Contribution of Wild Relatives to Durum Wheat (Triticum turgidum subsp. durum) Yield Stability across Contrasted Environments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Testing Environments and Experimental Design
2.3. Data Analysis
2.3.1. Analysis of Variance and Genotype by Environment Interaction
2.3.2. Analysis of Stability
3. Results
3.1. Analysis of Variance
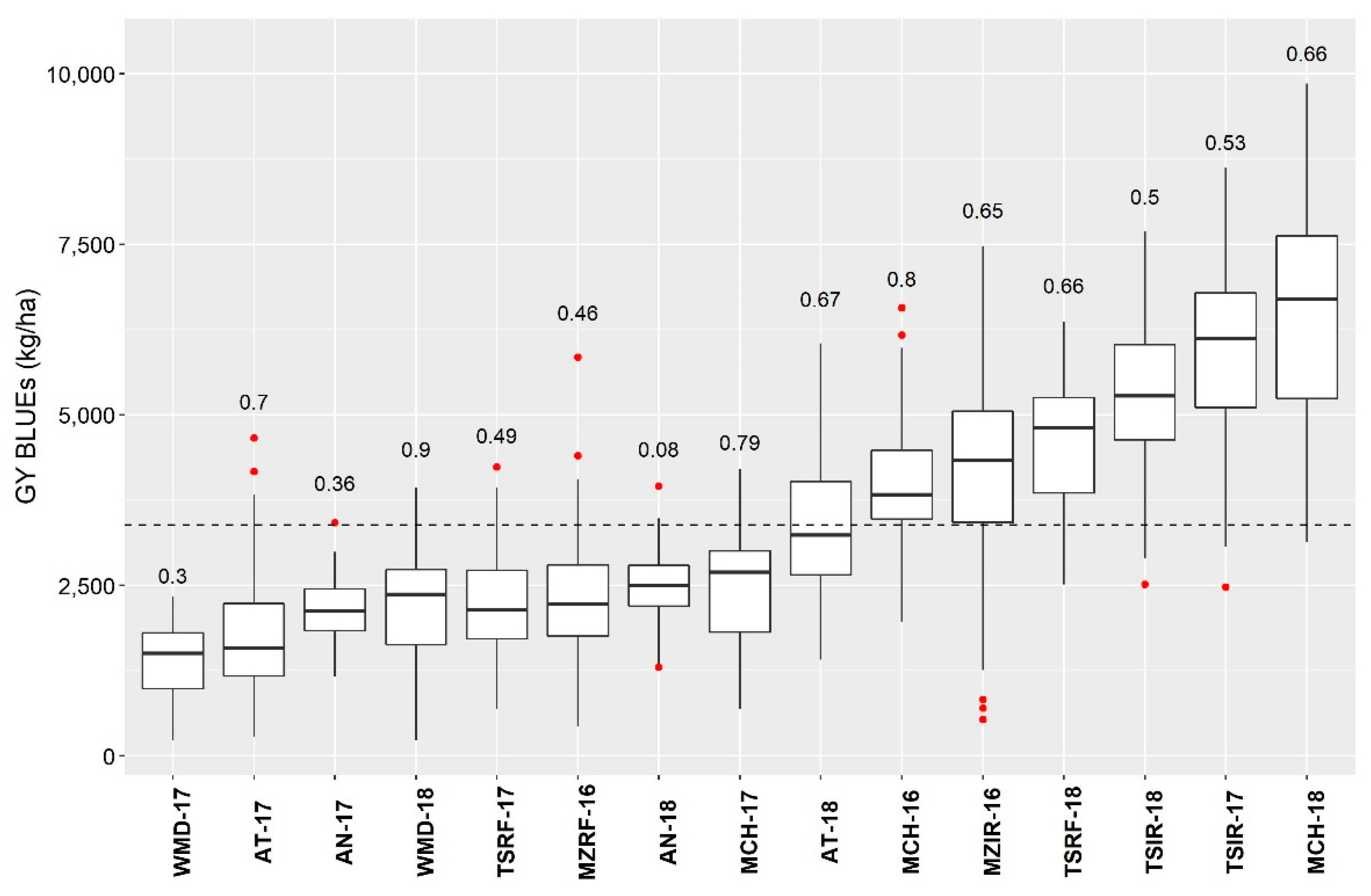
3.2. Characterization of the Testing Environments
3.2.1. Climatic Data
3.2.2. Environment Characterization for Yield
3.2.3. Association between the Environments
3.3. Yield Stability Assessment
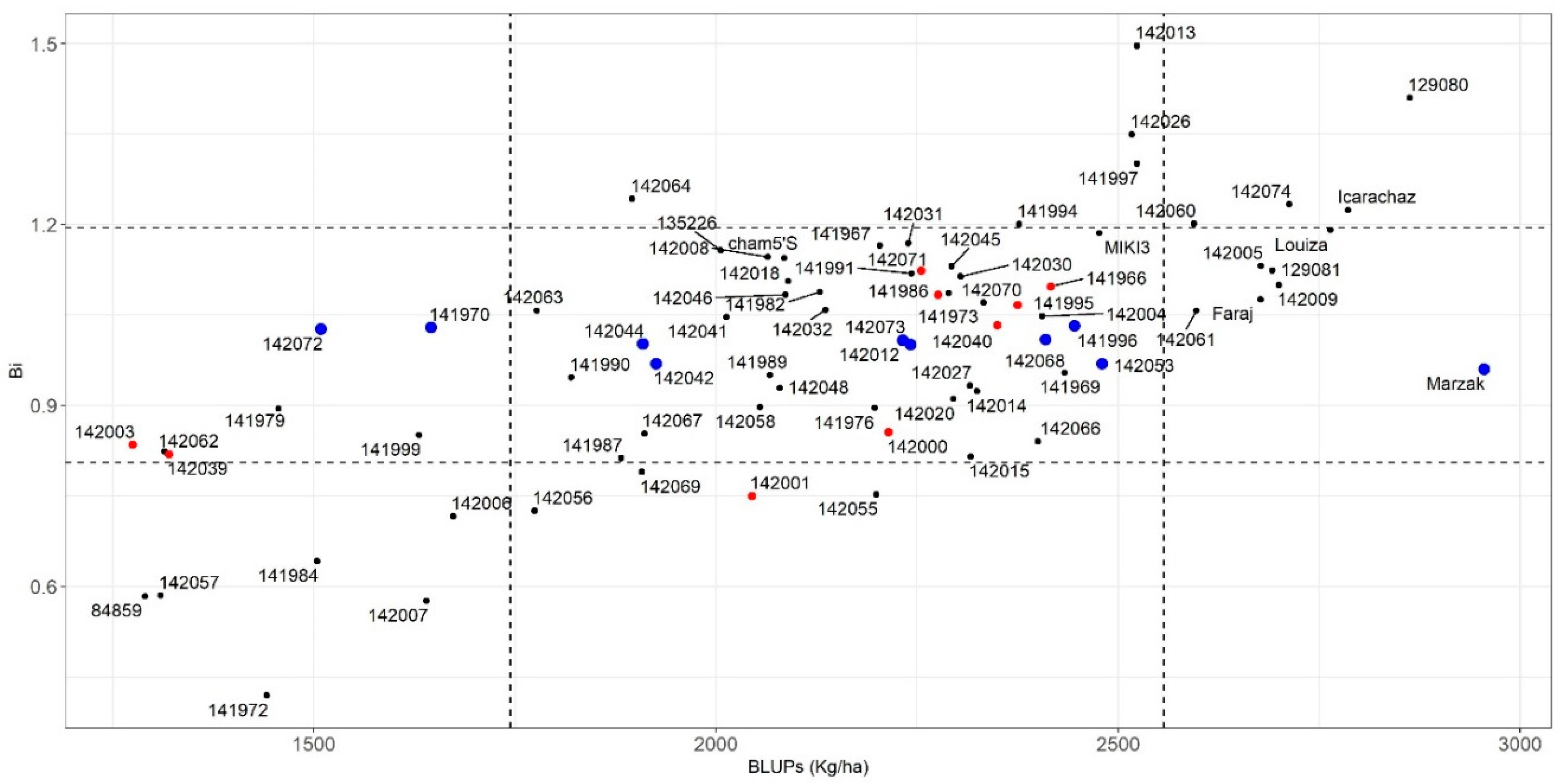
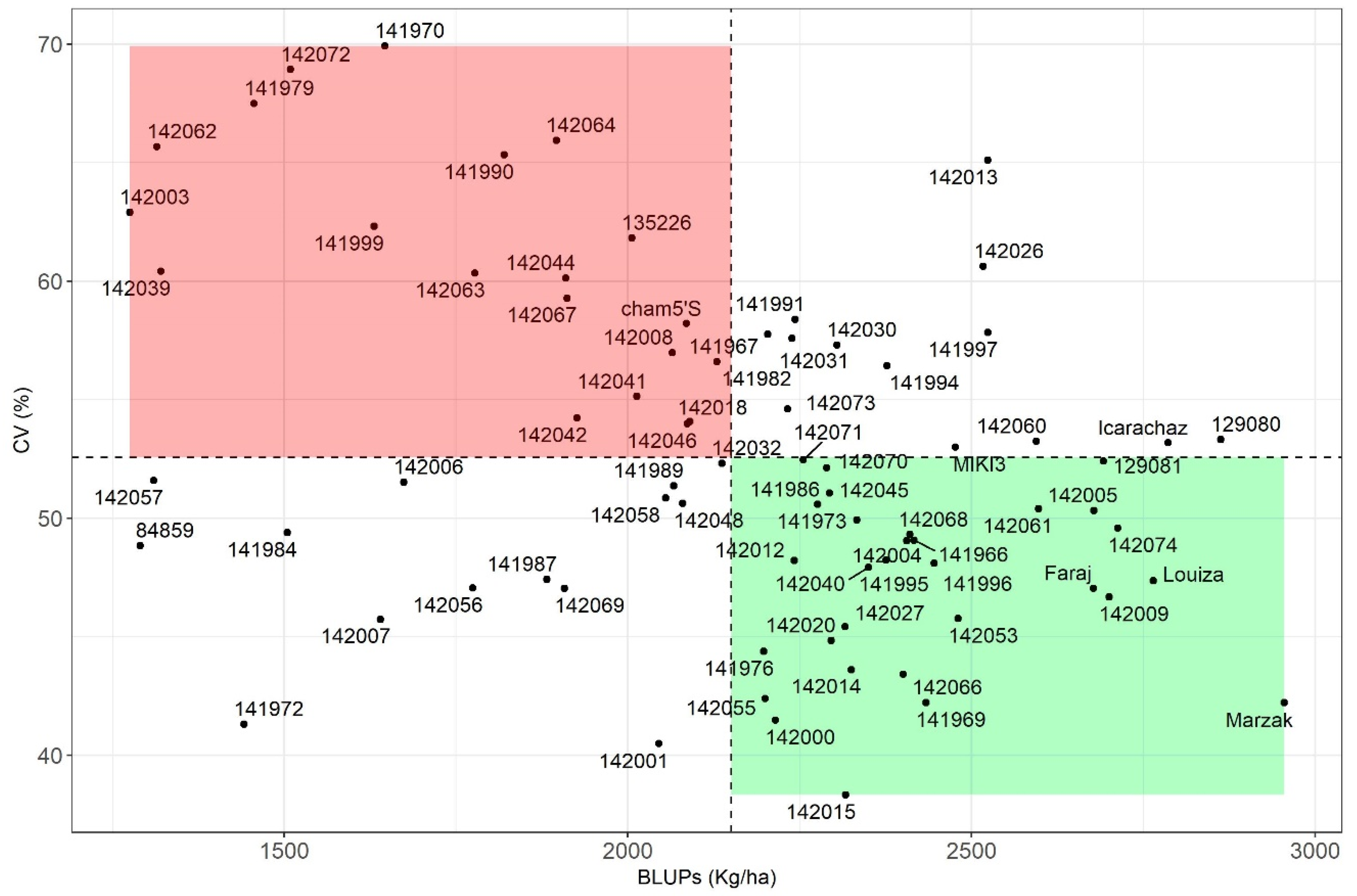
3.3.1. Parametric Stability Indices
3.3.2. Non-Parametric Stability Indices
3.3.3. AMMI Derived Stability
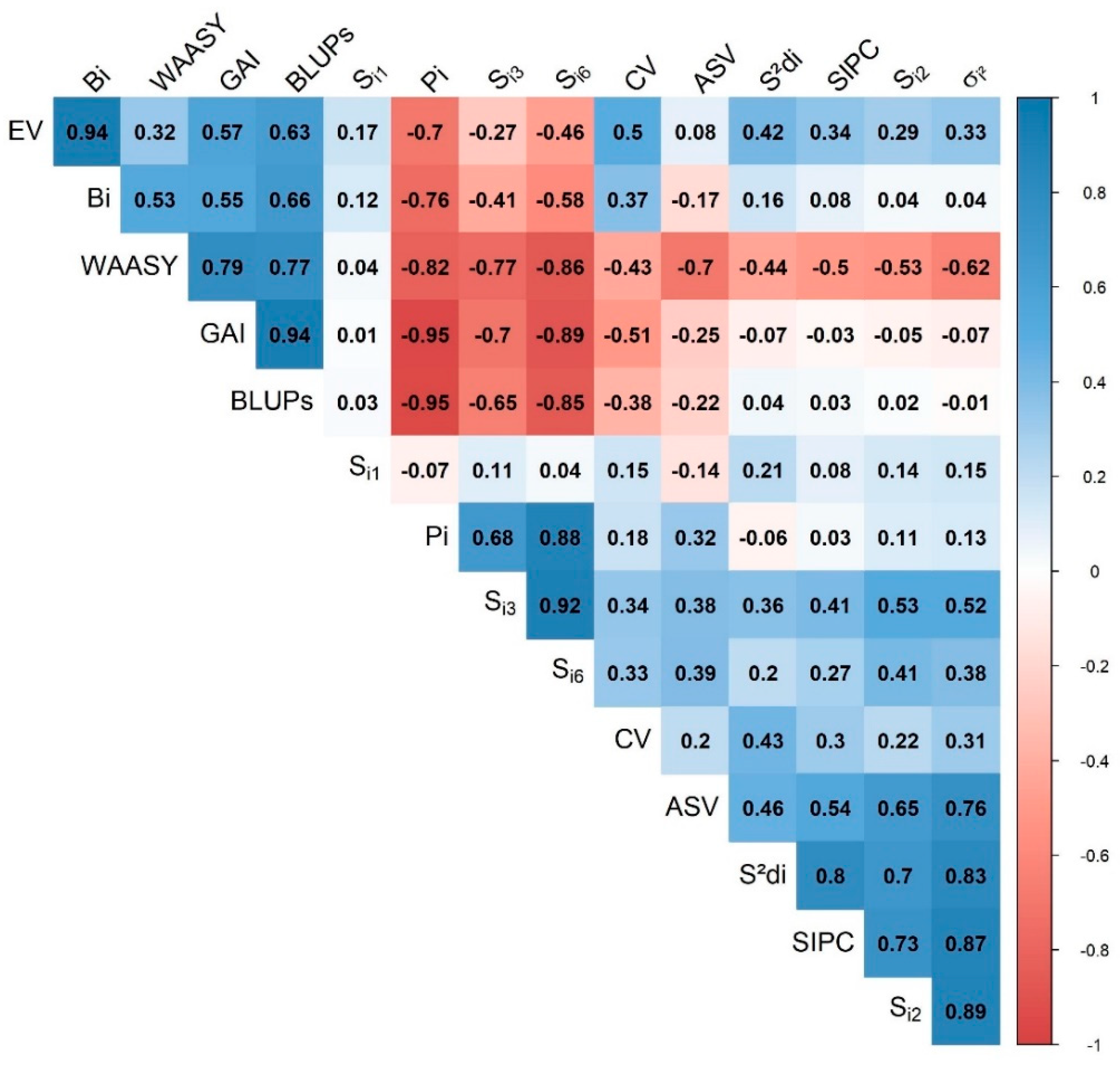
3.3.4. Association of the Stability Indices
4. Discussion
4.1. Dissection of the Genotype by Environment Interaction
4.2. Considerations for the Use of Durum Wheat Wild Relatives
4.3. Impact of Wild Relatives on Yield Potential
4.4. Yield Stability of the Durum Wheat Derivatives
4.5. Association between Different Stability Parameters
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Beres, B.L.; Rahmani, E.; Clarke, J.M.; Grassini, P.; Pozniak, C.J.; Geddes, C.M.; Porker, K.D.; May, W.E.; Ransom, J.K. A Systematic Review of Durum Wheat: Enhancing Production Systems by Exploring Genotype, Environment, and Management (G × E × M) Synergies. Front. Plant Sci. 2020, 11, 568657. [Google Scholar] [CrossRef]
- Sissons, M. Role of Durum Wheat Composition on the Quality of Pasta and Bread. Food 2008, 2, 75–90. [Google Scholar]
- Lidon, F.; Almeida, A.; Leitao, A.; Silva, M.; Pinheiro, N.; Macas, B.; Costa, R. A Synoptic Overview of Durum Wheat Production in the Mediterranean Region and Processing Following the European Union Requirements. Emir. J. Food Agric. 2014, 26, 693. [Google Scholar] [CrossRef] [Green Version]
- Able, J.; Sissons, M. Durum Wheat for the Future: Challenges, Research and Prospects in the 21st Century. Crop Pasture Sci. 2014, 65. [Google Scholar] [CrossRef]
- Lhaloui, S.; Buschman, L.; Bouhssini’, M.E.; Starks, K.; Kfith, D.; Houssaini, K.E. Control of Mayetiola Species (Diptera: Cecidomyiidae) with Carbofuran in Bread Wheat, Durum Wheat and Barley, with Yield Loss Assessment and Its Economic Analysis. Awamia 1992, 77, 55–73. [Google Scholar]
- Bassi, F.M.; Brahmi, H.; Sabraoui, A.; Amri, A.; Nsarellah, N.; Nachit, M.M.; Al-Abdallat, A.; Chen, M.S.; Lazraq, A.; El Bouhssini, M. Genetic Identification of Loci for Hessian Fly Resistance in Durum Wheat. Mol. Breed. 2019, 39, 24. [Google Scholar] [CrossRef] [Green Version]
- Rajaram, S.; Saari, E.E.; Hettel, G.P.; International Maize and Wheat Improvement Center (Eds.) Durum Wheats: Challenges and Opportunities. In Proceedings of the International Workshop, Ciudad Obregon, Mexico, 23–25 March 1992; International Maize and Wheat Improvement Center: Ciudad Obregon, Mexico, 1992. ISBN 978-968-6127-77-5. [Google Scholar]
- Marasas, C.N.; Smale, M.; Singh, R.P. The Economic Impact in Developing Countries of Leaf Rust Resistance Breeding in CIMMYT-Related Spring Bread Wheat; Economics Program Paper 04-01; CIMMYT: México, Mexico, 2003. [Google Scholar]
- Herrera-Foessel, S.A.; Singh, R.P.; Huerta-Espino, J.; Crossa, J.; Yuen, J.; Djurle, A. Effect of Leaf Rust on Grain Yield and Yield Traits of Durum Wheats with Race-Specific and Slow-Rusting Resistance to Leaf Rust. Plant Dis. 2006, 90, 1065–1072. [Google Scholar] [CrossRef]
- Ordoñez, M.E.; Kolmer, J.A. Virulence Phenotypes of a Worldwide Collection of Puccinia triticina from Durum Wheat. Phytopathology 2007, 97, 344–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergara-Diaz, O.; Kefauver, S.C.; Elazab, A.; Nieto-Taladriz, M.T.; Araus, J.L. Grain Yield Losses in Yellow-Rusted Durum Wheat Estimated Using Digital and Conventional Parameters under Field Conditions. Crop J. 2015, 3, 200–210. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Ye, W.; Wang, M.; Yan, X. Climate Change and Drought: A Risk Assessment of Crop-Yield Impacts. Clim. Res. 2009, 39, 31–46. [Google Scholar] [CrossRef]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate Trends and Global Crop Production Since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [Green Version]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.P.; Lobell, D.B.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising Temperatures Reduce Global Wheat Production. Nat. Clim Chang. 2015, 5, 143–147. [Google Scholar] [CrossRef]
- Turner, N.C.; Blum, A.; Cakir, M.; Steduto, P.; Tuberosa, R.; Young, N. Strategies to Increase the Yield and Yield Stability of Crops under Drought—Are We Making Progress? Funct. Plant Biol. 2014, 41, 1199. [Google Scholar] [CrossRef]
- Zhang, A.; Hochman, Z.; Horan, H.; Navarro, J.G.; Das, B.T.; Waldner, F. Socio-Psychological and Management Drivers Explain Farm Level Wheat Yield Gaps in Australia. Agron. Sustain. Dev. 2019, 39, 10. [Google Scholar] [CrossRef] [Green Version]
- Padovan, G. Understanding Effects of Genotype × Environment × Sowing Window Interactions for Durum Wheat in the Mediterranean Basin. Field Crop. Res. 2020, 259, 107969. [Google Scholar] [CrossRef]
- Xiong, W.; Reynolds, M.; Crossa, J.; Payne, T.; Schulthess, U.; Sonder, K.; Addimando, N.; Singh, R.; Ammar, K.; Gerard, B. Climate Change Has Increased Genotype-Environment Interactions in Wheat Breeding. Res. Sq. 2020. In Review. [Google Scholar]
- De Vita, P.; Mastrangelo, A.M.; Matteu, L.; Mazzucotelli, E.; Virzì, N.; Palumbo, M.; Storto, M.L.; Rizza, F.; Cattivelli, L. Genetic Improvement Effects on Yield Stability in Durum Wheat Genotypes Grown in Italy. Field Crop. Res. 2010, 119, 68–77. [Google Scholar] [CrossRef]
- Cattivelli, L.; Franco, M.; Alessandro, Z.; Fulvia, R.; Mastrangelo, A.M.; Pasquale, D.V.; Elisabetta, M. Adaptation of Durum Wheat to a Changing Environment. In Proceedings of the International Symposium on Genetics and Breeding of Durum Wheat; CIHEAM: Bari, Italy, 2014; pp. 279–282. [Google Scholar]
- Mohammadi, R.; Amri, A. Genotype x Environment Interaction Implication: A Case Study of Durum Wheat Breeding in Iran. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 515–558. ISBN 978-3-319-22517-3. [Google Scholar]
- Cattivelli, L.; Rizza, F.; Badeck, F.-W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought Tolerance Improvement in Crop Plants: An Integrated View from Breeding to Genomics. Field Crop. Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Mickelbart, M.V.; Hasegawa, P.M.; Bailey-Serres, J. Genetic Mechanisms of Abiotic Stress Tolerance That Translate to Crop Yield Stability. Nat. Rev. Genet. 2015, 16, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.A.; Nicotra, A.B.; Kruuk, L.E.B. Sparse Evidence for Selection on Phenotypic Plasticity in Response to Temperature. Phil. Trans. R. Soc. B 2019, 374, 20180185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajjar, R.; Hodgkin, T. The Use of Wild Relatives in Crop Improvement: A Survey of Developments over the Last 20 Years. Euphytica 2007, 156, 1–13. [Google Scholar] [CrossRef]
- Mondal, S.; Rutkoski, J.E.; Velu, G.; Singh, P.K.; Crespo-Herrera, L.A.; Guzmán, C.; Bhavani, S.; Lan, C.; He, X.; Singh, R.P. Harnessing Diversity in Wheat to Enhance Grain Yield, Climate Resilience, Disease and Insect Pest Resistance and Nutrition Through Conventional and Modern Breeding Approaches. Front. Plant Sci. 2016, 7, 991. [Google Scholar] [CrossRef] [Green Version]
- Anikster, Y.; Manisterski, J.; Long, D.L.; Leonard, K.J. Leaf Rust and Stem Rust Resistance in Triticum dicoccoides Populations in Israel. Plant Dis. 2005, 89, 55–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peleg, Z.; Saranga, Y.; Yazici, A.; Fahima, T.; Ozturk, L.; Cakmak, I. Grain Zinc, Iron and Protein Concentrations and Zinc-Efficiency in Wild Emmer Wheat under Contrasting Irrigation Regimes. Plant Soil 2008, 306, 57–67. [Google Scholar] [CrossRef] [Green Version]
- Suneja, Y.; Gupta, A.K.; Bains, N.S. Stress Adaptive Plasticity: Aegilops tauschii and Triticum dicoccoides as Potential Donors of Drought Associated Morpho-Physiological Traits in Wheat. Front. Plant Sci. 2019, 10, 211. [Google Scholar] [CrossRef] [Green Version]
- Marais, G.F.; Pretorius, Z.A.; Wellings, C.R.; McCallum, B.; Marais, A.S. Leaf Rust and Stripe Rust Resistance Genes Transferred to Common Wheat from Triticum dicoccoides. Euphytica 2005, 143, 115–123. [Google Scholar] [CrossRef]
- Vikal, Y.; Chhuneja, P.; Singh, R.; Dhaliwal, H.S. Tagging of an Aegilops speltoides Derived Leaf Rust Resistance Gene Lr 28 with a Microsatellite Marker in Wheat. J. Plant Biochem. Biotechnol. 2004, 13, 47–49. [Google Scholar] [CrossRef]
- Tanguy, A.-M.; Coriton, O.; Abélard, P.; Dedryver, F.; Jahier, J. Structure of Aegilops ventricosa Chromosome 6N, the Donor of Wheat Genes Yr17, Lr37, Sr38, and Cre5. Genome 2005, 48, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Marais, G.F.; McCallum, B.; Snyman, J.E.; Pretorius, Z.A.; Marais, A.S. Leaf Rust and Stripe Rust Resistance Genes Lr54 and Yr37 Transferred to Wheat from Aegilops kotschyi. Plant Breed. 2005, 124, 538–541. [Google Scholar] [CrossRef]
- Hovhannisyan, N.A.; Dulloo, M.E.; Yesayan, A.H.; Knüpffer, H.; Amri, A. Tracking of Powdery Mildew and Leaf Rust Resistance Genes in Triticum boeoticum and T. urartu, Wild Relatives of Common Wheat. Czech J. Genet. Plant Breed. 2011, 47, 45–57. [Google Scholar] [CrossRef] [Green Version]
- Rouse, M.N.; Olson, E.L.; Gill, B.S.; Pumphrey, M.O.; Jin, Y. Stem Rust Resistance in Aegilops tauschii Germplasm. Crop Sci. 2011, 51, 2074–2078. [Google Scholar] [CrossRef]
- Millet, E.; Manisterski, J.; Ben-Yehuda, P.; Distelfeld, A.; Deek, J.; Wan, A.; Chen, X.; Steffenson, B.J. Introgression of Leaf Rust and Stripe Rust Resistance from Sharon Goatgrass (Aegilops sharonensis Eig) into Bread Wheat (Triticum aestivum L.). Genome 2014, 57, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Cruz, C.D.; Peterson, G.L.; Bockus, W.W.; Kankanala, P.; Dubcovsky, J.; Jordan, K.W.; Akhunov, E.; Chumley, F.; Baldelomar, F.D.; Valent, B. The 2NS Translocation from Aegilops Ventricosa Confers Resistance to the Triticum Pathotype of Magnaporthe oryzae. Crop Sci. 2016, 56, 990–1000. [Google Scholar] [CrossRef] [Green Version]
- El Bouhssini, M.; Benlhabib, O.; Nachit, M.M.; Houari, A.; Bentika, A.; Nsarellah, N.; Lhaloui, S. Identification in Aegilops Species of Resistant Sources to Hessian Fly (Diptera: Cecidomyiidae) in Morocco. Genet. Resour. Crop Evol. 1998, 45, 343–345. [Google Scholar] [CrossRef]
- Nsarellah, N.; Amri, A.; Nachit, M.M.; El Bouhssini, M.; Lhaloui, S. New Durum Wheat with Hessian Fly Resistance from Triticum araraticum and T. carthlicum in Morocco. Plant Breed. 2003, 122, 435–437. [Google Scholar] [CrossRef]
- Gororo, N.N.; Eagles, H.A.; Eastwood, R.F.; Nicolas, M.E.; Flood, R.G. Use of Triticum tauschii to Improve Yield of Wheat in Low-Yielding Environments. Euphytica 2001, 123, 241–254. [Google Scholar] [CrossRef]
- Pradhan, G.P.; Prasad, P.V.V.; Fritz, A.K.; Kirkham, M.B.; Gill, B.S. High Temperature Tolerance in Aegilops Species and Its Potential Transfer to Wheat. Crop Sci. 2012, 52, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Nachit, M.M.; Elouafi, I. Durum Wheat Adaptation in the Mediterranean Dryland: Breeding, Stress Physiology, and Molecular Markers. In CSSA Special Publications; Rao, S.C., Ryan, J., Eds.; Crop Science Society of America and American Society of Agronomy: Madison, WI, USA, 2004; pp. 203–218. ISBN 978-0-89118-611-3. [Google Scholar]
- Dreisigacker, S.; Kishii, M.; Lage, J.; Warburton, M. Use of Synthetic Hexaploid Wheat to Increase Diversity for CIMMYT Bread Wheat Improvement. Aust. J. Agric. Res. 2008, 59, 413. [Google Scholar] [CrossRef]
- Finlay, K.; Wilkinson, G. The Analysis of Adaptation in a Plant-Breeding Programme. Aust. J. Agric. Res. 1963, 14, 742. [Google Scholar] [CrossRef] [Green Version]
- Eberhart, S.A.; Russell, W.A. Stability Parameters for Comparing Varieties. Crop Sci. 1966, 6, 36–40. [Google Scholar] [CrossRef] [Green Version]
- Wricke, G. On a Method of Understanding the Biological Diversity in Field Research. Z. Pfl.-Zucht 1962, 92–146. [Google Scholar]
- Shukla, G.K. Some Statistical Aspects of Partitioning Genotype-Environmental Components of Variability. Heredity 1972, 29, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Becker, H.C.; Leon, J. Stability Analysis in Plant Breeding. Plant Breed. 1988, 101, 1–23. [Google Scholar] [CrossRef]
- Francis, T.R.; Kannenberg, L.W. Yield stability studies in short-season maize. I. A descriptive method for grouping genotypes. Can. J. Plant Sci. 1978, 58, 1029–1034. [Google Scholar] [CrossRef]
- Roemer, J. Sinde Die Ertagdreichen Sorten Ertagissicherer? Mitt DLG 1917, 32, 87–89. [Google Scholar]
- Lin, C.S.; Binns, M.R. A superiority measure of cultivar performance for cultivar × location data. Can. J. Plant Sci. 1988, 68, 193–198. [Google Scholar] [CrossRef]
- Huehn, M. Beitrage Zur Erfassung Der Phanotypischen Stabilitat. EDV Med. Biol. 1979, 10, 112–117. [Google Scholar]
- Nassar, R.; Huhn, M. Studies on Estimation of Phenotypic Stability: Tests of Significance for Nonparametric Measures of Phenotypic Stability. Biometrics 1987, 43, 45. [Google Scholar] [CrossRef]
- Zobel, R.W.; Wright, M.J.; Gauch, H.G. Statistical Analysis of a Yield Trial. Agron. J. 1988, 80, 388–393. [Google Scholar] [CrossRef]
- Gauch, H.G. Model Selection and Validation for Yield Trials with Interaction. Biometrics 1988, 44, 705. [Google Scholar] [CrossRef]
- Sneller, C.H.; Kilgore-Norquest, L.; Dombek, D. Repeatability of Yield Stability Statistics in Soybean. Crop Sci. 1997, 37, 383–390. [Google Scholar] [CrossRef]
- Purchase, J.L.; Hatting, H.; van Deventer, C.S. Genotype × Environment Interaction of Winter Wheat (Triticum aestivum L.) in South Africa: II. Stability Analysis of Yield Performance. S. Afr. J. Plant Soil 2000, 17, 101–107. [Google Scholar] [CrossRef]
- Zali, H.; Farshadfar, E.; Sabaghpour, S.; Karimizadeh, R. Evaluation of Genotype × Environment Interaction in Chickpea Using Measures of Stability from AMMI Model. Ann. Biol. Res. 2012, 3, 3126–3136. [Google Scholar]
- Olivoto, T.; Lúcio, A.D.C.; Silva, J.A.G.; Marchioro, V.S.; Souza, V.Q.; Jost, E. Mean Performance and Stability in Multi-Environment Trials I: Combining Features of AMMI and BLUP Techniques. Agron. J. 2019, 111, 2949–2960. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-Assisted Prediction of Quantitative Traits Using the R Package Sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Olivoto, T.; Lúcio, A.D. Metan: An R Package for Multi-Environment Trial Analysis. Methods Ecol. Evol. 2020, 11, 783–789. [Google Scholar] [CrossRef]
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A Software to Analyze Data from Multi-Environment Plant Breeding Trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Yan, W.; Hunt, L.A.; Sheng, Q.; Szlavnics, Z. Cultivar Evaluation and Mega-Environment Investigation Based on the GGE Biplot. Crop Sci. 2000, 40, 597–605. [Google Scholar] [CrossRef]
- Dumble, S. GGEBiplots: GGE Biplots with “Ggplot2”. R Package Version 0.1.1. 2017. Available online: https://CRAN.R-project.org/package=GGEBiplots (accessed on 1 September 2021).
- Cooper, M.; DeLacy, I.H. Relationships among Analytical Methods Used to Study Genotypic Variation and Genotype-by-Environment Interaction in Plant Breeding Multi-Environment Experiments. Theoret. Appl. Genet. 1994, 88, 561–572. [Google Scholar] [CrossRef]
- Cheshkova, A. Agrostab: Stability Analysis for Agricultural Research. R Package Version 0.1.0. 2019. Available online: https://CRAN.R-Project.Org/Package=agrostab (accessed on 1 September 2021).
- Mohammadi, R.; Amri, A. Comparison of Parametric and Non-Parametric Methods for Selecting Stable and Adapted Durum Wheat Genotypes in Variable Environments. Euphytica 2008, 159, 419–432. [Google Scholar] [CrossRef]
- Harrell, F.; Dupont, C. Hmisc: Harrell Miscellaneous. R Package Version 4.4-1. 2020. Available online: https://CRAN.R-Proj.Org/Package=Hmisc (accessed on 1 September 2021).
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix. R Package Version 0.90. 2021. Available online: https://github.com/taiyun/corrplot (accessed on 1 September 2021).
- Ceccarelli, S.; Erskine, W. Genotype by Environment Interaction and International Breeding Programs. Exp. Agric. 1994, 30, 177–187. [Google Scholar] [CrossRef]
- Oliveira, I.C.M.; Guilhen, J.H.S.; de Ribeiro, P.C.O.; Gezan, S.A.; Schaffert, R.E.; Simeone, M.L.F.; Damasceno, C.M.B.; de Carneiro, J.E.S.; Carneiro, P.C.S.; da Parrella, R.A.C.; et al. Genotype-by-Environment Interaction and Yield Stability Analysis of Biomass Sorghum Hybrids Using Factor Analytic Models and Environmental Covariates. Field Crop. Res. 2020, 257, 107929. [Google Scholar] [CrossRef]
- de Leon, N.; Jannink, J.-L.; Edwards, J.W.; Kaeppler, S.M. Introduction to a Special Issue on Genotype by Environment Interaction. Crop Sci. 2016, 56, 2081–2089. [Google Scholar] [CrossRef] [Green Version]
- Ferrise, R.; Toscano, P.; Pasqui, M.; Moriondo, M.; Primicerio, J.; Semenov, M.; Bindi, M. Monthly-to-Seasonal Predictions of Durum Wheat Yield over the Mediterranean Basin. Clim. Res. 2015, 65, 7–21. [Google Scholar] [CrossRef] [Green Version]
- Chairi, F.; Aparicio, N.; Serret, M.D.; Araus, J.L. Breeding Effects on the Genotype × Environment Interaction for Yield of Durum Wheat Grown after the Green Revolution: The Case of Spain. Crop J. 2020, 8, 623–634. [Google Scholar] [CrossRef]
- Bassi, F.M.; Nachit, M.M. Genetic Gain for Yield and Allelic Diversity over 35 Years of Durum Wheat Breeding at ICARDA. Crop Breed. Genet. Genom. 2019, 1, e190004. [Google Scholar] [CrossRef] [Green Version]
- Aberkane, H.; Amri, A.; Belkadi, B.; Filali-Maltouf, A.; Kehel, Z.; Tahir, I.S.A.; Meheesi, S.; Tsivelikas, A. Evaluation of Durum Wheat Lines Derived from Interspecific Crosses under Drought and Heat Stress. Crop Sci. 2021, 61, 119–136. [Google Scholar] [CrossRef]
- Hao, M.; Zhang, L.; Ning, S.; Huang, L.; Yuan, Z.; Wu, B.; Yan, Z.; Dai, S.; Jiang, B.; Zheng, Y.; et al. The Resurgence of Introgression Breeding, as Exemplified in Wheat Improvement. Front. Plant Sci. 2020, 11, 252. [Google Scholar] [CrossRef] [Green Version]
- Warburton, M.L.; Crossa, J.; Franco, J.; Kazi, M.; Trethowan, R.; Rajaram, S.; Pfeiffer, W.; Zhang, P.; Dreisigacker, S.; Ginkel, M. van Bringing Wild Relatives Back into the Family: Recovering Genetic Diversity in CIMMYT Improved Wheat Germplasm. Euphytica 2006, 149, 289–301. [Google Scholar] [CrossRef]
- Kameswara Rao, N.; Reddy, L.J.; Bramel, P.J. Potential of Wild Species for Genetic Enhancement of Some Semi-Arid Food Crops. Genet. Resour. Crop Evol. 2003, 50, 707–721. [Google Scholar] [CrossRef]
- Kishii, M. An Update of Recent Use of Aegilops Species in Wheat Breeding. Front. Plant Sci. 2019, 10, 585. [Google Scholar] [CrossRef] [Green Version]
- Valkoun, J. Wheat Pre-Breeding Using Wild Progenitors. Euphytica 2001, 119, 17–23. [Google Scholar] [CrossRef]
- El Haddad, N.; Kabbaj, H.; Zaïm, M.; El Hassouni, K.; Tidiane Sall, A.; Azouz, M.; Ortiz, R.; Baum, M.; Amri, A.; Gamba, F.; et al. Crop Wild Relatives in Durum Wheat Breeding: Drift or Thrift? Crop Sci. 2021, 61, 37–54. [Google Scholar] [CrossRef]
- Li, J.; Wan, H.; Yang, W. Synthetic Hexaploid Wheat Enhances Variation and Adaptive Evolution of Bread Wheat in Breeding Processes. J. Sytemat. Evol. 2014, 52, 735–742. [Google Scholar] [CrossRef]
- Aberkane, H.; Belkadi, B.; Kehel, Z.; Filali-Maltouf, A.; Tahir, I.S.A.; Meheesi, S.; Amri, A. Assessment of Drought and Heat Tolerance of Durum Wheat Lines Derived from Interspecific Crosses Using Physiological Parameters and Stress Indices. Agronomy 2021, 11, 695. [Google Scholar] [CrossRef]
- Khanna-Chopra, R.; Viswanathan, C. Evaluation of Heat Stress Tolerance in Irrigated Environment of T. Aestivum and Related Species. I. Stability in Yield and Yield Components. Euphytica 1999, 106, 169–180. [Google Scholar] [CrossRef]
- Ullah, S.; Bramley, H.; Daetwyler, H.; He, S.; Mahmood, T.; Thistlethwaite, R.; Trethowan, R. Genetic Contribution of Emmer Wheat (Triticum dicoccon Schrank) to Heat Tolerance of Bread Wheat. Front. Plant Sci. 2018, 9, 1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaïm, M.; El Hassouni, K.; Gamba, F.; Filali-Maltouf, A.; Belkadi, B.; Sourour, A.; Amri, A.; Nachit, M.; Taghouti, M.; Bassi, F.M. Wide Crosses of Durum Wheat (Triticum durum Desf.) Reveal Good Disease Resistance, Yield Stability, and Industrial Quality across Mediterranean Sites. Field Crop. Res. 2017, 214, 219–227. [Google Scholar] [CrossRef]
- Simmonds, J.R.; Fish, L.J.; Leverington-Waite, M.A.; Wang, Y.; Howell, P.; Snape, J.W. Mapping of a Gene (Vir) for a Non-Glaucous, Viridescent Phenotype in Bread Wheat Derived from Triticum dicoccoides, and Its Association with Yield Variation. Euphytica 2008, 159, 333–341. [Google Scholar] [CrossRef]
- Farooq, S. Production of Low Input and Stress Tolerant Wheat Germplasm through the Use of Biodiversity Residing in the Wild Relatives. Hereditas 2004, 135, 211–215. [Google Scholar] [CrossRef] [Green Version]
- Peleg, Z.; Fahima, T.; Abbo, S.; Krugman, T.; Nevo, E.; Yakir, D.; Saranga, Y. Genetic Diversity for Drought Resistance in Wild Emmer Wheat and Its Ecogeographical Associations. Plant Cell Environ. 2005, 28, 176–191. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, M.J.; Slafer, G.A.; Berry, P.; Parry, M.A.J.; Snape, J.W.; Angus, W.J. Raising Yield Potential in Wheat. J. Exp. Bot. 2009, 60, 1899–1918. [Google Scholar] [CrossRef] [Green Version]
- Lopes, M.S.; Reynolds, M.P. Drought Adaptive Traits and Wide Adaptation in Elite Lines Derived from Resynthesized Hexaploid Wheat. Crop Sci. 2011, 51, 1617–1626. [Google Scholar] [CrossRef]
- Peng, J.; Sun, D.; Peng, Y.; Nevo, E. Gene Discovery in Triticum dicoccoides, the Direct Progenitor of Cultivated Wheats. Cereal Res. Commun. 2013, 41, 1–22. [Google Scholar] [CrossRef]
- Djanaguiraman, M.; Prasad, P.V.V.; Kumari, J.; Sehgal, S.K.; Friebe, B.; Djalovic, I.; Chen, Y.; Siddique, K.H.M.; Gill, B.S. Alien Chromosome Segment from Aegilops speltoides and Dasypyrum Villosum Increases Drought Tolerance in Wheat via Profuse and Deep Root System. BMC Plant Biol. 2019, 19, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuzmanović, L.; Giovenali, G.; Ruggeri, R.; Rossini, F.; Ceoloni, C. Small “Nested” Introgressions from Wild Thinopyrum Species, Conferring Effective Resistance to Fusarium Diseases, Positively Impact Durum Wheat Yield Potential. Plants 2021, 10, 579. [Google Scholar] [CrossRef]
- Friesen, L.F.; Brûlé-Babel, A.L.; Crow, G.H.; Rothenburger, P.A. Mixed Model and Stability Analysis of Spring Wheat Genotype Yield Evaluation Data from Manitoba, Canada. Can. J. Plant Sci. 2016, 96, 305–320. [Google Scholar] [CrossRef]
- Mehari, M.; Alamerew, S.; Lakew, B.; Yirga, H.; Tesfay, M. Parametric Stability Analysis of Malt Barley Genotypes for Grain Yield in Tigray, Ethiopia. World J. Agric. Sci. 2014, 10, 210–215. [Google Scholar] [CrossRef]
- Temesgen, T.; Keneni, G.; Sefera, T.; Jarso, M. Yield Stability and Relationships among Stability Parameters in Faba Bean (Vicia Faba L.) Genotypes. Crop J. 2015, 3, 258–268. [Google Scholar] [CrossRef] [Green Version]
- Fasahat, P. An Overview on the Use of Stability Parameters in Plant Breeding. Biom. Biostat. Int. J. 2015, 2, 00043. [Google Scholar] [CrossRef] [Green Version]



| Pedigree/Name | Wild Parent Genome | Number of Lines |
|---|---|---|
| Cham5*2/T. dicoccoides IG 118178 | AuAuBB | 6 |
| Cham5*3/T. dicoccoides IG 118178 | AuAuBB | 11 |
| Haurani*2/T. urartu IG 45489 | AuAu | 4 |
| Cham5*3/T. urartu IG 45488 | AuAu | 2 |
| Cham5*3/T. aegilopoides IG 118180 | AmAm | 19 |
| Cham5*2/T. urartu IG 118184 | AuAu | 2 |
| Cham5*3/T. aegilopoides IG 118181 | AmAm | 1 |
| Cham5*4/Ae. speltoides IG 47843 | SS (BB) | 6 |
| Cham5*2/T. aegilopoides IG 118180 | AmAm | 3 |
| Cham5*3/T. dicoccoides IG 118179 | AuAuBB | 2 |
| Haurani*2/T. aegilopoides IG 118185 | AmAm | 2 |
| Haurani*2/T. urartu IG 45475 | AuAu | 2 |
| Cham5*3/T. urartu IG 118182 | AuAu | 3 |
| Cham5*3/T. urartu IG 118184 | AuAu | 1 |
| Cham5*4/Ae. speltoides IG 47844 | SS (BB) | 1 |
| Cham5*2/T. urartu IG 118182 | AuAu | 1 |
| Haurani*3/T. dicoccoides IG 118178 | AuAuBB | 1 |
| Checks | - | 8 |
| Recurrent parents | - | 2 |
| Location | Country | Long | Lat | Season | ENV | Sowing Date | Treatment |
|---|---|---|---|---|---|---|---|
| Allal Tazi | Morocco | 34°31′ N | 6°14′ W | 2016–17 | AT-17 | 2016/11/12 | RFD |
| 2017–18 | AT-18 | 2017/11/21 | RFD | ||||
| Annoceur | Morocco | 33°41′ N | 4°51′ W | 2016–17 | AN-17 | 2016/12/02 | RFD |
| 2017–18 | AN-18 | 2017/11/21 | RFD | ||||
| Marchouch | Morocco | 33°36′ N | 6°42′ W | 2015–16 | MCH-16 | 2015/12/16 | SIR |
| 2016–17 | MCH-17 | 2016/11/19 | RFD | ||||
| 2017–18 | MCH-18 | 2017/11/29 | RFD | ||||
| Melk Zher | Morocco | 30°02′ N | 9°33′ W | 2015–16 | MZIR-16 | 2015/12/21 | FIR |
| 2015–16 | MZRF-16 | 2015/12/21 | SIR | ||||
| Tessaout | Morocco | 31°49′ N | 7°25′ W | 2016–17 | TSIR-17 | 2016/12/14 | FIR |
| 2016–17 | TSRF-17 | 2016/12/14 | RFD | ||||
| 2017–18 | TSIR-18 | 2017/12/22 | FIR | ||||
| 2017–18 | TSRF-18 | 2017/12/22 | RFD | ||||
| Wad Medani | Sudan | 14°24′ N | 33°31′ E | 2016–17 | WMD-17 | 2016/12/18 | FIR |
| 2017–18 | WMD-18 | 2017/12/07 | FIR |
| Source of Variation | DF | Variance Component |
|---|---|---|
| ENV Mean Sq (f) | 14 | 536,126,241 *** |
| Genotypic variance (r) | - | 221,131 *** |
| GE interaction variance (r) | - | 213,091 *** |
| AN-17 residuals (r) | - | 264,403 |
| AN-18 residuals (r) | - | 314,508 |
| AT-17 residuals (r) | - | 466,464 |
| AT-18 residuals (r) | - | 722,924 |
| MCH-16 residuals (r) | - | 1,268,491 |
| MCH-17 residuals (r) | - | 332,655 |
| MCH-18 residuals (r) | - | 2,107,375 |
| MZIR-16 residuals (r) | - | 2,075,582 |
| MZRF-16 residuals (r) | - | 795,924 |
| TSIR-17 residuals (r) | - | 1,469,300 |
| TSIR-18 residuals (r) | - | 1,269,044 |
| TSRF-17 residuals (r) | - | 404,398 |
| TSRF-18 residuals (r) | - | 566,288 |
| WMD-17 residuals (r) | - | 297,060 |
| WMD-18 residuals (r) | - | 203,483 |
| Source | Df | Mean Sq | % TSS | GEI Proportion (%) | Accumulated |
|---|---|---|---|---|---|
| ENV | 14 | 383,602,598 *** | 66.93 | - | - |
| REP(ENV) | 15 | 10,556,057 *** | 1.973 | - | - |
| BLOCK(REP*ENV) | 300 | 1,055,308 *** | 3.94 | - | - |
| GEN | 76 | 8,857,688 *** | 8.39 | - | - |
| GEN:ENV | 1061 | 1,417,790 *** | 18.74 | - | - |
| Residuals | 808 | 716,543 | - | ||
| PC1 | 89 | 3,871,018 | - | 22.6 | 22.6 |
| PC2 | 87 | 2,840,469 | - | 16.2 | 38.8 |
| PC3 | 85 | 2,148,503 | - | 12 | 50.7 |
| PC4 | 83 | 1,854,171 | - | 10.1 | 60.8 |
| PC5 | 81 | 1,343,685 | - | 7.1 | 68 |
| PC6 | 79 | 1,361,599 | - | 7 | 75 |
| PC7 | 77 | 1,111,644 | - | 5.6 | 80.6 |
| ENV | Sowing Date | Treatment | Max T | Min T | Mean T | Prec (mm) |
|---|---|---|---|---|---|---|
| AT-17 | 2016/11/12 | RFD | - | - | - | 592 |
| AT-18 | 2017/11/21 | RFD | - | - | - | 602 |
| AN-17 | 2016/12/02 | RFD | 23.87 | 7.14 | 15.05 | 306 |
| AN-18 | 2017/11/21 | RFD | 26.66 | 1 | 12.69 | 611 |
| MCH-16 | 2015/12/16 | SIR | 25.24 | 10 | 14 | 204.8 |
| MCH-17 | 2016/11/19 | RFD | 32.2 | 3.63 | 15.96 | 275.9 |
| MCH-18 | 2017/11/29 | RFD | 26.54 | 2.68 | 14.38 | 514.6 |
| MZIR-16 | 2015/12/21 | FIR | 24.63 | 10.86 | 17.75 | 85.8 |
| MZRF-16 | 2015/12/21 | SIR | ||||
| TSIR-17 | 2016/12/14 | FIR | 25.76 | 10.19 | 17.98 | 207 |
| TSRF-17 | 2016/12/14 | RFD | ||||
| TSIR-18 | 2017/12/22 | FIR | 26.17 | 10.31 | 18.24 | 294 |
| TSRF-18 | 2017/12/22 | RFD | ||||
| WMD-17 | 2016/12/18 | FIR | 37 | 18 | 27 | 0 |
| WMD-18 | 2017/12/07 | FIR | 37.3 | 18.6 | 27.9 | 0 |
| Desirable | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Pi | 129080 | 129081 | 142005 | Marzak | MIKI3 | Faraj | Icarachaz | 142060 | 142074 | 142009 |
| EV | 141972 | 84859 | 141984 | 142007 | 142057 | 142001 | 142056 | 142015 | 142055 | 142039 |
| GAI | Marzak | 129080 | 129081 | 142005 | Faraj | Louiza | Icarachaz | 142009 | 142074 | 142060 |
| σi2 | 141995 | 141966 | 142000 | 141986 | 142071 | 142048 | 142027 | 142039 | 142045 | 142040 |
| BLUPs | Marzak | 129080 | Icarachaz | Louiza | 142074 | 142009 | 129081 | 142005 | Faraj | 142061 |
| Bi | 142012 | 142044 | 142073 | 142068 | 142072 | 141970 | 142042 | Marzak | 142053 | 141996 |
| S2di | 142001 | 142000 | 141995 | 141966 | 142071 | 142039 | 142003 | 141986 | 142040 | 142069 |
| CV | 142015 | 142001 | 141972 | 142000 | Marzak | 141969 | 142055 | 142066 | 142014 | 141976 |
| WAASY | 141986 | 142045 | Louiza | 141966 | 142009 | 142027 | 141995 | Marzak | 142040 | 129080 |
| ASV | 142045 | 142063 | 142032 | 142040 | 141986 | 142046 | 142027 | 142012 | 141989 | 142053 |
| SIPC | 142008 | 142071 | 142001 | 142068 | 142015 | 141995 | 141966 | 142032 | 141986 | 142040 |
| Undesirable | ||||||||||
| Rank | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 |
| Pi | 142067 | 142007 | 142072 | 142003 | 142062 | 142039 | 142057 | 141984 | 84859 | 141972 |
| EV | 142005 | 141994 | 142064 | 142060 | 129081 | Icarachaz | 141997 | 129080 | 142026 | 142013 |
| GAI | 141972 | 142057 | 142039 | 142072 | 142062 | 142003 | 84859 | 141999 | 141970 | 141979 |
| σi2 | 142005 | 142061 | 141990 | 142007 | 142026 | 129081 | 142067 | 141972 | Icarachaz | 142013 |
| BLUPs | 141999 | 142072 | 141984 | 141979 | 141972 | 142039 | 142062 | 142057 | 84859 | 142003 |
| Bi | 142056 | 141997 | 141984 | 142013 | 142007 | 142001 | 129080 | 142057 | 84859 | 141972 |
| S2di | 142064 | 142026 | 142044 | 142073 | 141999 | Icarachaz | 141990 | 142061 | 129081 | 142067 |
| CV | 135226 | 141999 | 142003 | 142013 | 141990 | 142062 | 142064 | 141979 | 142072 | 141970 |
| WAASY | 141970 | 141984 | 142062 | 141999 | 141979 | 142057 | 84859 | 142067 | 142007 | 141972 |
| ASV | 129080 | 142060 | 142056 | 142013 | 142005 | 141984 | 142007 | 129081 | 141972 | 142067 |
| SIPC | 129081 | 142026 | Marzak | 142064 | 141991 | 142007 | 142013 | 142067 | 142061 | Icarachaz |
| Desirable | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
| Si1 | 141984 | 142044 | 142053 | cham5’S | 142056 | 142067 | 142071 | 141976 | 142055 | 141969 |
| Si2 | 141966 | 141995 | 142000 | 142071 | 141986 | 141996 | 142048 | 142069 | 142053 | 142018 |
| Si3 | 141995 | 142009 | 129080 | Marzak | 142039 | Faraj | 141996 | 142053 | 142060 | 142005 |
| Si6 | 129080 | Marzak | 142053 | 142009 | 141995 | 142060 | Faraj | 142005 | 141996 | 141966 |
| Undesirable | ||||||||||
| Rank | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 |
| Si1 | 142040 | Icarachaz | Marzak | 142008 | 142045 | 142063 | 142068 | 141972 | 141990 | 142032 |
| Si2 | 142007 | 142066 | 129081 | 141984 | 141990 | 142026 | Icarachaz | 141972 | 142067 | 142013 |
| Si3 | 142063 | 142006 | 141984 | 142064 | 142057 | 142067 | 142062 | 142072 | 142007 | 141972 |
| Si6 | 142003 | 141990 | 84859 | 142067 | 142057 | 142072 | 142062 | 142007 | 141972 | 141984 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aberkane, H.; Amri, A.; Belkadi, B.; Filali-Maltouf, A.; Valkoun, J.; Kehel, Z. Contribution of Wild Relatives to Durum Wheat (Triticum turgidum subsp. durum) Yield Stability across Contrasted Environments. Agronomy 2021, 11, 1992. https://doi.org/10.3390/agronomy11101992
Aberkane H, Amri A, Belkadi B, Filali-Maltouf A, Valkoun J, Kehel Z. Contribution of Wild Relatives to Durum Wheat (Triticum turgidum subsp. durum) Yield Stability across Contrasted Environments. Agronomy. 2021; 11(10):1992. https://doi.org/10.3390/agronomy11101992
Chicago/Turabian StyleAberkane, Hafid, Ahmed Amri, Bouchra Belkadi, Abdelkarim Filali-Maltouf, Jan Valkoun, and Zakaria Kehel. 2021. "Contribution of Wild Relatives to Durum Wheat (Triticum turgidum subsp. durum) Yield Stability across Contrasted Environments" Agronomy 11, no. 10: 1992. https://doi.org/10.3390/agronomy11101992
APA StyleAberkane, H., Amri, A., Belkadi, B., Filali-Maltouf, A., Valkoun, J., & Kehel, Z. (2021). Contribution of Wild Relatives to Durum Wheat (Triticum turgidum subsp. durum) Yield Stability across Contrasted Environments. Agronomy, 11(10), 1992. https://doi.org/10.3390/agronomy11101992







