Genome-Wide Association Mapping for Stomata and Yield Indices in Bread Wheat under Water Limited Conditions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotyping
2.3. Statistical Analysis
2.4. Genotyping of Bread Wheat Genotypes
2.5. Population Structure
2.6. GWAS Analysis
3. Results
3.1. Phenotypic Evaluation
3.2. Population Structure
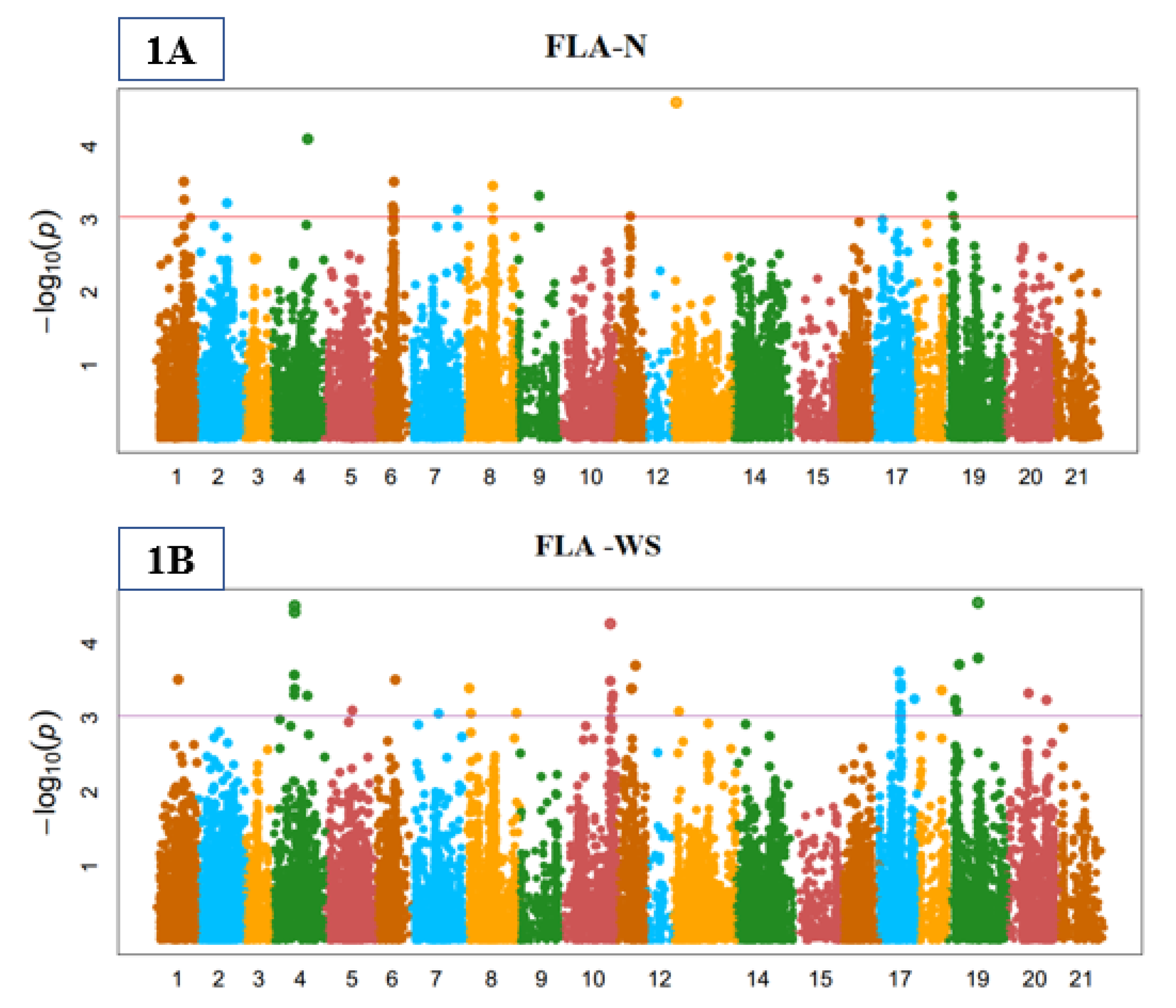
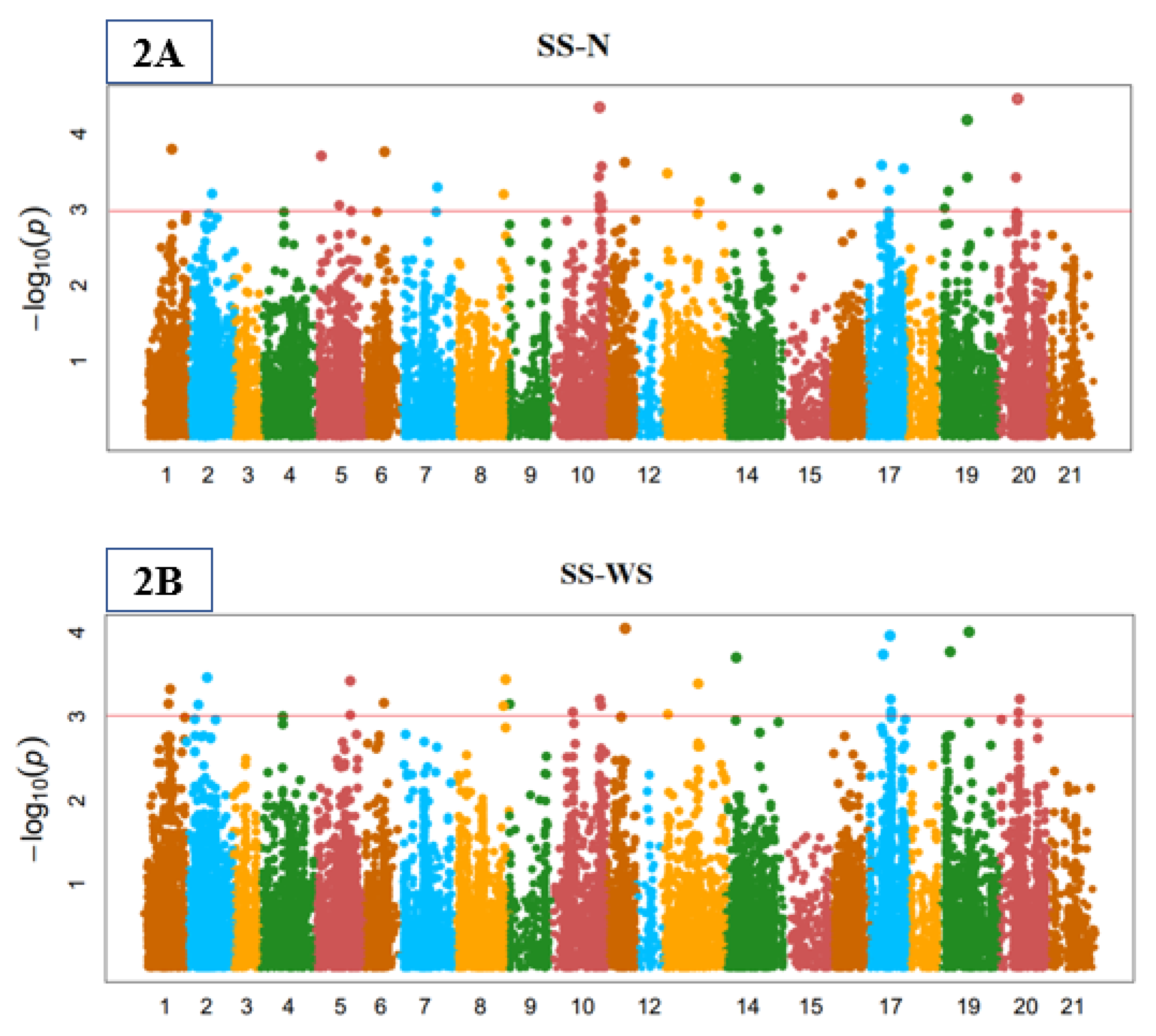
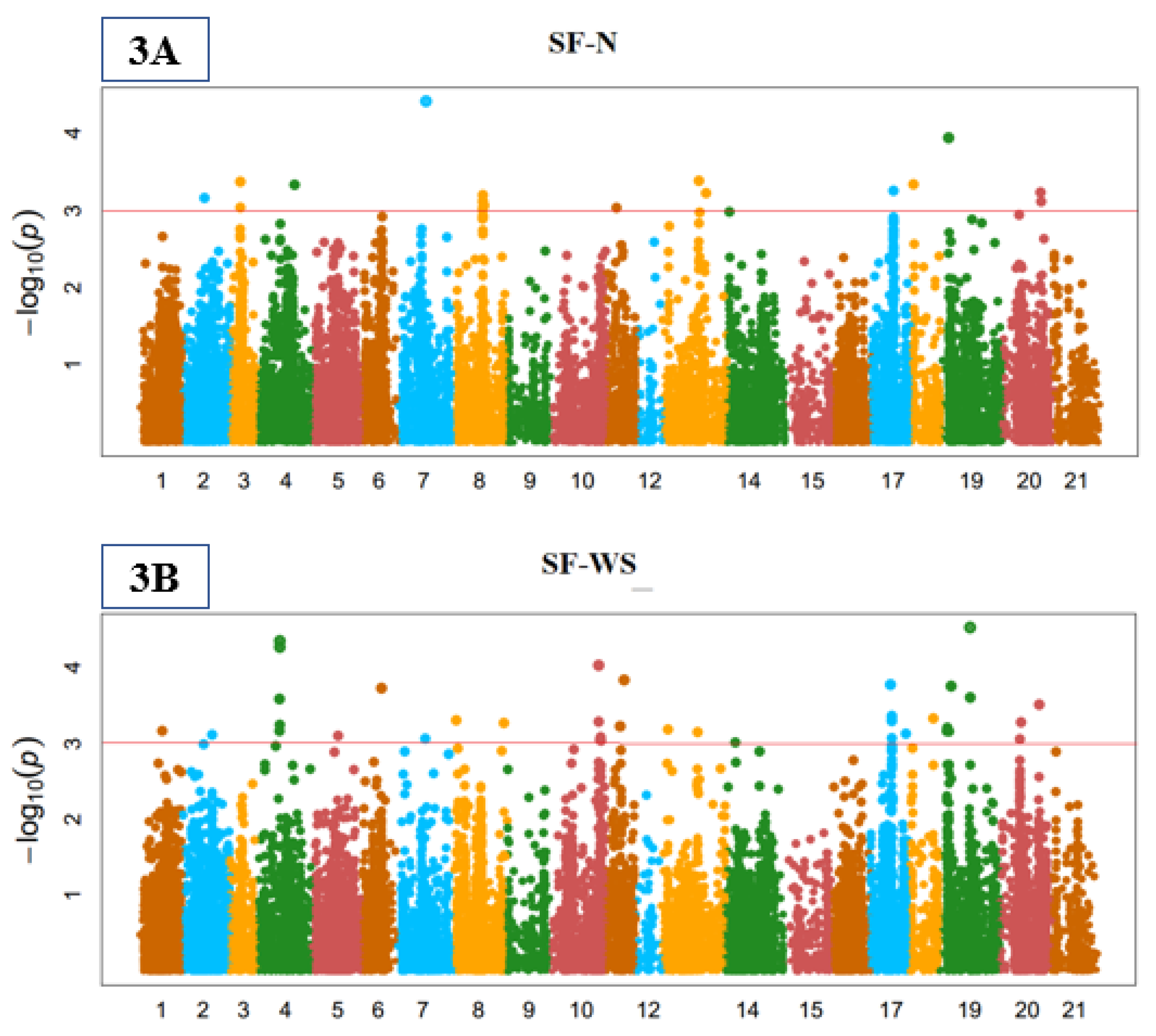
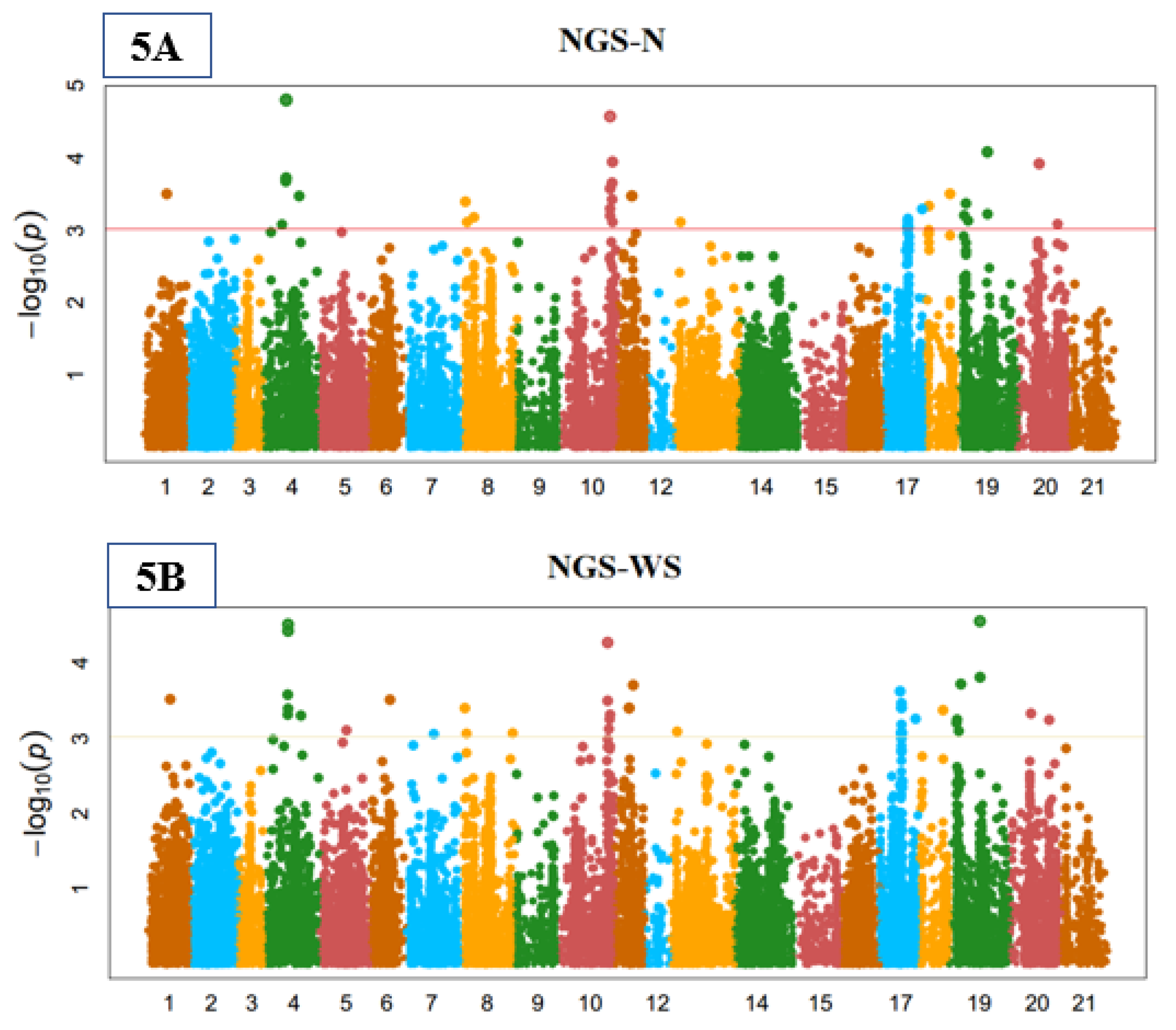
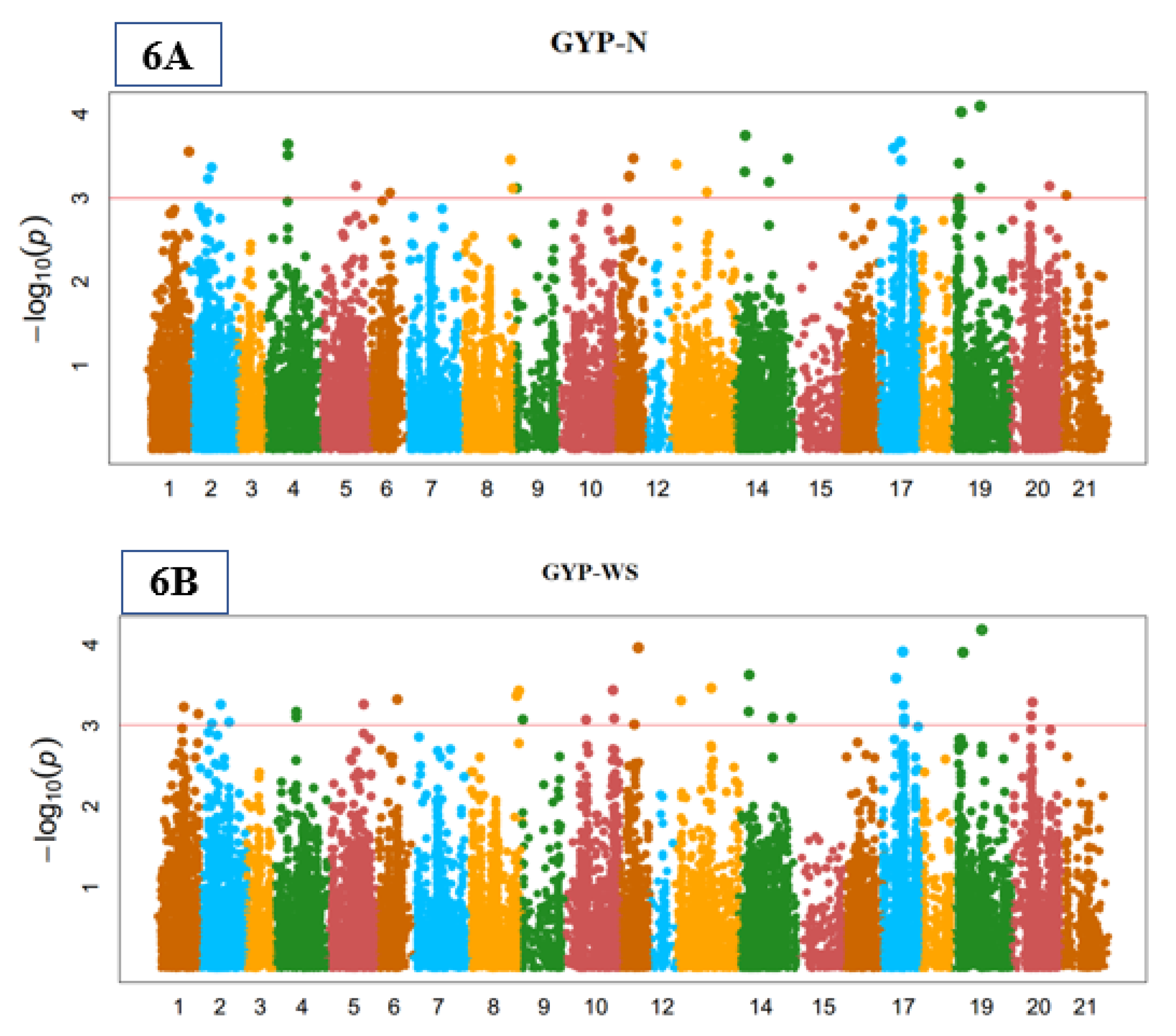
3.3. Genome-Wide Marker-Trait Associations (MTA)
3.4. Stomata Indices
3.5. Yield-Related Indices
3.6. Genome-Wide Multiple Traits Loci Associations
4. Discussions
4.1. Phenotypic Evaluation
4.2. Population Structure
4.3. Genome-Wide Marker-Traits Associations
4.4. Genome-Wide Multiple Traits Loci Associations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Ayalew, H.; Liu, H.; Börner, A.; Kobiljski, B.; Liu, C.; Yan, G. Genome-Wide Association Mapping of Major Root Length QTLs Under PEG Induced Water Stress in Wheat. Front. Plant Sci. 2018, 9, 1759. [Google Scholar] [CrossRef]
- Zhao, T.; Dai, A. The Magnitude and Causes of Global Drought Changes in the Twenty-First Century under a Low–Moderate Emissions Scenario. J. Clim. 2015, 28, 4490–4512. [Google Scholar] [CrossRef]
- Ahmed, H.G.M.-D.; Sajjad, M.; Zeng, Y.; Iqbal, M.; Khan, S.H.; Ullah, A.; Akhtar, M.N. Genome-Wide Association Mapping through 90K SNP Array for Quality and Yield Attributes in Bread Wheat against Water-Deficit Conditions. Agriculture 2020, 10, 392. [Google Scholar] [CrossRef]
- Dixon, J.; Braun, H.-J.; Kosina, P.; Crouch, J.H. Wheat Facts and Futures 2009; CIMMYT: Mexico-Veracruz, Mexico, 2009. [Google Scholar]
- Ahmed, H.G.M.-D.; Khan, A.S.; Kashif, M.; Khan, S.H. Genetic analysis of yield and physical traits of spring wheat grain. J. Natl. Sci. Found. Sri Lanka 2018, 46, 23. [Google Scholar] [CrossRef]
- Ahmed, H.G.M.-D.; Zeng, Y.; Yang, X.; Anwaar, H.A.; Mansha, M.Z.; Hanif, C.M.S.; Ikram, K.; Ullah, A.; Alghanem, S.M.S. Conferring Drought-Tolerant Wheat Genotypes through Morpho-Physiological and Chlorophyll Indices at Seedling Stage. Saudi J. Biol. Sci. 2020, 27, 2116–2123. [Google Scholar] [CrossRef]
- Toker, C.; Canci, H.; Yildirim, T. Evaluation of perennial wild Cicer species for drought resistance. Genet. Resour. Crop. Evol. 2007, 54, 1781–1786. [Google Scholar] [CrossRef]
- Ahmed, H.; Khan, A.S.; Muhammad, K.; Khan, S.H. Genetic Mechanism of Leaf Venation and Stomatal Traits for Breeding Drought Tolerant Lines in Wheat. Bangladesh J. Bot. 2017, 46, 35–41. [Google Scholar]
- Mir, R.R.; Zaman-Allah, M.; Sreenivasulu, N.; Trethowan, R.; Varshney, R.K. Integrated genomics, physiology and breeding approaches for improving drought tolerance in crops. Theor. Appl. Genet. 2012, 125, 625–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, I.; Khaliq, I.; Khan, A.S.; Farooq, M. Screening of Spring Wheat (Triticum Aestivum L.) Genotypes for Drought Tolerance on the Basis of Seedling Traits. Pak. J. Agric. Sci. 2014, 51, 367–372. [Google Scholar]
- Ashfaq, W.; Ul-Allah, S.; Kashif, M.; Sattar, A.; Nabi, H.G. Genetic Variability Study among Wheat Genotypes under Normal and Drought Conditions. J. Glob. Innov. Agric. Soc. Sci. 2016, 4, 111–116. [Google Scholar] [CrossRef]
- Noorka, I.R.; Batool, A.; Rauf, S.; Teixeira da Silva, J.; Ashraf, E. Estimation of Heterosis in Wheat (Triticum Aestivum L.) under Contrasting Water Regimes. Int. J. Plant Breed. 2013, 7, 55–60. [Google Scholar]
- Ahmed, H.; Sajjad, M.; Li, M.; Azmat, M.; Rizwan, M.; Maqsood, R.; Khan, S. Selection Criteria for Drought-Tolerant Bread Wheat Genotypes at Seedling Stage. Sustainability 2019, 11, 2584. [Google Scholar] [CrossRef] [Green Version]
- Adopted, I. Climate Change 2014 Synthesis Report; IPCC: Geneva, Switzerland, 2014. [Google Scholar]
- Merchuk-Ovnat, L.; Fahima, T.; Ephrath, J.E.; Krugman, T.; Saranga, Y. Ancestral QTL Alleles from Wild Emmer Wheat Enhance Root Development under Drought in Modern Wheat. Front. Plant Sci. 2017, 8, 703. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Khaliq, I.; Saddam, M.S.; Zafar, S.; Aslam, W.; Muhammad, D. Estimation of Genetic Mechanism of Stomatal Traits Influencing Grain Yield in Hexaploid Wheat (Triticum Aestivum L.). Int. J. Biosci. 2015, 6, 72–77. [Google Scholar]
- Shahinnia, F.; Le Roy, J.; Laborde, B.; Sznajder, B.; Kalambettu, P.; Mahjourimajd, S.; Tilbrook, J.; Fleury, D. Genetic association of stomatal traits and yield in wheat grown in low rainfall environments. BMC Plant Biol. 2016, 16, 1–14. [Google Scholar] [CrossRef]
- Ain, Q.-U.; Rasheed, A.; Anwar, A.; Mahmood, T.; Imtiaz, M.; Xia, X.; He, Z.; Quraishi, U.M. Genome-wide association for grain yield under rainfed conditions in historical wheat cultivars from Pakistan. Front. Plant Sci. 2015, 6, 743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, A.; Khaliq, I.; Ahmad, M.; Ahmed, H.; Khan, A.G.; Farooq, M.S. Comparative Performance of Spring Wheat (Triticum Aestivum L.) through Heat Stress Indices. Pak. J. Bot. 2018, 50, 481–488. [Google Scholar]
- Ahmed, H.; Khan, A.S.; Khan, S.H.; Kashif, M. Genome Wide Allelic Pattern and Genetic Diversity of Spring Wheat Genotypes through Ssr Markers. Int. J. Agric. Biol. 2017, 19, 1559–1565. [Google Scholar]
- Rahimi, Y.; Bihamta, M.R.; Taleei, A.; Alipour, H.; Ingvarsson, P. Genome-wide association study of agronomic traits in bread wheat reveals novel putative alleles for future breeding programs. BMC Plant Biol. 2019, 19, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.C.; Crossa, J.; Velu, G.; Huerta-Espino, J.; Vargas, M.; Payne, T.S.; Singh, R.P. Genetic Gains for Grain Yield in CIMMYT Spring Bread Wheat across International Environments. Crop. Sci. 2012, 52, 1522–1533. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.; Lopes, M.; Dreisigacker, S.; Reynolds, M.P. Correction to: Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor. Appl. Genet. 2018, 131, 999. [Google Scholar] [CrossRef] [Green Version]
- Aisawi, K.A.B.; Reynolds, M.P.; Singh, R.P.; Foulkes, J. The Physiological Basis of the Genetic Progress in Yield Potential of CIMMYT Spring Wheat Cultivars from 1966 to 2009. Crop. Sci. 2015, 55, 1749–1764. [Google Scholar] [CrossRef]
- Sukumaran, S.; Dreisigacker, S.; Lopes, M.; Chavez, P.; Reynolds, M.P. Genome-wide association study for grain yield and related traits in an elite spring wheat population grown in temperate irrigated environments. Theor. Appl. Genet. 2015, 128, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, W.; Ogbonnaya, F.C.; Jighly, A.; Sanchez-Garcia, M.; Sohail, Q.; Rajaram, S.; Baum, M. Genome-Wide Association Mapping of Yield and Grain Quality Traits in Winter Wheat Genotypes. PLoS ONE 2015, 10, e0141339. [Google Scholar] [CrossRef] [Green Version]
- Sorkheh, K.; Malysheva-Otto, L.V.; Wirthensohn, M.G.; Tarkesh-Esfahani, S.; Martínez-Gómez, P. Linkage Disequilibrium, Genetic Association Mapping and Gene Localization in Crop Plants. Genetics Mol. Biol. 2008, 31, 805–814. [Google Scholar] [CrossRef]
- Noorka, I.R.; Teixeira da Silva, J.A. Physical and Morphological Markers for Adaptation of Drought-Tolerant Wheat to Arid Environments. Pak. J. Agric. Sci. 2014, 51, 943–952. [Google Scholar]
- Muller, J. Determining Leaf Surface Area by Means of a Wheat Osmoregulation Water Use: The Challenge. Agric. Meteorol. 1991, 14, 311–320. [Google Scholar]
- Sheehan, D.; Hrapchak, B. Theory and Practice of Histotechnology, 2nd ed.; Mosby, C.V., Ed.; The CV Mosby Company: St. Louis, MO, USA, 1980. [Google Scholar]
- Payne, R. A Guide to Anova and Design in Genstat; VSN International: Hempstead, UK, 2008. [Google Scholar]
- Spss, I. Ibm Spss Statistics Version 21; International Business Machines Corp: Boston, MA, USA, 2012; p. 126. [Google Scholar]
- Qaseem, M.F.; Qureshi, R.; Muqaddasi, Q.H.; Shaheen, H.; Kousar, R.; Röder, M. Genome-wide association mapping in bread wheat subjected to independent and combined high temperature and drought stress. PLoS ONE 2018, 13, e0199121. [Google Scholar] [CrossRef] [Green Version]
- Dreisigacker, S.; Tiwari, R.; Sheoran, S. Laboratory Manual: Icar-Cimmyt Molecular Breeding Course in Wheat; CIMMYT: Mexico-Veracruz, Mexico, 2013. [Google Scholar]
- Wang, S.; Wong, D.; Forrest, K.; Allen, A.; Chao, S.; Huang, B.E.; Maccaferri, M.; Salvi, S.; Milner, S.G.; Cattivelli, L.; et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 2014, 12, 787–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, D.A.; Vonholdt, B.M. Structure Harvester: A Website and Program for Visualizing Structure Output and Implementing the Evanno Method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.; Buckler, E.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Zhang, Z.; Zhu, C.; Tabanao, D.A.; Pressoir, G.; Tuinstra, M.R.; Kresovich, S.; Todhunter, R.J.; Buckler, E. Simulation Appraisal of the Adequacy of Number of Background Markers for Relationship Estimation in Association Mapping. Plant Genome 2009, 2, 63–77. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Pressoir, G.; Briggs, W.H.; Bi, I.V.; Yamasaki, M.; Doebley, J.F.; McMullen, M.D.; Gaut, B.S.; Nielsen, D.M.; Holland, J.B.; et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat. Genet. 2006, 38, 203–208. [Google Scholar] [CrossRef]
- Dodig, D.; Zoric, M.; Kobiljski, B.; Savić, J.; Kandić, V.; Quarrie, S.; Barnes, J. Genetic and Association Mapping Study of Wheat Agronomic Traits Under Contrasting Water Regimes. Int. J. Mol. Sci. 2012, 13, 6167–6188. [Google Scholar] [CrossRef] [Green Version]
- Zampieri, M.; Ceglar, A.; Dentener, F.; Toreti, A. Wheat yield loss attributable to heat waves, drought and water excess at the global, national and subnational scales. Environ. Res. Lett. 2017, 12, 064008. [Google Scholar] [CrossRef]
- ElBasyoni, I.; Saadalla, M.; Baenziger, S.; Bockelman, H.; Morsy, S. Cell Membrane Stability and Association Mapping for Drought and Heat Tolerance in a Worldwide Wheat Collection. Sustainability 2017, 9, 1606. [Google Scholar] [CrossRef] [Green Version]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-Wide Association Mapping of Yield and Yield Components of Spring Wheat under Contrasting Moisture Regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef]
- Daba, S.D.; Tyagi, P.; Brown-Guedira, G.; Mohammadi, M.M. Genome-Wide Association Studies to Identify Loci and Candidate Genes Controlling Kernel Weight and Length in a Historical Us Wheat Population. Front. Plant Sci. 2018, 9, 1045. [Google Scholar]
- Su, J.; Zhang, F.; Chong, X.; Song, A.; Guan, Z.; Fang, W.; Chen, F. Genome-wide association study identifies favorable SNP alleles and candidate genes for waterlogging tolerance in chrysanthemums. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, X.; Wang, J.; Li, M.; Wang, Q.; Tian, F.; Su, Z.; Pan, Y.; Liu, D.; Lipka, A.E.; et al. GAPIT Version 2: An Enhanced Integrated Tool for Genomic Association and Prediction. Plant Genome 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Huang, M.; Fan, B.; Buckler, E.; Zhang, Z. Iterative Usage of Fixed and Random Effect Models for Powerful and Efficient Genome-Wide Association Studies. PLoS Genet. 2016, 12, e1005767. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-G.; Jia, S.-S.; Sun, D.-Z.; Fan, H.; Chang, X.-P.; Jing, R.-L. Mapping QTLs for stomatal density and size under drought stress in wheat (Triticum aestivum L.). J. Integr. Agric. 2016, 15, 1955–1967. [Google Scholar] [CrossRef] [Green Version]
- ShuguangWang, F.D.; Sun, Y.C.D.; Yan, R.J.X. QTL analysis for stomatal density and size in wheat spike organ. Emir. J. Food Agric. 2018, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Thomelin, P.; Bonneau, J.; Taylor, J.; Choulet, F.; Sourdille, P.; Langridge, P. Positional Cloning of a Qtl, Qdhy. 3bl, on Chromosome 3bl for Drought and Heat Tolerance in Bread Wheat. In Proceedings of the Plant and Animal Genome Conference (PAG XXIV), San Diego, CA, USA, 9 January 2016. [Google Scholar]
- Mwadzingeni, L.; Shimelis, H.; Rees, J.; Tsilo, T.J. Genome-wide association analysis of agronomic traits in wheat under drought-stressed and non-stressed conditions. PLoS ONE 2017, 12, e0171692. [Google Scholar] [CrossRef] [Green Version]
- Gizaw, S.A. Genome-Wide Association Studies of Drought Resistance and Yield Potential in Wheat (Triticum Aestivum L.) Using Agronomic and Remotely Sensed Traits; ProQuest LLC: Ann Arbor, MI, USA, 2015. [Google Scholar]
- Lozada, D.N.; Mason, R.E.; Babar, A.; Carver, B.F.; Guedira, G.-B.; Merrill, K.; Arguello, M.N.; Acuna, A.; Vieira, L.; Holder, A.; et al. Association mapping reveals loci associated with multiple traits that affect grain yield and adaptation in soft winter wheat. Euphytica 2017, 213, 222. [Google Scholar] [CrossRef]
- Ogbonnaya, F.C.; Rasheed, A.; Okechukwu, E.C.; Jighly, A.; Makdis, F.; Wuletaw, T.; Hagras, A.; Uguru, M.I.; Agbo, C.U. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor. Appl. Genet. 2017, 130, 1819–1835. [Google Scholar] [CrossRef]
- Abbas, M.; Sheikh, A.; Sabir, H.M.; Nighat, S. Factors Responsible for Low Wheat Productivity in Central Punjab. Pak. J. Agric. Sci. 2005, 42, 3–4. [Google Scholar]
- Bordes, J.; Goudemand, E.; Duchalais, L.; Chevarin, L.; Oury, F.X.; Heumez, E.; Lapierre, A.; Perretant, M.R.; Rolland, B.; Béghin, D.; et al. Genome-wide association mapping of three important traits using bread wheat elite breeding populations. Mol. Breed. 2013, 33, 755–768. [Google Scholar] [CrossRef]
- Zhang, W.; Dubcovsky, J. Association between allelic variation at the Phytoene synthase 1 gene and yellow pigment content in the wheat grain. Theor. Appl. Genet. 2008, 116, 635–645. [Google Scholar] [CrossRef] [Green Version]

| Source | DF/ Season | FLA | SS | SF | LV | NGS | GYP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | S2 | S1 | S2 | S1 | S2 | S1 | S2 | S1 | S2 | S1 | S2 | ||
| REP | 2 | 958 | 805 ** | 6.53 | 7.41 | 4.04 | 2.77 | 5.65 | 6.48 | 43.36 | 42.18 | 40.48 | 62.85 |
| GET | 95 | 46.8 ** | 56.0 ** | 36.2 ** | 34.6 ** | 36.66 ** | 36.05 ** | 36.30 ** | 35.46 ** | 297.18 ** | 280.91 ** | 147.58 ** | 143.89 ** |
| LEL | 1 | 9331 ** | 9121 ** | 4090.8 * | 4196.9 | 3457.44 * | 3559.24 * | 581.77 * | 490.11 ** | 570.25 * | 415.06 | 441 ** | 424.61 * |
| GET × LEL | 95 | 6.44 ** | 8.22 ** | 18.43 ** | 18.09 ** | 16.38 ** | 20.29 ** | 17.95 ** | 19.28 ** | 137.13 ** | 149 ** | 100.08 ** | 106.65 ** |
| Error | 382 | 2.91 | 5.01 | 4.99 | 5.19 | 5.4 | 4.32 | 5.06 | 5.2 | 48.27 | 49.66 | 28.63 | 27.84 |
| Total | 575 | ||||||||||||
| Heritability | 73.52 | 81.19 | 69.21 | 65.38 | 54.67 | 61.00 | 68.29 | 65.98 | 64.33 | 60.82 | 57.70 | 56.26 | |
| Table | FLA | SS | SF | LV | NGS | GYP | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | WS | N | WS | N | WS | N | WS | N | WS | N | WS | |
| Mean | 33.2 | 18.2 | 3699 | 3480 | 13.25 | 12.7 | 7.81 | 7.54 | 51.5 | 33 | 22.50 | 15.4 |
| Variance | 4.13 | 5.91 | 10.69 | 13.16 | 10.69 | 10.71 | 16.76 | 10.96 | 89.97 | 99.9 | 49.39 | 52.31 |
| SE | 0.21 | 0.25 | 0.33 | 0.37 | 0.33 | 0.36 | 0.41 | 0.32 | 0.96 | 0.99 | 0.71 | 0.74 |
| Traits | Env. | FLA | SS | SF | LV | NGS |
|---|---|---|---|---|---|---|
| SS | N | −0.22 | ||||
| WS | −0.20 | |||||
| SF | N | 0.36 * | 0.39 * | |||
| WS | 0.38 * | 0.41 * | ||||
| LV | N | 0.45 * | 0.34 | 0.91 ** | ||
| WS | 0.37 * | 0.33 | 0.84 ** | |||
| NGS | N | 0.76 ** | −0.22 | 0.56 ** | 0.56 ** | |
| WS | 0.77 ** | 0.18 | 0.91 ** | 0.89 ** | ||
| GYP | N | 0.88 ** | −0.09 | 0.88 ** | 0.92 ** | 0.56 ** |
| WS | 0.81 ** | −0.12 | 0.84 ** | 0.83 ** | 0.89 ** |
| Trait | Normal Conditions | Water Shortage |
|---|---|---|
| FLA | G-9244 followed by G-9610, G-9883, C-586642 and G-9493 | G-586642 followed by G-9610, G-9493, G-9883and G-9244 |
| SS | G-9610 followed by G-9244, G-9883, C-586642 and G-9493 | G-9610 followed by C-586642, G-9883, G-9244 and G-9493 |
| SF | G-9610 followed by G-9244, G-9883 C-586642 and G-9493 | G-9610 followed by G-9244, G-9883, C-586642 and G-9493 |
| LV | G-9610 followed by C-586642, G-9610, G-9244 and G-9493 | G-9610 followed by G-9244, G-9883, C-586642 and G-9493 |
| NGS | G-9244 followed by G-9610, G-9493, C-586642 and G-9883 | G-9610 followed by G-9244, G-9883, C-586642 and G-9493 |
| GYP | G-9493 followed by G-9796, G-9610, G-9883 and G-9244 | G-9493 followed by G-9796, G-9610, G-9244 and G-9883 |
| Significant MTAs | |||||
|---|---|---|---|---|---|
| Genome and Chromosome Wise Distribution | Characteristics Wise Distribution | ||||
| Genome | Normal | Water Shortage | Traits | Normal | Water Shortage |
| A-Genome | Total 75 MTAs (1A = 8, 2A = 14, 3A = 3, 4A = 16, 5A = 9, 6A = 2, 7A = 23) | Total 92 MTAs (1A = 9, 2A = 22, 3A = 3, 4A = 24, 5A = 10, 7A = 24) | FLA | 19 | 50 |
| SS | 30 | 29 | |||
| B-Genome | Total 86 MTAs (1B = 8, 2B = 4, 3B = 16, 4B = 16, 5B = 11, 6B = 24, 7B = 7) | Total 140 MTAs (1B = 9, 2B = 7, 3B = 14, 4B = 28, 5B = 12, 6B = 57, 7B = 13) | SF | 19 | 41 |
| LV | 32 | 37 | |||
| D-genome | Total 17 MTAs (1D = 2, 2D = 9, 3D = 3, 6D = 2, 7D = 1) | Total 12 MTAs (2D = 6, 3D = 3, 6D = 3) | NGS | 47 | 50 |
| GYP | 31 | 37 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, H.G.M.-D.; Iqbal, M.N.; Iqbal, M.A.; Zeng, Y.; Ullah, A.; Iqbal, M.; Raza, H.; Yar, M.M.; Sarwar, N.; Imran, M.; et al. Genome-Wide Association Mapping for Stomata and Yield Indices in Bread Wheat under Water Limited Conditions. Agronomy 2021, 11, 1646. https://doi.org/10.3390/agronomy11081646
Ahmed HGM-D, Iqbal MN, Iqbal MA, Zeng Y, Ullah A, Iqbal M, Raza H, Yar MM, Sarwar N, Imran M, et al. Genome-Wide Association Mapping for Stomata and Yield Indices in Bread Wheat under Water Limited Conditions. Agronomy. 2021; 11(8):1646. https://doi.org/10.3390/agronomy11081646
Chicago/Turabian StyleAhmed, Hafiz Ghulam Muhu-Din, Muhammad Nouman Iqbal, Muhammad Arslan Iqbal, Yawen Zeng, Aziz Ullah, Muhammad Iqbal, Humayun Raza, Muhammad Majid Yar, Nadeem Sarwar, Muhammad Imran, and et al. 2021. "Genome-Wide Association Mapping for Stomata and Yield Indices in Bread Wheat under Water Limited Conditions" Agronomy 11, no. 8: 1646. https://doi.org/10.3390/agronomy11081646
APA StyleAhmed, H. G. M.-D., Iqbal, M. N., Iqbal, M. A., Zeng, Y., Ullah, A., Iqbal, M., Raza, H., Yar, M. M., Sarwar, N., Imran, M., & Hussain, S. (2021). Genome-Wide Association Mapping for Stomata and Yield Indices in Bread Wheat under Water Limited Conditions. Agronomy, 11(8), 1646. https://doi.org/10.3390/agronomy11081646









