Abstract
Estimating foliar damage is essential in agricultural processes to provide proper crop management, such as monitoring the defoliation level to take preventive actions. Furthermore, it is helpful to avoid the reduction of plant energy production, nutrition decrement, and consequently, the reduction of the final production of the crop and economic losses. In this sense, numerous proposals support the defoliation estimate task, ranging from traditional methodologies to computational solutions. However, subjectivity characteristics, reproducibility limitations, and imprecise results persist. Then, these circumstances justify the search for new solutions, especially in defoliation assessments. The main goal of this paper consists of developing an automatic method to estimate the percentage of damaged leaf areas consumed by insects. As a novelty, our method provides high precision in calculating defoliation severity caused by insect predation on the leaves of various plant species and works effectively to estimate leaf loss in leaves with border damage. We describe our method and evaluate its performance concerning 12 different plant species. Our experimental results demonstrate high accuracy in the determination of leaf area loss with a correlation coefficient superior to 0.84 for apple, blueberry, cherry, corn, grape, bell pepper, potato, raspberry, soybean, and strawberry leaves, and mean absolute error (MAE) less than 4% in defoliation levels up to 54% in soybean, strawberry, potato, and corn leaves. In addition, the method maintains a mean error of less than 50%, even for severe defoliation levels up to 99%.
1. Introduction
Leaf damage affects the correct quantification of the foliar area, and due to various causes, this is a non-trivial problem. For example, wind can whip the foliage causing tears in the leaves, and hailstorms can damage them by creating holes or even causing total defoliation. Besides, leaf skeletonization and insect predation also promote the appearance of even more harmful external damage. The reasons for leaf skeletonization may result from insects or diseases and occasionally chemical damage, leading to a visual pattern of plant deformities. In contrast, the types of damage induced by maxillary insects are more diverse due to the different developmental stages of insects such as larval, nymph, and adult stages [1].
Estimating leaf loss is a crucial tool for planning sustainable agricultural practices. As the leaves are inputs for monitoring, evaluation, and decision-making, when the leaves are damaged, the deformities can be used to guide the proper management of a crop. In this sense, to increase productivity, pest control based on leaf analysis is mandatory in crop management. Predatory insects have caused significant economic impacts in recent decades, and an average annual loss of US$ 11.40 billion in agricultural production is estimated. From 1960 to 2020, the economic loss has progressively increased, reaching the mark of US$ 165.01 billion in 2020 [2]. The main consequence is related to the functional reduction of the total leaf surface, namely defoliation [3], which reduces the energy capacity of the plant, light interception, plant growth rate, and dry mass accumulation [4]. Consequently, leaf injury caused by insect herbivory negatively affects crop grain yield [5]. Therefore, estimating leaf loss is a primary practice for conducting inspection methodologies and performing control services in crop fields.
A wide range of proposals, from traditional methodologies to computer-based solutions, addresses this issue, aiming to reduce subjectivity, ensure reproducibility, and increase accuracy. Some of the defoliation estimate methods use human expertise to perform visual evaluation and manual quantification [6,7], predictive models of linear leaf dimensions [8,9], integrative method of leaf area [10,11], digital image processing for mathematical models generation [12,13], and deep learning algorithms [3,14]. Nevertheless, many techniques and methods are applied to a specific type of plant and do not generalize in order to be consistent with various plant species and issues [14].
In this context, manual, semi-automated, and fully automated methodologies address leaf area monitoring. Although their contributions are relevant to agricultural processes, they must overcome some limitations. Visual assessment may increase the error due to its subjective characteristics. Manual quantification requires extensive work, expertise, and extended time for analysis and evaluation. Automatic meter devices are expensive, require technical support, and demand maintenance. The results of computer-based solutions that require user interaction depend heavily on prior training and proper application handling. Additionally, automated computer-aided approaches require many leaf samples to generalize the construction of statistical models or the intensive application of feature engineering in formulating mathematical models.
In addition, solutions for use in agriculture need to consider devices with limited computing power, such as embedded systems, Internet of Things (IoT) ecosystems, and intelligent agricultural machinery. As application processing requires efficient solutions, it is crucial to consider lightweight processes and energy efficiency to not overload systems. In this sense, we investigated image processing techniques that guarantee high performance while being simple to understand and implement and using few computational resources.
To contribute to this area, we present an automatic method to measure the percentage of insect predation on leaves. As a novelty, our method provides high precision for various targeted species such as tomato, strawberry, soybean, raspberry, potato, bell pepper, peach, grape, corn, cherry, blueberry, and apple. Furthermore, it effectively estimates leaf loss in leaves with border damage, converges quickly, and does not require human interaction. Our paper presents the processing steps of the defoliation estimate method, which is based on leaf properties, image processing techniques and statistical measurements. We emphasize that our method uses comprehensive steps that are easy to implement and suitable for environments with limited computing power.
The remainder of the paper is organized as follows. The work related to leaf defoliation methods is presented in Section 2. In Section 3, we present details of our method. Section 4 provides information about the test settings, image data set, and experimental design. In Section 5, we present experimental tests and discussion about the results. Then, in Section 6, we conclude the paper and present some future work.
2. Related Work
Mallof et al. [15] implemented a software component called LeafJ to be added to ImageJ popular software, a popular computer program that is used to classify leaf shapes and compute leaf silhouettes. The LeafJ plugin is a semi-automatic tool for studies on leaf morphology that was designed to increase the features of ImageJ by providing petiole length measurement and leaf blade parameters. Although it supports leaf area measurements, it was not prepared to estimate biomass loss in damaged leaves. Therefore, its use is recommended only for healthy leaves.
Easlon and Bloom [16] estimated leaf surface through different color thresholds and morphological operation to connect image components. Their proposal used a red calibration landmark with a known area as a visual reference point to calibrate leaf area estimation, thereby removing the need to estimate camera distance and focal length. The authors used a leaf area meter to be compared with their method. Additionally, they used the ImageJ software in the comparative analysis. An adequate precision was shown in both leaf segmentation and leaf area estimation. However, their application is sensitive to illumination changes and perspective distortion. Additionally, it requires user interaction and intervention for the best results, and it does not measure insect predation on leaves.
Kaur et al. [17] proposed an elementary computer program for calculating the plant leaf area in which a digital scanner and a threshold segmentation method are settled to separate the leaves from the image background. Likewise, Jadon et al. [18] proposed a simple method that applies digital image processing to estimate foliar area in which a digital camera and a white background paper suppress the use of a scanner device in the image acquisition step. However, as these methods do not deal with damaged leaves, none can adequately address the problem of leaf herbivory along the borders.
In order to address the problem of measuring leaf area with deformation caused by insects, Machado et al. [5] developed a computer application that uses image processing techniques to estimate herbivory. They compared their computer application with manual quantification of injured leaves and a leaf area electronic integrator (considered a standard method of leaf area analysis). The results showed that the defoliation estimate made by the computer program developed by the authors was close to the values measured by the other methods. However, specialized intervention is still required to draw manually the edges of the leaves that have been compromised; therefore, depending on the user’s expertise, assertiveness may be better or worse achieved.
Liang et al. [13] proposed some instructions to calculate the area, border, and defoliation estimate of soybean leaves. Their proposal requires the selection of image samples to build a representative canopy statistical model, which is used to distinguish leaves from non-leaves and backgrounds. Although they have shown adequate results, the proposed methodology may be affected by the image acquisition stage, in which unrepresentative samples may reduce the potential of their method.
In da Silva et al. [3], the authors compared deep learning models to estimate defoliation levels and elaborated an automatic method to compute damage in injured leaves. In addition, the authors have proposed strategies to generate images with artificial defoliation to deal with the number of data examples to feed the neural networks. Although their proposal creates new possibilities for calculating the leaf area in which deep neural networks can be used for defoliation analysis, it does require a significant amount of data samples to improve the overall learning procedure (in their experiments, three data sets were prepared, each one with 10,000 labeled data). Likewise, Silva et al. [14] investigated leaf damage estimation using deep neural networks and prepared an artificial random damage generation method to create a synthetic database. However, as in [3], their method requires an image data set with many image samples (the authors used more than 22,000 samples during the training step.) In this sense, the processing of large databases can be restrictive for devices with limited computing power, as in smart farming ecosystems, or demand equipment with specific hardware configurations to reduce energy consumption and provide timely responses.
In the same way, Zhang et al. [19] used images taken from unmanned aerial vehicles to determine soybean defoliation in crop fields. This work presented a computer learning model to estimate crop defoliation. Additionally, the authors prepared computer models to characterize defoliated crops so that wrong characterizations of healthy crops could be avoided. Although promising, the author’s method does not include defoliation estimation for isolated leaf samples. Manso et al. [20] presented a method to detect damage in coffee leaves that uses image segmentation and an artificial neural network to identify and classify leaf damage. However, their method only works for leaves with visible leaf damage, i.e., for damage that can be identified by the difference in color between the healthy and diseased areas. Thus, this method does not include the estimate of defoliation in which the leaf area was consumed, for example, by chewing or cutting insects. Liang et al. [13] presented a method to determine the soybean canopy defoliation using RGB images to provide informative data in pest management. Although these researchers show promising results for soybean leaves, they do not look at their solutions in a broader context to determine whether their solutions generalize to different plant forms and species [14].
With the advancement of machine learning algorithms, deep learning models have been widely used to support agricultural management [21,22,23]. However, training stages on supervised models demand large data sets with data annotation, which can be challenging to prepare [24,25,26]. Moreover, they are not efficient in predicting unexpected scenarios in data sets that have not been used for training, and the learning steps of these networks are time-consuming [27,28]. In this regard, computer vision has addressed pattern recognition in digital images so that the models can be less dependent on the image data sets [29,30]. Furthermore, lightweight models have been designed to consider the characteristics of agricultural environments, such as reduced computing resources, limited processing power, and embedded device systems [13,31,32,33,34].
Our method differs from the related work in some aspects as it requires fewer image samples relative to the deep learning methods (only 60 samples) and does not demand samples of herbivory in preparing the image models. Also, the method is stable against image transformation, such as rotation and scale, and is a fully-automated method that uses data input to construct image models for template matching. Moreover, the method measures the percentage of the foliar damage area by counting the pixels, uses a global thresholding method to detach the leaves from the image background, and uses digital image processing techniques in its design.
In this sense, our method can automatically estimate the leaf area consumed by insects using digital images without requiring large volumes of data to build the templates. Additionally, our method indicates the severity level of defoliation regardless of whether the leaf damage occurs in inner regions or at the edge of the leaves, something that only semi-automatic methods or deep learning models were able to address.
3. Method
Our method is organized into three steps to handle background removal to highlight leaf regions, adjust images based on their geometric shape, and estimate defoliation severity. The adjusted images are used to prepare a database to retrieve leaf models similar to query images. Then, the retrieved images are used to estimate the percentage of damage in the query images. Figure 1 presents the flowchart of the method.
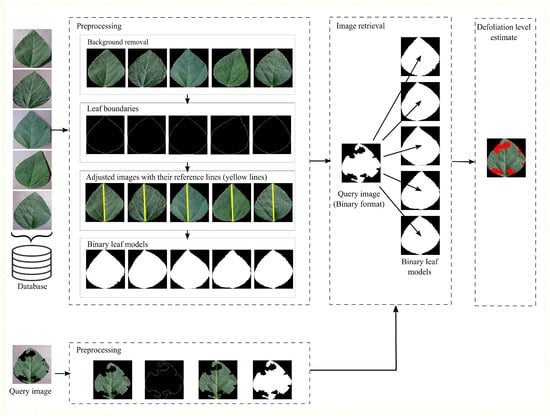
Figure 1.
Flowchart of the presented method.
3.1. Preprocessing
The input RGB images are processed with a median filter. From the resulting images, the channel G (Green color) is updated by multiplying its current channel values by 2 with the subtraction of the R (Red color) and B (Blue color) channel values. Then, the Otsu segmentation method [35] is applied to detach the leaves from the background, and binary images are obtained where the inner pixels are removed, leaving only the boundary pixels. The distances between the remaining pixels are measured, and the two most distant points draw a line that we call the reference line. The reference line is used to rotate the images to the vertical position. Then, the images are surrounded by bounding boxes, their outer areas are cropped, and the images are resized to their original sizes. Thus, the resulting images are binarized, and the leaf model data set is prepared.
3.2. Image Retrieval
Considering a binary query image also obtained after the previous step, a comparison is made between the query image and the binary target models from the model data set. The binary leaf model with the slightest difference from the query image is selected and used to determine the damaged leaf area to estimate the leaf damage area In Equation (1), the damage region is obtained by the intersection of two sets.
where m and n represent the numbers of rows and columns of the images, respectively.
3.3. Defoliation Level Estimate
In Equation (4), the retrieved image model () is compared with the damaged input leaf () through logical conjunction. After this operation, a logical image is obtained, which presents the missing leaf areas.
After that, the percentage of pixels in is calculated according to Equation (5),
where and m and n denote the number, in terms of image dimensionality (rows and columns), of the image .
4. Materials
4.1. Database Description
We use the database prepared by [36], which is available online as a result of [37]. In obtaining the samples, technicians collected leaves by removing them from plants and placing them against a sheet of paper that provided a gray or black background to begin the process of acquiring digital images. They obtained the image samples considering various lighting conditions, leaf shape, and foliar position. Based on this, we randomly selected healthy leaves from the dataset, which include samples of tomato, strawberry, soybean, raspberry, potato, bell pepper, peach, grape, corn, cherry, blueberry, and apple. The images are in RGB format and have a size equal to pixels.
4.2. Experiment Design
We randomly selected 120 images for each of the 12 types of plants, totaling 1440 images for use in the experiments. We divided the data into two groups where 60 images of each plant species are used to construct the leaf model data set, and the other 60 images of each plant species are used to validate the method. The leaf images contained in these groups are different, and therefore, a leaf sample belongs exclusively to just one of them.
In the data used to validate the proposal, we apply a synthetic defoliation strategy to simulate insect predation. We manually segmented insect bite traces from images with leaves consumed by the insects Spodoptera frugiperda and Chrysodeixis includens. Then, we used the bite segments to simulate real cases of herbivory in healthy leaves. Our artificial defoliation program has four random parameters. The first determines the insects used to simulate defoliation. The second specifies the number of bite segments. The third applies rotation transformations to the bite segments. Finally, the fourth parameter resizes the bite segments. In addition, the user can specify the required defoliation level from 1 to 99%. After applying the synthetic defoliation, the defoliation level is computed and the reference data (ground-truth) is prepared.
This approach is necessary because the data set used does not include cases of defoliation caused by insect herbivores such as chewing or cutting insects. Additionally, data sets containing samples in this category are private and not publicly available, as in [5,38]. Therefore, related works such as [3,14] used artificial defoliation strategies. Unlike other works, we used actual defoliation simulation, which is the novelty of our study.
4.3. Execution Time
A notebook with Core i7-9750H (2.6 GHz; 12 MB Cache) and 16 GB RAM was used to perform the experimental tests. The execution time of the leaf model with 60 images was s. In addition, the average time to compute the defoliation estimate was s. Our source code is freely available for download and was written in MATLAB.
4.4. Evaluation
The accuracy of our proposal is measured according to the linear correlation [39], root mean square error (RMSE), and mean absolute error (MAE) between the results and the reference data (ground-truth) following Equations (6)–(8):
where x contains the reference data values obtained from the synthetic defoliation strategy, and is the average value of these defoliation levels. y contains the estimated leaf damage values, and is the average value of the estimated defoliation levels. n represents the number of images used to validate the method.
5. Results and Discussion
The method’s accuracy is evaluated concerning the query images transformed with the defoliation strategy. As we compare the ground-truth images with the outputs of our method, we verify the quantitative and visual results. Therefore, the performance of our method is measured according to the severity of the estimated and actual defoliation.
Figure 2 presents visual results of our method with a leaf sample for each of the twelve plant species under study. After the segmentation and defoliation processes, the query images are obtained from the input images, and the final result highlights the damaged leaf regions. As noted, the preprocessing steps adjust the images to a position that reduces the effects of scaling and image rotation in evaluating the correspondence between a query image and leaf models. Thus, our method can present consistent results even if the images were not acquired following a strict standard form of leaves and camera positioning. Figure 2 also illustrates the diversity of samples contained in the data set we used, showing that our method has the potential for generalization as it deals with different types of plant species, leaf shapes, and different levels of leaf damage.
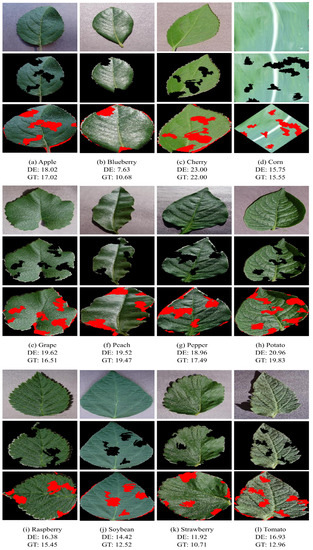
Figure 2.
Visual results of our method. The first row of each figure panel shows the images as they are in the data set, the second row presents the query images after segmentation and defoliation, and the third row presents the final result with defoliation estimate (DE) and ground truth (GT).
Additionally, we evaluate the proposal considering different defoliation levels. We consider a gradual increase in the defoliation level from which we apply progressive jumps of 3% until reaching 99% leaf damage. Figure 3 presents the outcomes where the best results occurred when the defoliation level was between 0% and 60%. Although the error increases after 60% defoliation, the error is still less than 50% even when the leaves are destroyed, such as after 90% defoliation. It is worth mentioning that this pattern is repeated in all plant species investigated in this study. In this way, it shows the generalization potential of the method.
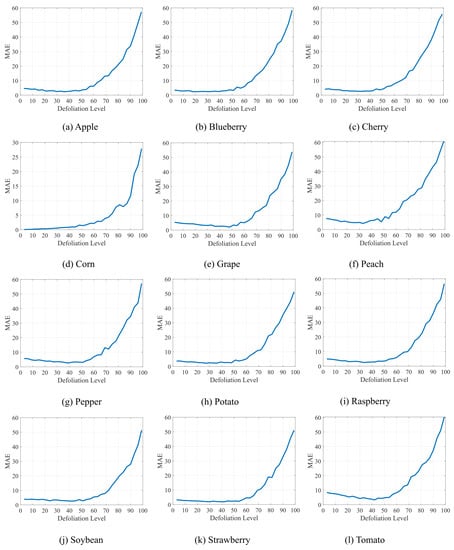
Figure 3.
Results of our method concerning different levels of defoliation.
Table 1 shows the linear correlation and the mean error between the damaged leaves for each of the 12 plant species considering a maximum defoliation level of 30%. A solid linear correlation can be noted, especially for corn, strawberry, grape, potato, blueberry, and soybean, whose correlation was equal to , , , , , and , respectively. Tomato and Peach showed the lowest positive result, mainly due to the diversity of leaf shapes caused by the collection of different cultivars, different stages of leaf growth, and the irregular pattern that plant species can produce. Tomato has samples of various tomato species with early and adult leaves, and peach has leaves with minor to moderate curvature and a wide to narrow leaf canopy. Additionally, the intensity of shading on peach leaves reduced the capability of the segmentation process, justifying the weak correlation that was obtained. Effective treatment of shadows is open to be addressed in future work. Although our method presents consistent results, we point out that excessive leaf deformation can compromise the correct identification of the reference line (Section 3), generating an inadequate leaf positioning. Additionally, when the images used to build the models are significantly different from the query images, assertiveness may decrease.

Table 1.
Results of our method on different plant species.
Figure 4 presents the scatter plot between the actual and estimated defoliation levels considering the correlation results of different plant leaves for the maximum defoliation level of 30%. The correlation coefficient above for apple, blueberry, cherry, corn, grape, bell pepper, potato, raspberry, soybean, and strawberry shows a positive linear correlation between the variables and indicates that the estimated damage closely follows the actual damage. In addition, a large part of the data is close to the regression line, which emphasizes the strong correlation between them. Since the fit line is above the reference line (Y = T), the estimated data are partially overestimated.
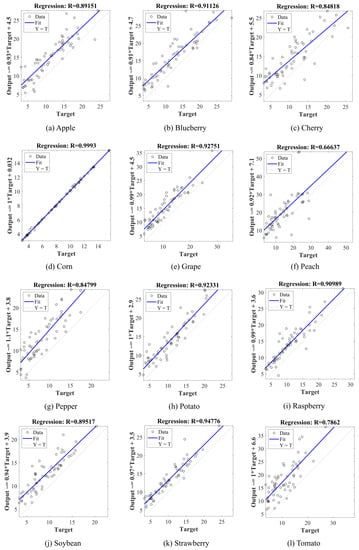
Figure 4.
Regression line between ground-truth defoliation levels and estimated damage by our method concerning different plant species. Target T refers to the reference data, while Y refers to the estimated value.
Complementarily, Figure 5 demonstrates the behavior of the method concerning the estimated defoliation level. As can be observed, the estimated values are more significant than the actual damage, i.e., the method overestimates the defoliation level, indicating percentages of leaf damage above the actual values. However, these values are not discrepant since the curves have similar shapes. In the real scenario, this behavior is preferable to underestimated results that could induce the reduction or interruption of management operations. Furthermore, as this behavior is repeated in other plant species, the overestimation can be measured and used in leaf analysis. For example, in soybean leaves, the mean overestimated value (mean absolute error, Table 1) is . Based on this knowledge, the curves that characterize the actual and estimated defoliation level (Figure 5) can be adjusted to present a more assertive result.
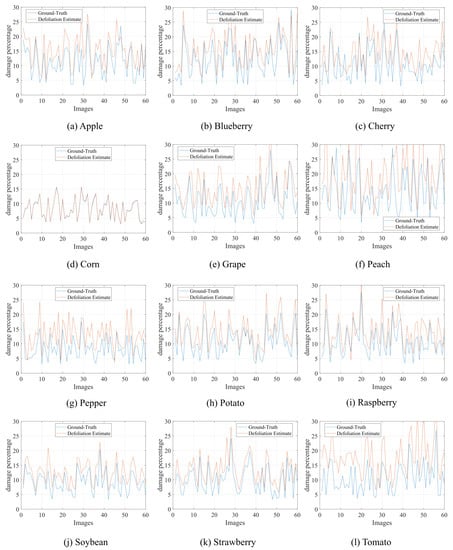
Figure 5.
Comparison between ground-truth defoliation levels and estimated damage by our method in different plant species.
Table 2 presents a comparison between our method and some related work. These methods used the automatic estimation of leaves or human intervention to delineate the leaves in the images (semi-automatic approach). Some of them were not prepared to deal with leaf losses. In opposition, other methods consider leaf damage, including the destruction in border regions with defoliation levels between 0 and 65%. Except for our method, the automatic approaches were developed using deep learning models, which require many image samples. Additionally, only our method and Silva et al.’s [40] method were evaluated exclusively with public databases. The other authors built their bases for evaluation or used local and public data sets.

Table 2.
Some relevant information about our method and related work.
Table 3 presents a quantitative comparison considering our method and some related works. Although each work has applied its own experimental evaluations, they have some characteristics in common, such as performance measurement (r and RMSE) and type of plant under analysis (soybean leaf). In this sense, this comparison shows that our proposal presents results close to semi-automatic methods that demand human intervention and automatic methods that use deep learning. For example, Bradshaw et al. [38] and Machado et al. [5] used local databases and compared their results with leaf area measurement devices (LI-COR). On the other hand, da Silva et al. [3] used local and public databases and synthetic defoliation methods to train computational models. In contrast, our method is evaluated with a public database that contains different plant species and variability in leaf shape. Furthermore, our method involves a few processing steps, which makes it suitable for systems with reduced computing power that generally qualify applications for smart farms.

Table 3.
Quantitative results provided by related work and our method considering soybean leaves.
6. Conclusions
This study presented a new method to calculate the percentage of damage caused by insect herbivores on plant leaves. The method uses a few processing steps, making it suitable for intelligent farm environments with limited computing power. Based on the experimental evaluation, our method is assertive in measuring the loss of leaf area, considering different shapes, sizes, and morphology of leaves. The results were best for defoliation percentages of a maximum of 60% as the predictions deteriorated beyond 60%. For defoliation below 60%, the mean absolute error was lower or close to 10% for all plant species under study, showing the generalization potential of our method. Additionally, when verifying the correlation between leaf damage and estimated damage, we observed a positive relationship between the variables with correlation values between and for apple, cherry, pepper, and soybean, and above for blueberry, corn, grape, potato, raspberry, and strawberry.
As a novelty, our method provides high precision for diverse plant species, works effectively to estimate leaf loss in leaves with border damage, converges quickly, and does not require human interaction. Furthermore, it does not require samples of herbivory to prepare the image models and a massive amount of data to converge appropriately. Thus, we present a comprehensive solution for crop monitoring and decision-making through quantitative and visual analysis.
In this sense, we conclude that our proposal is a valuable option to support agricultural management based on analytical information where insect predation can be addressed before it compromises the entire plantation. The source code of our method is publicly available for use at no monetary cost. As part of future work, we intend to consider other image data sets and plant species and prepare a data set with actual herbivory cases. Moreover, we intend to add a new processing layer to our method so that we can handle the shadows that are generated during the image acquisition process.
Author Contributions
Conceptualization, G.S.V. and A.U.F.; methodology, G.S.V.; software, G.S.V. and A.U.F.; validation, B.M.R. and N.M.S.; formal analysis, J.C.F. and J.P.F.; investigation, G.S.V.; resources, F.S.; data curation, J.C.L.; writing—original draft preparation, G.S.V.; writing—review and editing, G.S.V.; visualization, G.S.V. and A.U.F.; supervision, F.S.; project administration, F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Image database and code used in this research are freely available at https://github.com/digitalepidemiologylab/plantvillage_deeplearning_paper_dataset (accessed on 27 September 2022), Image Database; and https://github.com/pixellab-ufg/leaf-analysis (accessed on 27 September 2022), Code.
Acknowledgments
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior-Brasil (CAPES)—Finance Code 001. Also, we would like to thank and acknowledge partial support for this research from INF/UFG (Federal University of Goias), and IF Goiano (Federal Institute Goiano).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| MAE | Mean Absolute Error |
| IoT | internet of things |
| GT | Ground-Truth |
| RAM | Random Access Memory |
| GHz | Gigahertz |
| MB | Megabyte |
| GB | Gigabyte |
| RMSE | Root Mean Square Error |
| DE | Defoliation Estimate |
| LD | Local data set |
| SA | Semi-Automatic |
| AU | Automatic |
References
- Carvalho, M.R.; Wilf, P.; Barrios, H.; Windsor, D.M.; Currano, E.D.; Labandeira, C.C.; Jaramillo, C.A. Insect leaf-chewing damage tracks herbivore richness in modern and ancient forests. PLoS ONE 2014, 9, e94950. [Google Scholar]
- Renault, D.; Angulo, E.; Cuthbert, R.N.; Haubrock, P.J.; Capinha, C.; Bang, A.; Kramer, A.M.; Courchamp, F. The magnitude, diversity, and distribution of the economic costs of invasive terrestrial invertebrates worldwide. Sci. Total. Environ. 2022, 835, 155391. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, L.A.; Bressan, P.O.; Gonçalves, D.N.; Freitas, D.M.; Machado, B.B.; Gonçalves, W.N. Estimating soybean leaf defoliation using convolutional neural networks and synthetic images. Comput. Electron. Agric. 2019, 156, 360–368. [Google Scholar] [CrossRef]
- Fernandes, E.T.; Ávila, C.J.; da Silva, I.F. Effects of different levels of artificial defoliation on the vegetative and reproductive stages of soybean. EntomoBrasilis 2022, 15, e991. [Google Scholar] [CrossRef]
- Machado, B.B.; Orue, J.P.; Arruda, M.S.; Santos, C.V.; Sarath, D.S.; Goncalves, W.N.; Silva, G.G.; Pistori, H.; Roel, A.R.; Rodrigues, J.F., Jr. BioLeaf: A professional mobile application to measure foliar damage caused by insect herbivory. Comput. Electron. Agric. 2016, 129, 44–55. [Google Scholar] [CrossRef]
- Kvet, J.; Marshall, J. Assessment of leaf area and other assimilating plant surfaces. In Plant Photosynthetic Production—Manual of Methods; Sestak, Z., Catsky, J., Jarvis, P.G., Eds.; Junk: The Hague, The Netherlands, 1971; pp. 517–555. [Google Scholar]
- Kogan, M.; Turnipseed, S.; Shepard, M.; De Oliveira, E.; Borgo, A. Pilot insect pest management program for soybean in southern Brazil. J. Econ. Entomol. 1977, 70, 659–663. [Google Scholar] [CrossRef]
- Santos, J.; Costa, R.; Silva, D.; Souza, A.; Moura, F.; Junior, J.; Silva, J. Use of allometric models to estimate leaf area in Hymenaea courbaril L. Theor. Exp. Plant Physiol. 2016, 28, 357–369. [Google Scholar] [CrossRef]
- Carvalho, J.O.D.; Toebe, M.; Tartaglia, F.L.; Bandeira, C.T.; Tambara, A.L. Leaf area estimation from linear measurements in different ages of Crotalaria juncea plants. An. Acad. Bras. Cienc. 2017, 89, 1851–1868. [Google Scholar] [CrossRef]
- LI-COR. LI-3000C Area Meter. 2022. Available online: https://www.licor.com/env/products/leaf_area/LI-3100C (accessed on 27 September 2022).
- ADC. AM350 Portable Leaf Area Meter. 2022. Available online: https://www.adc.co.uk/products/am350-portable-leaf-area-meter (accessed on 27 September 2022).
- Carrasco-Benavides, M.; Mora, M.; Maldonado, G.; Olguín-Cáceres, J.; von Bennewitz, E.; Ortega-Farías, S.; Gajardo, J.; Fuentes, S. Assessment of an automated digital method to estimate leaf area index (LAI) in cherry trees. N. Z. J. Crop. Hortic. Sci. 2016, 44, 247–261. [Google Scholar] [CrossRef]
- Liang, W.; Kirk, K.R.; Greene, J.K. Estimation of soybean leaf area, edge, and defoliation using color image analysis. Comput. Electron. Agric. 2018, 150, 41–51. [Google Scholar] [CrossRef]
- Silva, M.; Ribeiro, S.; Bianchi, A.; Oliveira, R. An Improved Deep Learning Application for Leaf Shape Reconstruction and Damage Estimation. In Proceedings of the 23rd International Conference on Enterprise Information Systems—ICEIS, INSTICC, SciTePress, Online, 26–28 April 2021; Volume 1, pp. 484–495. [Google Scholar] [CrossRef]
- Maloof, J.N.; Nozue, K.; Mumbach, M.R.; Palmer, C.M. LeafJ: An ImageJ plugin for semi-automated leaf shape measurement. J. Vis. Exp. 2013, 71, e50028. [Google Scholar] [CrossRef] [PubMed]
- Easlon, H.M.; Bloom, A.J. Easy Leaf Area: Automated digital image analysis for rapid and accurate measurement of leaf area. Appl. Plant Sci. 2014, 2, 1400033. [Google Scholar] [CrossRef]
- Kaur, G.; Din, S.; Brar, A.S.; Singh, D. Scanner image analysis to estimate leaf area. Int. J. Comput. Appl. 2014, 107, 3. [Google Scholar] [CrossRef]
- Jadon, M.; Agarwal, R.; Singh, R. An easy method for leaf area estimation based on digital images. In Proceedings of the 2016 International Conference on Computational Techniques in Information and Communication Technologies (ICCTICT), New Delhi, India, 11–13 March 2016; pp. 307–310. [Google Scholar]
- Zhang, Z.; Khanal, S.; Raudenbush, A.; Tilmon, K.; Stewart, C. Assessing the efficacy of machine learning techniques to characterize soybean defoliation from unmanned aerial vehicles. Comput. Electron. Agric. 2022, 193, 106682. [Google Scholar] [CrossRef]
- Manso, G.L.; Knidel, H.; Krohling, R.A.; Ventura, J.A. A smartphone application to detection and classification of coffee leaf miner and coffee leaf rust. arXiv 2019, arXiv:1904.00742. [Google Scholar]
- Alves, A.N.; Souza, W.S.; Borges, D.L. Cotton pests classification in field-based images using deep residual networks. Comput. Electron. Agric. 2020, 174, 105488. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Dang, L.M.; Sadeghi-Niaraki, A.; Moon, H. Crop pest recognition in natural scenes using convolutional neural networks. Comput. Electron. Agric. 2020, 169, 105174. [Google Scholar] [CrossRef]
- Liu, J.; Wang, X. Tomato diseases and pests detection based on improved Yolo V3 convolutional neural network. Front. Plant Sci. 2020, 11, 898. [Google Scholar] [CrossRef]
- Gomes, J.C.; Borges, D.L. Insect Pest Image Recognition: A Few-Shot Machine Learning Approach including Maturity Stages Classification. Agronomy 2022, 12, 1733. [Google Scholar] [CrossRef]
- Tian, X.; Jiao, L.; Liu, X.; Zhang, X. Feature integration of EODH and Color-SIFT: Application to image retrieval based on codebook. Signal Process. Image Commun. 2014, 29, 530–545. [Google Scholar] [CrossRef]
- Niu, D.; Zhao, X.; Lin, X.; Zhang, C. A novel image retrieval method based on multi-features fusion. Signal Process. Image Commun. 2020, 87, 115911. [Google Scholar] [CrossRef]
- Chu, K.; Liu, G.H. Image retrieval based on a multi-integration features model. Math. Probl. Eng. 2020, 2020, 1461459. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, G.H. Image retrieval using the intensity variation descriptor. Math. Probl. Eng. 2020, 2020, 6283987. [Google Scholar] [CrossRef]
- Vieira, G.S.; Rocha, B.M.; Fonseca, A.U.; Sousa, N.M.; Ferreira, J.C.; Cabacinha, C.D.; Soares, F. Automatic detection of insect predation through the segmentation of damaged leaves. Smart Agric. Technol. 2022, 2, 100056. [Google Scholar] [CrossRef]
- Vieira, G.S.; Sousa, N.M.; Rocha, B.M.; Fonseca, A.U.; Soares, F. A Method for the Detection and Reconstruction of Foliar Damage caused by Predatory Insects. In Proceedings of the 2021 IEEE 45th Annual Computers, Software, and Applications Conference (COMPSAC), Madrid, Spain, 12–16 July 2021; pp. 1502–1507. [Google Scholar]
- Intaravanne, Y.; Sumriddetchkajorn, S. Android-based rice leaf color analyzer for estimating the needed amount of nitrogen fertilizer. Comput. Electron. Agric. 2015, 116, 228–233. [Google Scholar] [CrossRef]
- Pivoto, D.; Waquil, P.D.; Talamini, E.; Finocchio, C.P.S.; Corte, V.F.D.; de Vargas Mores, G. Scientific development of smart farming technologies and their application in Brazil. Inf. Process. Agric. 2018, 5, 21–32. [Google Scholar] [CrossRef]
- Rocha, B.M.; Vieira, G.S.; Fonseca, A.U.; Sousa, N.M.; Pedrini, H.; Soares, F. Detection of Curved Rows and Gaps in Aerial Images of Sugarcane Field Using Image Processing Techniques. IEEE Can. J. Electr. Comput. Eng. 2022, 1–8. [Google Scholar] [CrossRef]
- Lin, J.; Chen, X.; Pan, R.; Cao, T.; Cai, J.; Chen, Y.; Peng, X.; Cernava, T.; Zhang, X. GrapeNet: A Lightweight Convolutional Neural Network Model for Identification of Grape Leaf Diseases. Agriculture 2022, 12, 887. [Google Scholar] [CrossRef]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics through machine learning and crowdsourcing. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.D.; Rice, M.E.; Hill, J.H. Digital analysis of leaf surface area: Effects of shape, resolution, and size. J. Kans. Entomol. Soc. 2007, 80, 339–347. [Google Scholar] [CrossRef][Green Version]
- Soares, F.A.; Flôres, E.L.; Cabacinha, C.D.; Carrijo, G.A.; Veiga, A.C. Recursive diameter prediction and volume calculation of eucalyptus trees using Multilayer Perceptron Networks. Comput. Electron. Agric. 2011, 78, 19–27. [Google Scholar] [CrossRef]
- Silva, A.d.L.; Alves, M.V.d.S.; Coan, A.I. Importance of anatomical leaf features for characterization of three species of Mapania (Mapanioideae, Cyperaceae) from the Amazon Forest, Brazil. Acta Amaz. 2014, 44, 447–456. [Google Scholar] [CrossRef]
- Novotnỳ, P.; Suk, T. Leaf recognition of woody species in Central Europe. Biosyst. Eng. 2013, 115, 444–452. [Google Scholar] [CrossRef]
- Wu, S.G.; Bao, F.S.; Xu, E.Y.; Wang, Y.X.; Chang, Y.F.; Xiang, Q.L. A leaf recognition algorithm for plant classification using probabilistic neural network. In Proceedings of the 2007 IEEE International Symposium on Signal Processing and Information Technology, Giza, Egypt, 15–18 December 2007; pp. 11–16. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).