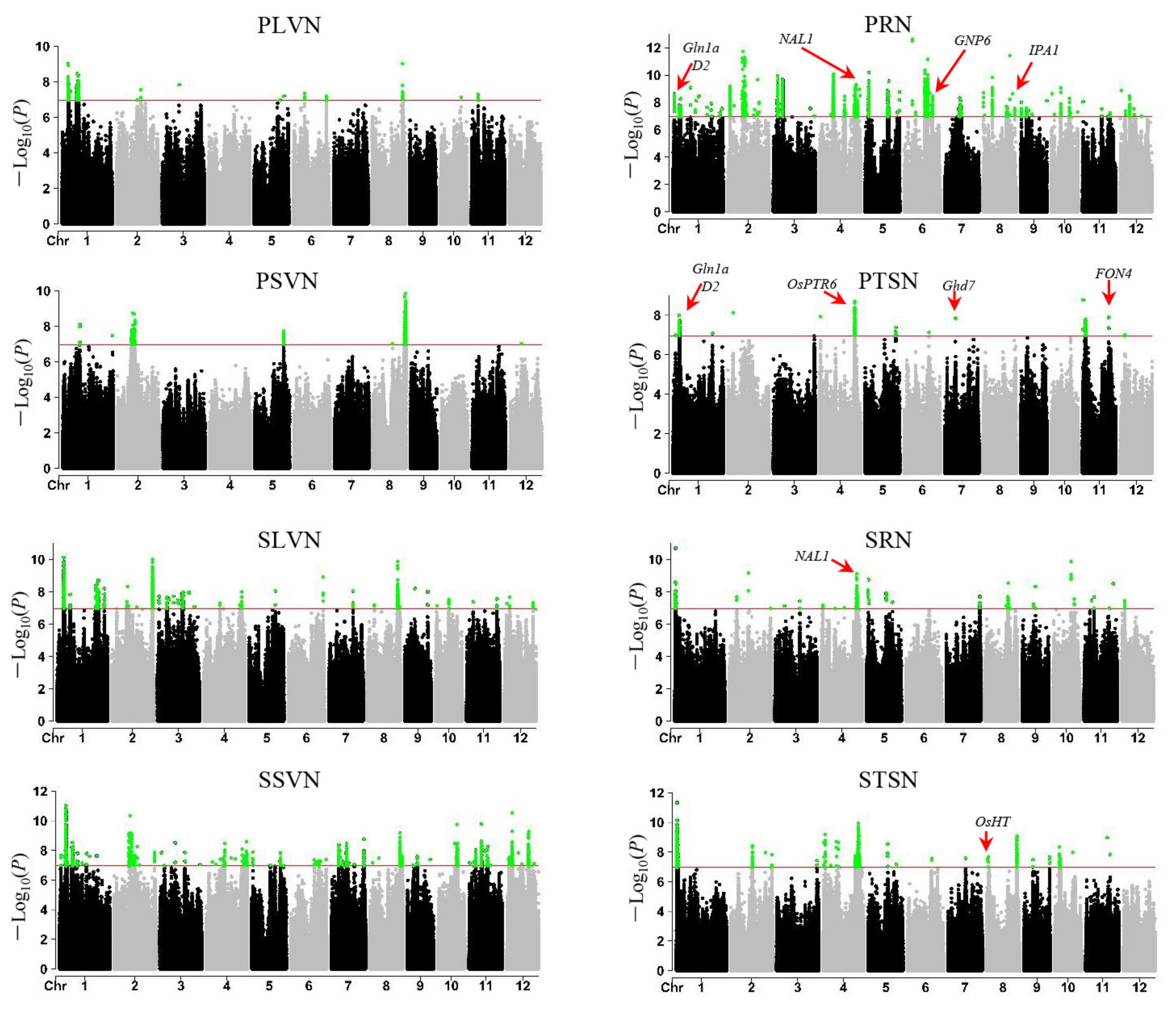
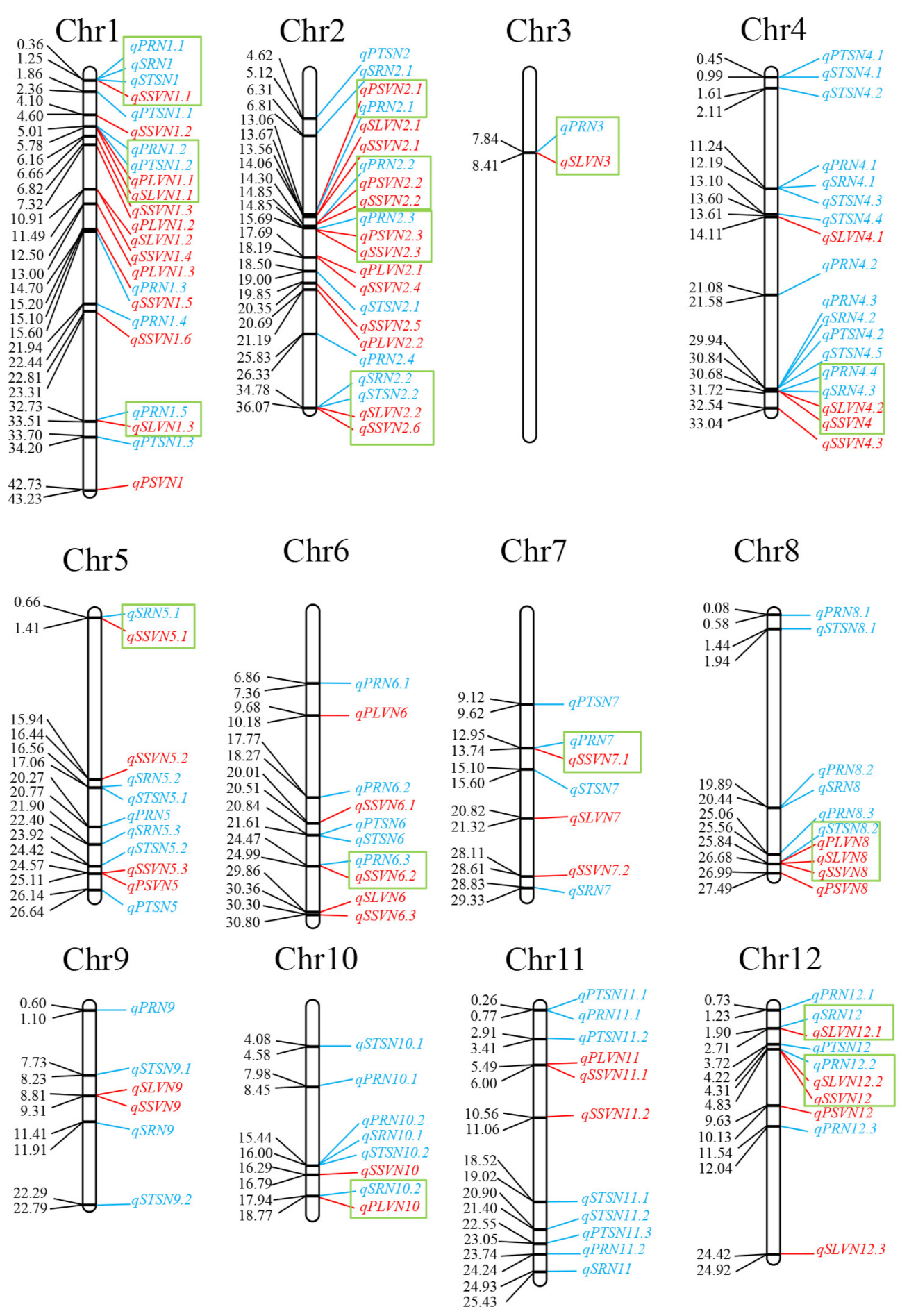
3.2. QTL Mapping
3.3. Coincidence of QTL for Sink- and Flow-Related Traits
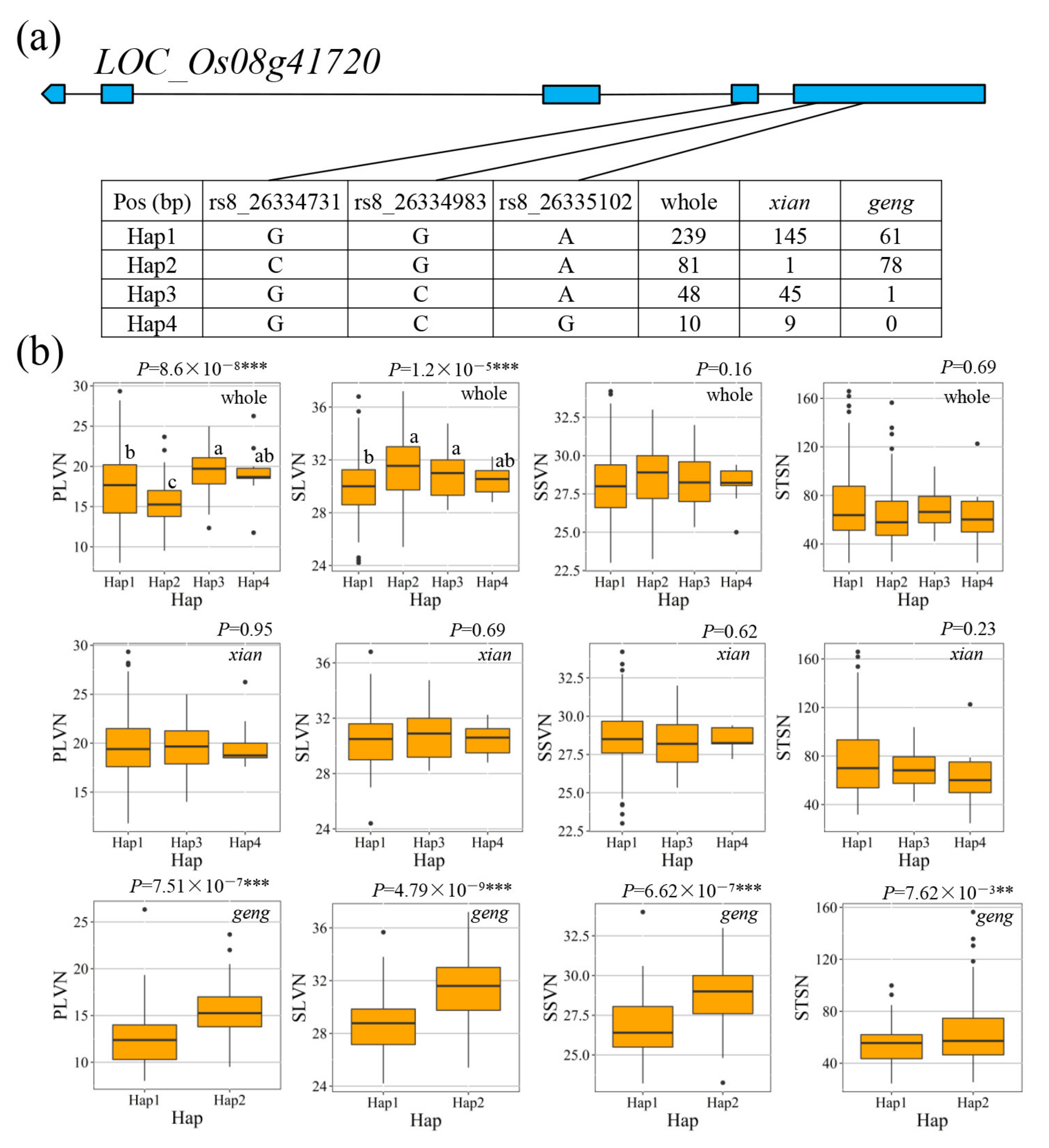
3.4. Candidate Gene Analysis for Important QTL
4. Discussion
4.1. Characteristics of Vascular Bundle between Xian and Geng Groups
4.2. Comparisons of QTL Detected in this Study with Previously Reported Cloned Genes
4.3. Candidate Gene Identification for Important QTL
4.4. Application in Rice Breeding for High Yield Potential
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ashraf, M.; Akbar, M.; Salim, M. Genetic improvement in physiological traits of rice yield. In Genetic Improvement of Field Crops; Slafer, G.A., Ed.; Marcel Dekker Incorporates: New York, NY, USA, 1994; pp. 413–455. [Google Scholar]
- Evans, L.T.; Dunstone, R.L.; Rawson, H.M.; Williams, R.F. The Phloem of the Wheat Stem in Relation to Requirements for Assimilate by The Ear. Aust. J. Biol. Sci. 1970, 23, 743–752. [Google Scholar] [CrossRef]
- Peterson, D.M.; Housley, T.L.; Luk, T.M. Oat stem vascular size in relation to kernel number and weight. II. Field Environment. Crop Sci. 1982, 22, 274–278. [Google Scholar] [CrossRef]
- Xu, Z.J.; Chen, W.F.; Chao, H.R. Relation between the characters of panicle and vascular bundle in neck-panicle if rice. Acta Agron. Sin. 1998, 24, 43–54. [Google Scholar]
- Fukuyama, T.; Takayama, T. Variations of the vascular bundle system in Asian rice cultivars. Euphytica 1995, 86, 227–231. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Li, P.; Wang, L.X.; Tan, C.J.; Hu, Z.L.; Zhu, Y.G.; Zhu, L.H. Identification of quantitative trait loci (QTLs) for the characters of vascular bundles in peduncle related to indica-japonica differentiation in rice (Oryza sativa L.). Euphytica 2002, 128, 279–284. [Google Scholar] [CrossRef]
- Cui, K.; Peng, S.; Xing, Y.; Yu, S.; Xu, C.; Zhang, Q. Molecular dissection of the genetic relationships of source, sink and transport tissue with yield traits in rice. Theor. Appl. Genet. 2003, 106, 649–658. [Google Scholar] [CrossRef]
- Bai, X.F.; Wu, B.; Xing, Y.Z. Yield-related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 2012, 54, 300–311. [Google Scholar] [CrossRef]
- Zhai, L.; Zheng, T.; Wang, X.; Wang, Y.; Chen, K.; Wang, S.; Wang, Y.; Xu, J.; Li, Z. QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice 2018, 11, 13. [Google Scholar] [CrossRef]
- Fei, C.; Geng, X.; Xu, Z.J.; Xu, Q. Multiple areas investigation reveals the genes related to vascular bundles in rice. Rice 2019, 12, 17. [Google Scholar] [CrossRef]
- Ren, D.; Xu, Q.; Qiu, Z.; Cui, Y.; Zhou, T.; Zeng, D.; Guo, L.; Qian, Q. FON4 prevents the multi-floret spikelet in rice. Plant Biotechnol. J. 2019, 17, 1007–1009. [Google Scholar] [CrossRef]
- Xue, W.; Xing, Y.; Weng, X.; Zhao, Y.; Tang, W.; Wang, L.; Zhou, H.; Yu, S.; Xu, C.; Li, X.; et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008, 40, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Fang, N.; Xu, R.; Huang, L.; Zhang, B.; Duan, P.; Li, N.; Luo, Y.; Li, Y. SMALL GRAIN 11 Controls Grain Size, Grain Number and Grain Yield in Rice. Rice 2016, 9, 64. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Gu, M.; Hu, J.; Qu, H.; Xu, G. Rice OsLHT1 Functions in Leaf-to-Panicle Nitrogen Allocation for Grain Yield and Quality. Front. Plant Sci. 2020, 11, 1150. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Sun, X.M.; Ma, X.Q.; Xu, B.X.; Zhao, X.; Ma, Z.Q.; Li, G.L.; Khan, N.U.; Pan, Y.H.; Liang, Y.T.; et al. GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J. 2021, 9, 57–67. [Google Scholar] [CrossRef]
- Terao, T.; Nagata, K.; Morino, K.; Hirose, T. A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 2010, 120, 875–893. [Google Scholar] [CrossRef]
- Fu, X.; Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu, C.; Li, J. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Tabangin, M.E.; Woo, J.G.; Martin, L.J. The effect of minor allele frequency on the likelihood of obtaining false positives. BMC Proc. 2009, 3 (Suppl. S7), S41. [Google Scholar] [CrossRef]
- Kang, H.M.; Sul, J.H.; Service, S.K.; Zaitlen, N.A.; Kong, S.; Freimer, N.B.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed]
- Li, M.X.; Yeung, J.M.Y.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2011, 131, 747–756. [Google Scholar] [CrossRef]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.H.; Blay, S.; Mcneney, B.; Graham, J. LDheatmap: An R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.C.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice Annotation Project Database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Alexandrov, N.; Tai, S.; Wang, W.; Mansueto, L.; Palis, K.; Fuentes, R.R.; Ulat, V.J.; Chebotarov, D.; Zhang, G.; Li, Z.; et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic. Acids Res. 2015, 43, D1023–D1027. [Google Scholar] [CrossRef]
- Wang, X.Q.; Pang, Y.L.; Wang, C.C.; Chen, K.; Zhu, Y.J.; Shen, C.; Ali, J.; Xu, J.L.; Li, Z.K. New candidate genes affecting rice grain appearance and milling quality detected by genome-wide and gene-based association analyses. Front. Plant Sci. 2017, 7, 1998. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, K.; Pang, Y.L.; Naveed, S.A.; Zhao, X.Q.; Wang, X.Q.; Wang, Y.; Dingkuhn, M.; Pasuquin, J.; Li, Z.K.; et al. QTL mapping and candidate gene analysis of ferrous iron and zinc toxicity tolerance at seedling stage in rice by genomewide association study. BMC Genom. 2017, 18, 828. [Google Scholar] [CrossRef] [PubMed]
- Fang, Z.; Bai, G.; Huang, W.; Wang, Z.; Wang, X.; Zhang, M. The Rice Peptide Transporter OsNPF7.3 Is Induced by Organic Nitrogen, and Contributes to Nitrogen Allocation and Grain Yield. Front. Plant Sci. 2017, 8, 1338. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Q.; Xu, Z.J.; Chen, W.F.; Xu, H.; Liu, H.G.; Zhu, C.J.; Wang, Y.; Wang, J.Y. The relation between the characters of vascular bundle and panicle in the filial generation from Indica and japonica Rice. Acta Agr Boreali-Sin. 2007, 5, 8–14. [Google Scholar]
- Fukushima, A.; Akita, S. Varietal differences of the course and differentiation time of large vascular bundles in the rachis of rice. Jpn. J. Crop Sci. 1997, 66, 24–28. [Google Scholar] [CrossRef][Green Version]
- Liu, T.S.; Bi, W.J.; Zhang, J.; Cui, Y.; Yan, Z.Q.; Wang, Y.Z.; Fei, C.; Xu, H.; Tang, L.; Chen, W.F.; et al. Characterization of the relationship between vascular bundles features and indica-allelic frequency using a seventh filial generations of indica × japonica rice crosses. Euphytica 2016, 209, 739–748. [Google Scholar] [CrossRef]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef]
- Nakamura, A. The role of OsBRI1 and its homologous genes, OsBRL1 and OsBRL3, in rice. Plant Physiol. 2006, 140, 580–590. [Google Scholar] [CrossRef]
- Caño-Delgado, A.; Yin, Y.H.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Nam, K.H.; Li, J.M.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef]
- Berleth, T.; Scarpella, E.; Prusinkiewicz, P. Towards the systems biology of auxin-transport-mediated patterning. Trends Plant Sci. 2007, 12, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Sang, X.; Zhang, T.; Yu, Z.; Li, Y.; Zhao, F.; Wang, Z.; Wang, Y.; Yu, P.; Wang, N.; et al. ABNORMAL VASCULAR BUNDLES regulates cell proliferation and procambium cell establishment during aerial organ development in rice. New Phytol. 2017, 213, 275–286. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhai, L.; Wang, Y.; Yan, A.; Chen, L.; Shao, K.; Zhang, W.; Xu, J. Genetic Bases of Flow- and Sink-Related Traits in Rice Revealed by Genome-Wide Association Study. Agronomy 2022, 12, 776. https://doi.org/10.3390/agronomy12040776
Zhai L, Wang Y, Yan A, Chen L, Shao K, Zhang W, Xu J. Genetic Bases of Flow- and Sink-Related Traits in Rice Revealed by Genome-Wide Association Study. Agronomy. 2022; 12(4):776. https://doi.org/10.3390/agronomy12040776
Chicago/Turabian StyleZhai, Laiyuan, Yun Wang, An Yan, Liqiang Chen, Kuitian Shao, Wenzhong Zhang, and Jianlong Xu. 2022. "Genetic Bases of Flow- and Sink-Related Traits in Rice Revealed by Genome-Wide Association Study" Agronomy 12, no. 4: 776. https://doi.org/10.3390/agronomy12040776
APA StyleZhai, L., Wang, Y., Yan, A., Chen, L., Shao, K., Zhang, W., & Xu, J. (2022). Genetic Bases of Flow- and Sink-Related Traits in Rice Revealed by Genome-Wide Association Study. Agronomy, 12(4), 776. https://doi.org/10.3390/agronomy12040776





