The Significance of Microbial Transformation of Nitrogen Compounds in the Light of Integrated Crop Management
Abstract
1. Introduction
2. Nitrogen Fertilization vs. Yield Potential
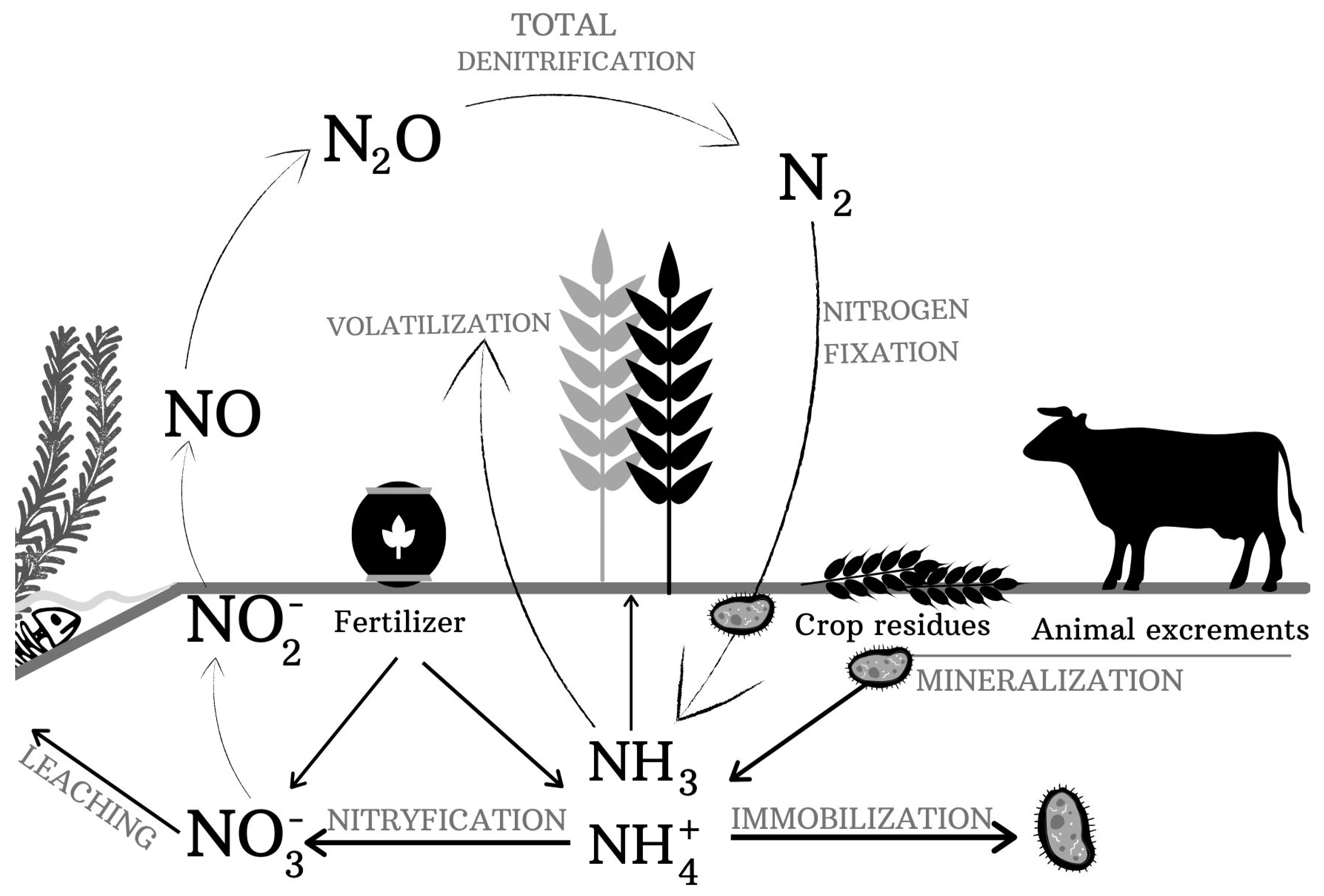
3. Microbial Transformations of Organic Nitrogen Forms in Soil
4. Microbial Transformations of Mineral Nitrogen in Soil
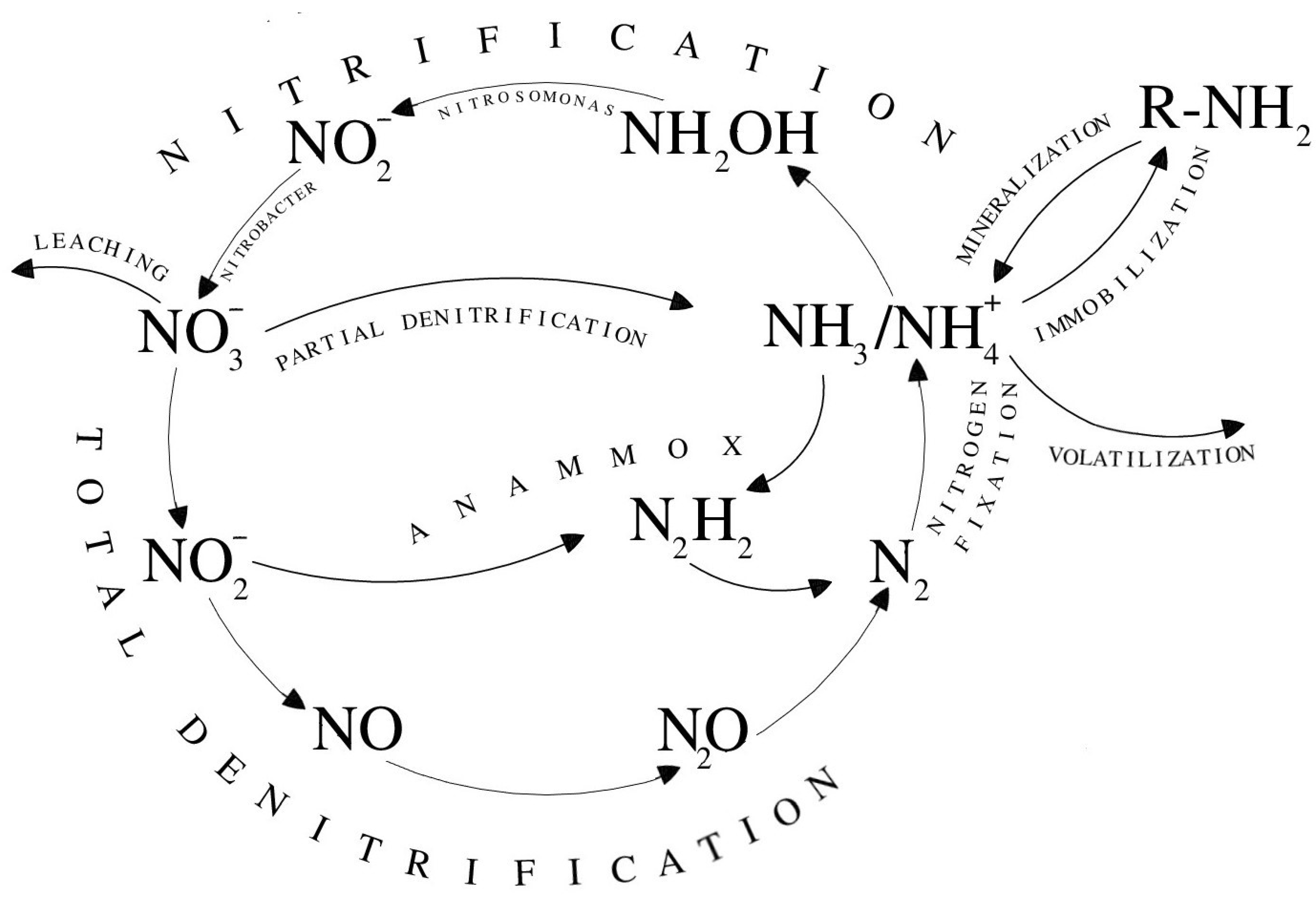
4.1. Immobilization
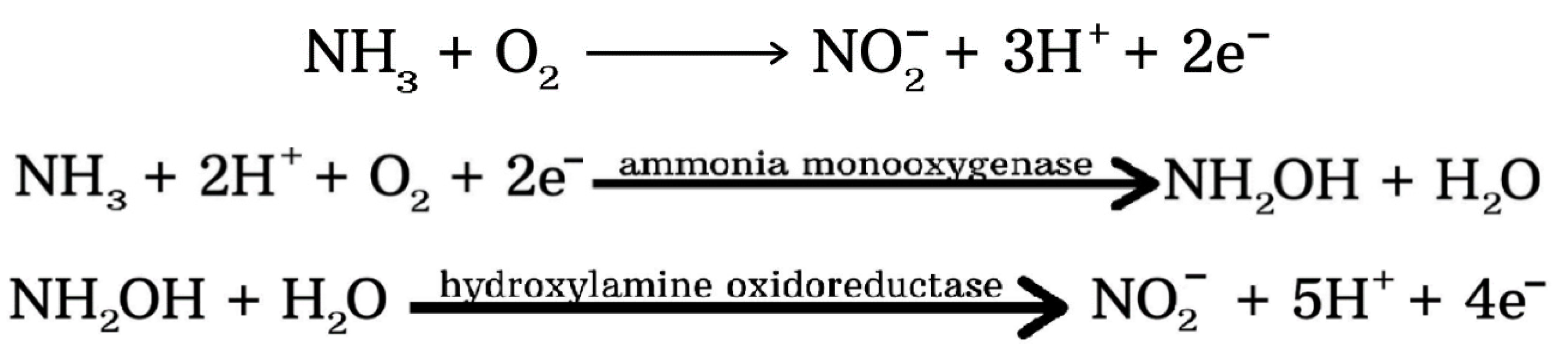
4.2. Nitrification
4.3. Denitrification
4.4. Codenitrification
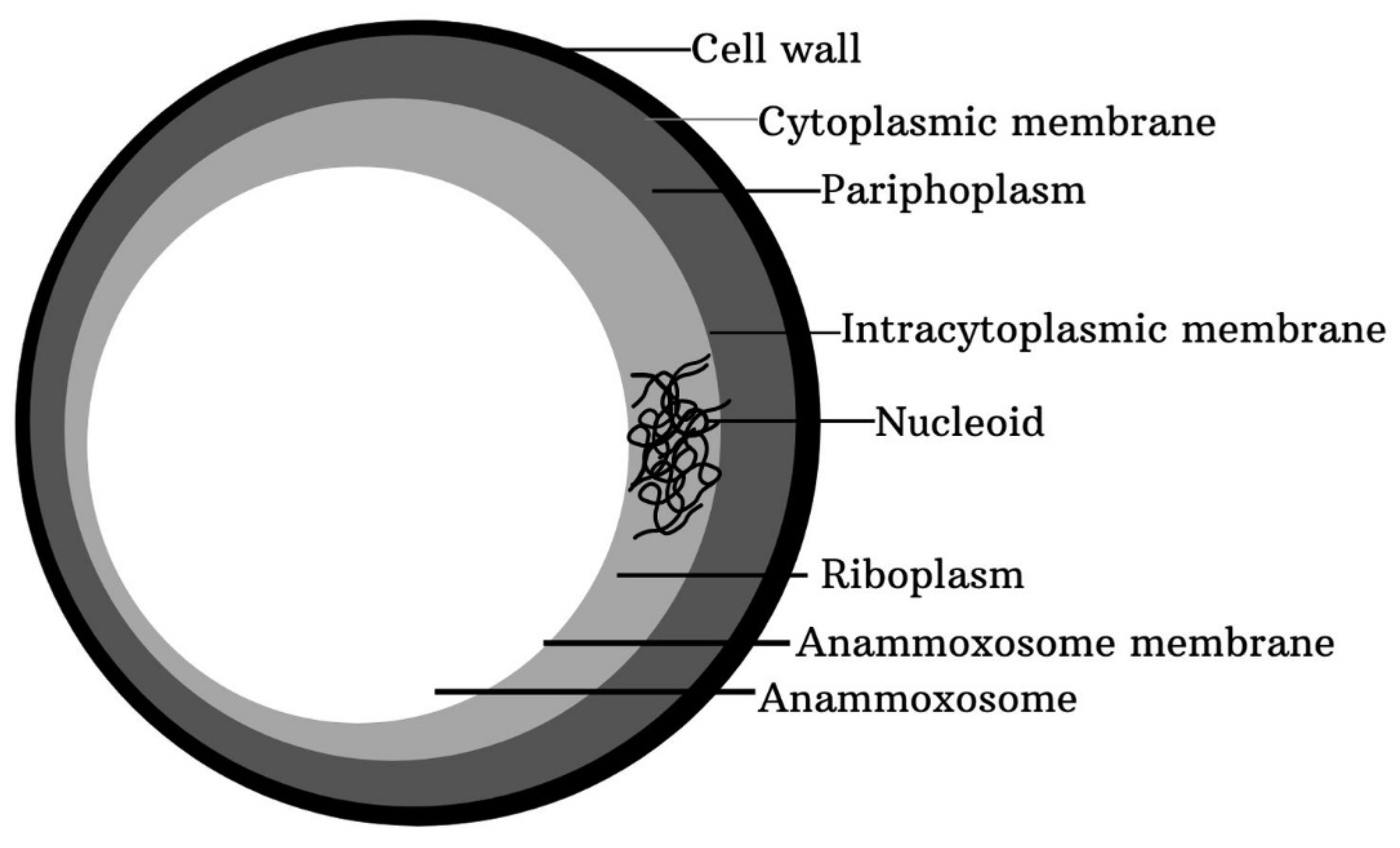
4.5. Anammox
4.6. Nitrogen-Fixing
4.7. Ammonia Volatilization
4.8. Leaching
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Hofman, G.; Van Cleemput, O. Soil and Plant Nitrogen; International Fertilizer Industry Association: Paris, France, 2004. [Google Scholar]
- Czekała, J. Effect of long-term cultivation of plants without the participation of cereals on the content of nitrogen forms in the humus level of the soil. J. Res. Appl. Agric. Eng. 2010, 55, 49–53. [Google Scholar]
- Ohyama, T. Nitrogen as a Major Essential Element of Plants. Nitr. Assim. Plants 2010, 37, 2–17. [Google Scholar]
- Robertson, G.P.; Groffman, P.M. Nitrogen transformations. In Soil Microbiology, Ecology and Biochemistry; Paul, E., Ed.; Elsevier: Amsterdam, The Netherlands, 2015; pp. 421–446. [Google Scholar]
- Kopcewicz, J.; Lewak, S.; Jaworski, K. Plant Physiology; Scientific Publisher PWN: Warsaw, Poland, 2019. [Google Scholar]
- Zboińska, M. How do plants take up and fix nitrogen? Eduk. Biol. Środowisk. 2018, 2, 19–31. [Google Scholar]
- Pajares, S.; Bohannan, B.J. Ecology of nitrogen fixing, nitrifying, and denitrifying microorganisms in tropical forest soils. Front. Microbiol. 2016, 7, 1045. [Google Scholar] [CrossRef] [PubMed]
- Staszewski, Z. Nitrogen in soil and its impact on the environment. Science notebooks. Civ. Water Eng. Environ. Manag. 2011, 50–58. [Google Scholar]
- Cabello, P.; Luque-Almagro, V.M.; Roldán, M.D.; Moreno-Vivián, C. Nitrogen cycle. In Encyclopedia of Microbiology; Schmidt, T., Ed.; Academic Press: Cambridge, MA, USA, 2004; pp. 301–310. [Google Scholar]
- Lamb, J.A.; Fernandez, F.G.; Kaiser, D.E. Understanding Nitrogen in Soils; University of Minnesota: Minneapolis, MN, USA, 2014; pp. 1–5. [Google Scholar]
- Frenk, S.; Hadar, Y.; Minz, D. Resilience of soil bacterial community to irrigation with water of different qualities under mediterranean climate. Environ. Microbiol. 2014, 16, 559–569. [Google Scholar] [CrossRef]
- García-Orenes, F.; Morugán-Coronado, A.; Zornoza, R.; Scow, K. Changes in soil microbial community structure influenced by agricultural management practices in a mediterranean agro-ecosystem. PLoS ONE 2013, 8, e80522. [Google Scholar]
- Morugán-Coronado, A.; García-Orenes, F.; McMillan, M.; Pereg, L. The effect of moisture on soil microbial properties and nitrogen cyclers in mediterranean sweet orange orchards under organic and inorganic fertilization. Sci. Total Environ. 2019, 655, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Hayatsu, M.; Tago, K.; Saito, M. Various players in the nitrogen cycle: Diversity and functions of the microorganisms involved in nitrification and denitrification. Soil Sci. Plant Nutr. 2008, 54, 33–45. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A global census of nitrogenase diversity. Environ. Microbiol. 2011, 13, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Henry, S.; Bru, D.; Stres, B.; Hallet, S.; Philippot, L. Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 2006, 72, 5181–5189. [Google Scholar] [CrossRef]
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Lücker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira: Functional and phylogenetic marker for Nitrospira. Environ. Microbiol. 2014, 16, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Prescott, C.E.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R.; Joergensen, R.G.; Ludwig, B. Pathways of nitrogen utilization by soil microorganisms—A review. Soil Biol. Biochem. 2010, 42, 2058–2067. [Google Scholar] [CrossRef]
- Elser, J.J.; Bracken, M.E.S.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef]
- FAO. World Fertilizer Trends and Outlook to 2022; Summary Report; FAO: Rome, Italy, 2019. [Google Scholar]
- Heffer, P. Assessment of Fertilizer Use by Crop at the Global Level; International Fertilizer Industry Association: Paris, France, 2013. [Google Scholar]
- Kopiński, J. Diversification of nitrogen fertilization in Polish agriculture. Pol. J. Agric. 2018, 32, 3–16. [Google Scholar]
- Barker, A.V.; Pilbeam, D.J. Handbook of Plant Nutrition; CRC Press: Bocca Raton, FL, USA, 2015. [Google Scholar]
- Dyśko, J.; Kaniszewski, K.; Kowalczyk, W.; Nowak, J.; Wójcik, P. Sustainable Fertilization of Horticultural Plants. Institute of Horticulture: Skierniewice, Poland, 2014. [Google Scholar]
- McCauley, A.; Jones, C.; Jacobsen, J. Functions and Deficiency and Toxicity Symptoms; Nutrient Management Module; Montana State University: Bozeman, MT, USA, 2009. [Google Scholar]
- Roy, R.N.; Finck, A.; Blair, G.J.; Tandon, H.I.S. Plant Nutrition for Food Security: A Guide for Integrated Nutrient Management; FAO Fertilizer and Plant Nutrition Bulletin; FAO: Rome, Italy, 2006. [Google Scholar]
- Wong, M. Visual symptoms of plant nutrient deficiencies in nursery and landscape plants. Soil Crop Manag. 2005, 10, 1–4. [Google Scholar]
- Martínez-Espinosa, R.M.; Cole, J.A.; Richardson, D.J.; Watmough, N.J. Enzymology and ecology of the nitrogen cycle. Biochem. Soc. Trans. 2011, 39, 175–178. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, H.; Yan, X.; Yagi, K. Estimations of emission factors for fertilizer-induced direct N2O emissions from agricultural soils in Japan: Summary of available data. Soil Sci. Plant Nutr. 2006, 52, 774–787. [Google Scholar] [CrossRef]
- Beaulieu, J.J.; Tank, J.L.; Hamilton, S.K.; Wollheim, W.M.; Hall, R.O.; Mulholland, P.J.; Peterson, B.J.; Ashkenas, L.R.; Cooper, L.W.; Dahm, C.N. Nitrous oxide emission from denitrification in stream and river networks. Proc. Natl. Acad. Sci. USA 2011, 108, 214–219. [Google Scholar] [CrossRef]
- Dodds, W.; Smith, V. Nitrogen, phosphorus, and eutrophication in streams. Inland Water 2016, 6, 155–164. [Google Scholar] [CrossRef]
- Howarth, R.W. Coastal nitrogen pollution: A review of sources and trends globally and regionally. Harmful Algae 2008, 8, 14–20. [Google Scholar] [CrossRef]
- Johnson, P.T.J.; Townsend, A.R.; Cleveland, C.C.; Glibert, P.M.; Howarth, R.W.; McKenzie, V.J.; Rejmankova, E.; Ward, M.H. Linking environmental nutrient enrichment and disease emergence in humans and wildlife. Ecol. Appl. 2010, 20, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Canqui, H.; Schlegel, A.J. Implications of inorganic fertilization of irrigated corn on soil properties: Lessons learned after 50 years. J. Environ. Qual. 2013, 42, 861–871. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, W.; Li, M.; Han, Y. Does fertilizer education program increase the technical efficiency of chemical fertilizer use? Evidence from wheat production in China. Sustainability 2019, 11, 543. [Google Scholar] [CrossRef]
- Han, S.; Zeng, L.; Luo, X.; Xiong, X.; Wen, S.; Wang, B.; Chen, W.; Huang, Q. Shifts in Nitrobacter-and Nitrospira-like Nitrite-oxidizing bacterial communities under long-term fertilization practices. Soil Biol. Biochem. 2018, 124, 118–125. [Google Scholar] [CrossRef]
- Li, J.H.; Yang, Y.J.; Li, B.W.; Li, W.J.; Wang, G.; Knops, J.M. Effects of nitrogen and phosphorus fertilization on soil carbon fractions in Alpine Meadows on the Qinghai-Tibetan Plateau. PLoS ONE 2014, 9, e103266. [Google Scholar] [CrossRef] [PubMed]
- Stockmann, U.; Adams, M.A.; Crawford, J.W.; Field, D.J.; Henakaarchchi, N.; Jenkins, M.; Minasny, B.; McBratney, A.B.; De Courcelles, V.d.R.; Singh, K. The knowns, known unknowns and unknowns of sequestration of soil organic carbon. Agric. Ecosyst. Environ. 2013, 164, 80–99. [Google Scholar] [CrossRef]
- Xiang, X.; He, D.; He, J.-S.; Myrold, D.D.; Chu, H. Ammonia-oxidizing bacteria rather than archaea respond to short-term urea amendment in an Alpine Grassland. Soil Biol. Biochem. 2017, 107, 218–225. [Google Scholar] [CrossRef]
- Zamanian, K.; Kuzyakov, Y. Contribution of soil inorganic carbon to atmospheric CO2: More Important than previously thought. Glob. Chang. Biol. 2019, 25, E1–E3. [Google Scholar] [CrossRef]
- Zhou, X.; Fornara, D.; Wasson, E.A.; Wang, D.; Ren, G.; Christie, P.; Jia, Z. Effects of 44 years of chronic nitrogen fertilization on the soil nitrifying community of permanent grassland. Soil Biol. Biochem. 2015, 91, 76–83. [Google Scholar] [CrossRef]
- Allison, S.D.; Lu, Y.; Weihe, C.; Goulden, M.L.; Martiny, A.C.; Treseder, K.K.; Martiny, J.B. Microbial abundance and composition influence litter decomposition response to environmental change. Ecology 2013, 94, 714–725. [Google Scholar] [CrossRef]
- Coolon, J.D.; Jones, K.L.; Todd, T.C.; Blair, J.M.; Herman, M.A. Long-term nitrogen amendment alters the diversity and assemblage of soil bacterial communities in tallgrass prairie. PLoS ONE 2013, 8, e67884. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, Y.; Zuo, Q.; Du, B.; Chen, W.; Wei, D.; Huang, Q. Contrasting responses of bacterial and fungal communities to aggregate-size fractions and long-term fertilizations in soils of northeastern China. Sci. Total Environ. 2018, 635, 784–792. [Google Scholar] [CrossRef]
- Ramirez, K.S.; Lauber, C.L.; Knight, R.; Bradford, M.A.; Fierer, N. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 2010, 91, 3463–3470. [Google Scholar] [CrossRef]
- Hallin, S.; Jones, C.M.; Schloter, M.; Philippot, L. Relationship between N-cycling communities and ecosystem functioning in a 50-year-old fertilization experiment. ISME J. 2009, 3, 597–605. [Google Scholar] [CrossRef]
- Wessén, E.; Nyberg, K.; Jansson, J.K.; Hallin, S. Responses of bacterial and archaeal ammonia oxidizers to soil organic and fertilizer amendments under long-term management. Appl. Ecol. 2010, 45, 193–200. [Google Scholar] [CrossRef]
- Treseder, K.K. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecol. Lett. 2008, 11, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Paungfoo-Lonhienne, C.; Yeoh, Y.K.; Kasinadhuni, N.R.P.; Lonhienne, T.G.; Robinson, N.; Hugenholtz, P.; Ragan, M.A.; Schmidt, S. Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 2015, 5, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Morrison, E.W.; Frey, S.D.; Sadowsky, J.J.; van Diepen, L.T.A.; Thomas, W.K.; Pringle, A. Chronic nitrogen additions fundamentally restructure the soil fungal community in a temperate forest. Fungal Ecol. 2016, 23, 48–57. [Google Scholar] [CrossRef]
- Afreh, D.; Zhang, J.; Guan, D.; Liu, K.; Song, Z.; Zheng, C.; Deng, A.; Feng, X.; Zhang, X.; Wu, Y. Long-term fertilization on nitrogen use efficiency and greenhouse gas emissions in a double maize cropping system in subtropical China. Soil Till. Res. 2018, 180, 259–267. [Google Scholar] [CrossRef]
- Miller, M.N.; Zebarth, B.; Dandie, C.E.; Burton, D.L.; Goyer, C.; Trevors, J.T. Crop residue influence on denitrification, n2o emissions and denitrifier community abundance in soil. Soil Biol. Biochem. 2008, 40, 2553–2562. [Google Scholar] [CrossRef]
- Saunders, O.E.; Fortuna, A.-M.; Harrison, J.H.; Cogger, C.G.; Whitefield, E.; Green, T. Gaseous Nitrogen and bacterial responses to raw and digested dairy manure applications in incubated soil. Environ. Sci. Technol. 2012, 46, 11684–11692. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-C.; Shamsi, I.H.; Xu, D.-T.; Wang, G.-H.; Lin, X.-Y.; Jilani, G.; Hussain, N.; Chaudhry, A.N. Chemical fertilizer and organic manure inputs in soil exhibit a vice versa pattern of microbial community structure. Appl. Soil Ecol. 2012, 57, 1–8. [Google Scholar] [CrossRef]
- Xiang, X.; Liu, J.; Zhang, J.; Li, D.; Xu, C.; Kuzyakov, Y. Divergence in fungal abundance and community structure between soils under long-term mineral and organic fertilization. Soil Till. Res. 2020, 196, 104491. [Google Scholar] [CrossRef]
- Bandyopadhyay, K.K.; Misra, A.K.; Ghosh, P.K.; Hati, K.M.; Mandal, K.G.; Moahnty, M. Effect of irrigation and nitrogen application methods on input use efficiency of wheat under limited water supply in a vertisol of central India. Irrig. Sci. 2010, 28, 285–299. [Google Scholar] [CrossRef]
- Herencia, J.F.; Garcia-Galavis, P.A.; Maqueda, C. Long-term effect of organic and mineral fertilization on soil physical properties under greenhouse and outdoor management practices. Pedosphere 2011, 21, 443–453. [Google Scholar] [CrossRef]
- Šimon, T. The Influence of long-term organic and mineral fertilization on soil organic matter. Soil Water Res. 2008, 3, 41–51. [Google Scholar] [CrossRef]
- Fließbach, A.; Oberholzer, H.-R.; Gunst, L.; Mäder, P. Soil organic matter and biological soil quality indicators after 21 years of organic and conventional farming. Agric. Ecosyst. Environ. 2007, 118, 273–284. [Google Scholar] [CrossRef]
- Bradley, D.; Christodoulou, M.; Caspari, C.; Di Luca, P. Integrated Crop Management Systems in the EU; Amended Final Report for European Commission DG Environment; Agra CEAS Consulting: Ashford, UK, 2002. [Google Scholar]
- Department of Agriculture and Rural Development. Regulation of the Minister of Agriculture and Rural Development in Poland on Integrated Agricultural Production of 16 December 2010 (Official Journal 2010, No. 256, Item 1722); Department of Agriculture and Rural Development: Warsaw, Poland, 2010.
- European Parliament and of the Council. Regulation (EU) 2019/1009 of the European Parliament and of the Council of 5 June 2019. Laying down rules on the making available on the market of EU fertilising products; European Parliament and of the Council: Strasbourg, France, 2019. [Google Scholar]
- Sejm and Senate Complex of Poland. Fertilizer and Fertilization Act of 10 July 2007 in Poland (Official Journal 2007, No. 147, Item 1033); Sejm and Senate Complex of Poland: Warsaw, Poland, 2007. [Google Scholar]
- Delgado, J.; Lemunyon, J. Nutrient management. In Encyclopedia of Soil Science; Lal, R., Ed.; Marcel Decker: New York, NY, USA, 2006; pp. 1157–1160. [Google Scholar]
- Prasad, R.; Hochmuth, G.J.; Boote, K.J. Estimation of nitrogen pools in irrigated potato production on sandy soil using the model SUBSTOR. PLoS ONE 2015, 10, e0117891. [Google Scholar] [CrossRef]
- Shober, A.L.; Hochmuth, G.; Wiese, C. An Overview of Nutrient Budgets for Use in Nutrient Management Planning; University of Florida: Gainesville, FL, USA, 2011. [Google Scholar]
- Cissé, L. Balanced fertilization for sustainable use of plant nutrients. In Fertilizer Best Management Practices; International Fertilizer Industry Association: Paris, France, 2007; pp. 33–46. [Google Scholar]
- Bhatt, B.; Chandra, R.; Ram, S.; Pareek, N. Long-term effects of fertilization and manuring on productivity and soil biological properties under rice (Oryza sativa)–wheat (Triticum aestivum) sequence in mollisols. Arch. Agron. Soil Sci. 2016, 62, 1109–1122. [Google Scholar] [CrossRef]
- Eo, J.; Park, K.-C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Blumenthal, J.; Baltensperger, D.D.; Cassman, K.G.; Mason, S.; Pavlista, A. Importance and effect of nitrogen on crop quality and health. In Nitrogen in the Environment; Hatfield, J.L., Follett, R.F., Eds.; Academic Press: Cambridge, MA, USA, 2008; pp. 51–70. [Google Scholar]
- D’Amico-Damião, V.; Nunes, H.D.; Couto, P.A.; Lemos, L.B. Straw type and nitrogen fertilization influence winter common bean yield and quality. Int. J. Plant Prod. 2020, 14, 703–712. [Google Scholar] [CrossRef]
- Pikuła, D. Effect of long-term straw fertilization on plant yield and soil fertility. Stud. Rep. 2015, 45, 85–96. [Google Scholar]
- Crohn, D. Nitrogen mineralization and its importance in organic waste recycling. In Proceedings of the National Alfalfa Symposium, San Diego, CA, USA, 13–15 December 2004; pp. 13–15. [Google Scholar]
- Bailey, K.L.; Lazarovits, G. Suppressing soil-borne diseases with residue management and organic amendments. Soil Till. Res. 2003, 72, 169–180. [Google Scholar] [CrossRef]
- Libudzisz, Z.; Kowal, K.; Żakowska, Z. Technical Microbiology. Microorganisms and Their Environment; Scientific Publisher PWN: Warsaw, Poland, 2007. [Google Scholar]
- Vranova, V.; Rejsek, K.; Formanek, P. Proteolytic activity in soil: A review. Appl. Soil Ecol. 2013, 70, 23–32. [Google Scholar] [CrossRef]
- Rahman, R.; Basri, M.; Salleh, A.B. Thermostable alkaline protease from Bacillus stearothermophilus f1—Nutritional factors affecting protease production. Ann. Microbiol. 2003, 53, 199–210. [Google Scholar]
- Ayyasamy, P.M.; Chun, S.; Lee, S. Desorption and dissolution of heavy metals from contaminated soil using Shewanella sp.(HN-41) Amended with various carbon sources and synthetic soil organic matters. J. Hazard. Mater. 2009, 161, 1095–1102. [Google Scholar] [CrossRef]
- Bijak, M.; Ponczek, M.B.; Nowak, P. Serine proteases and their classification according to the merops system. Kosmos 2015, 64, 31–45. [Google Scholar]
- Burchacka, E.; Witkowska, D. Serine proteases and their function in the pathogenesis of bacterial infections. Adv. Hyg. Exp. Med. 2016, 70, 678–694. [Google Scholar]
- Rudenskaya, G.N.; Pupov, D.V. Cysteine proteinases of microorganisms and viruses. Biochemistry 2008, 73, 1–13. [Google Scholar] [CrossRef]
- Wenig, K.; Chatwell, L.; von Pawel-Rammingen, U.; Bjorck, L.; Huber, R.; Sondermann, P. Structure of the Streptococcal endopeptidase IdeS, a cysteine proteinase with strict specificity for IgG. Proc. Natl. Acad. Sci. USA 2004, 101, 17371–17376. [Google Scholar] [CrossRef] [PubMed]
- Yegin, S.; Fernandez-Lahore, M.; Salgado, A.J.G.; Guvenc, U.; Goksungur, Y.; Tari, C. Aspartic proteinases from Mucor spp. in cheese manufacturing. Appl. Microbiol. Biotechnol. 2011, 89, 949–960. [Google Scholar] [CrossRef] [PubMed]
- Gradišar, H.; Friedrich, J.; Križaj, I.; Jerala, R. Similarities and specificities of fungal keratinolytic proteases: Comparison of keratinases of Paecilomyces marquandii and Doratomyces microsporus to some known proteases. Appl. Environ. Microbiol. 2005, 71, 3420–3426. [Google Scholar] [CrossRef]
- Sumantha, A.; Larroche, C.; Pandey, A. Microbiology and industrial biotechnology of food-grade proteases: A perspective. Food Technol. Biotechnol. 2006, 44, 211. [Google Scholar]
- Allison, S.D.; Vitousek, P.M. Responses of extracellular enzymes to simple and complex nutrient inputs. Soil Biol. Biochem. 2005, 37, 937–944. [Google Scholar] [CrossRef]
- Dong, S.; Brooks, D.; Jones, M.D.; Grayston, S.J. A Method for linking in situ activities of hydrolytic enzymes to associated organisms in forest soils. Soil Biol. Biochem. 2007, 39, 2414–2419. [Google Scholar] [CrossRef]
- Grandy, A.S.; Neff, J.C.; Weintraub, M.N. Carbon structure and enzyme activities in alpine and forest ecosystems. Soil Biol. Biochem. 2007, 39, 2701–2711. [Google Scholar] [CrossRef]
- Fuka, M.M.; Engel, M.; Hagn, A.; Munch, J.C.; Sommer, M.; Schloter, M. Changes of diversity pattern of proteolytic bacteria over time and space in an agricultural soil. Microb. Ecol. 2009, 57, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Fuka, M.M.; Engel, M.; Gattinger, A.; Bausenwein, U.; Sommer, M.; Munch, J.C.; Schloter, M. Factors influencing variability of proteolytic genes and activities in arable soils. Soil Biol. Biochem. 2008, 40, 1646–1653. [Google Scholar] [CrossRef]
- Näsholm, T.; Kielland, K.; Ganeteg, U. Uptake of organic nitrogen by plants: Tansley review. New Phytol. 2009, 182, 31–48. [Google Scholar] [CrossRef]
- Brzostek, E.R.; Finzi, A.C. Substrate supply, fine roots, and temperature control proteolytic enzyme activity in temperate forest soils. Ecology 2011, 92, 892–902. [Google Scholar] [CrossRef]
- Landi, L.; Renella, G.; Giagnoni, L.; Nannipieri, P. Activities of Proteolytic Enzymes. In Methods of Soil Enzymology; Dick, R.P., Ed.; Soil Science Society of America: Madison, WI, USA, 2015; pp. 247–260. [Google Scholar]
- Gardini, F.; Martuscelli, M.; Caruso, M.C.; Galgano, F.; Crudele, M.A.; Favati, F.; Guerzoni, M.E.; Suzzi, G. Effects of pH, temperature and NaCl concentration on the growth kinetics, proteolytic activity and biogenic amine production of Enterococcus faecalis. Int. J. Food Microbiol. 2001, 64, 105–117. [Google Scholar] [CrossRef]
- Bach, H.-J.; Munch, J.C. Identification of bacterial sources of soil peptidases. Biol. Fertil. Soils 2000, 31, 219–224. [Google Scholar] [CrossRef]
- Marx, M.-C.; Kandeler, E.; Wood, M.; Wermbter, N.; Jarvis, S.C. Exploring the enzymatic landscape: Distribution and kinetics of hydrolytic enzymes in soil particle-size fractions. Soil Biol. Biochem. 2005, 37, 35–48. [Google Scholar] [CrossRef]
- Niewiadomska, A.; Sulewska, H.; Wolna-Maruwka, A.; Klama, J. Effect of organic fertilization on development of proteolytic bacteria and activity of proteases in the soil for cultivation of maize (Zea mays L.). Arc. Environ. Prot. 2010, 36, 47–56. [Google Scholar]
- Nitu, T.T.; Milu, U.M.; Jahangir, M.M.R. Cover Crops and Soil Nitrogen Cycling. In Cover Crops and Sustainable Agriculture; Islam, R., Sherman, B., Eds.; CRC Press: Boca Raton, FL, USA, 2021; pp. 209–226. [Google Scholar]
- Nannipieri, P.; Kandeler, E.; Ruggiero, P. Enzyme activities and microbiological and biochemical processes in soil. In Enzymes in the Environment: Activity, Ecology and Applications; Burns, R.G., Dick, R.P., Eds.; Marcel Dekker: New York, NY, USA, 2002; pp. 1–33. [Google Scholar]
- Wyczółkowski, A.; Dąbek-Szreniawska, M. Enzymes involved in the mineralization of organic nitrogen. Acta Agrophys. 2005, 3, 37–61. [Google Scholar]
- Schloter, M.; Nannipieri, P.; Sørensen, S.J.; van Elsas, J.D. Microbial indicators for soil quality. Biol Fertil Soils 2018, 54, 1–10. [Google Scholar] [CrossRef]
- Alur, M.D. Metabolic pathways, nitrogen metabolism. In Metabolic Pathways, Nitrogen Metabolism; Robison, R., Batt, C.A., Patel, P.D., Eds.; Academic Press: Cambridge, MA, USA, 1999; pp. 1288–1298. [Google Scholar]
- Szostak, B.; Jezierska-Tys, S.; Bekier-Jaworska, E. The intensity of the ammonification and nitrification process in soil in pig farms. Acta Agrophys. 2005, 6, 251–260. [Google Scholar]
- Dąbek-Szreniawska, M.; Zimon, A.; Wyczółkowski, A.I. Activity of enzymes in the ammonification process in soil with the addition of nitrogenous organic substances. Acta Agroph. 2006, 8, 23–33. [Google Scholar]
- Stefanakis, A.; Akratos, C.S.; Tsihrintzis, V.A. Treatment Processes in VFCWs. In Vertical Flow Constructed Wetlands; Stefanakis, A., Akratos, C.S., Tsihrintzis, V.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 57–84. [Google Scholar]
- Vymazal, J. Removal of Nutrients in Various Types of Constructed Wetlands. Sci. Total Environ. 2007, 380, 48–65. [Google Scholar] [CrossRef]
- Vymazal, J.; Greenway, M.; Tonderski, K.; Brix, H.; Mander, Ü. Constructed wetlands for wastewater treatment. In Wetlands and Natural Resource Management; Verhoeven, J.T.A., Beltman, B., Bobbink, R., Whigham, D.F., Eds.; Springer: New York, NY, USA, 2006; pp. 69–96. [Google Scholar]
- Saeed, T.; Sun, G. A Review on nitrogen and organics removal mechanisms in subsurface flow constructed wetlands: Dependency on environmental parameters, operating conditions and supporting media. J. Environ. Manag. 2012, 112, 429–448. [Google Scholar] [CrossRef]
- Fujii, K.; Yamada, T.; Hayakawa, C.; Nakanishi, A.; Funakawa, S. Decoupling of protein depolymerization and ammonification in nitrogen mineralization of acidic forest soils. Appl. Soil Ecol. 2020, 153, 103572. [Google Scholar] [CrossRef]
- Yevdokimov, I.V.; Saha, S.; Blagodatsky, S.A.; Kudeyarov, V.N. Nitrogen immobilization by soil microorganisms depending on nitrogen application rates. Eur. Soil Sci. 2005, 38, 516–523. [Google Scholar]
- Bottomley, P.; Taylor, A.E.; Myrold, D.D. A Consideration of the relative contributions of different microbial subpopulations to the soil n cycle. Front. Microbiol. 2012, 3, 373. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; He, H.; Zhang, X.; Kazanci, C.; Li, Z.; Necpalova, M.; Ma, Q. Calculation of fungal and bacterial inorganic nitrogen immobilization rates in soil. Soil Biol. Biochem. 2021, 153, 108114. [Google Scholar] [CrossRef]
- Myrold, D.D.; Posavatz, N.R. Potential importance of bacteria and fungi in nitrate assimilation in soil. Soil Biol. Biochem. 2007, 39, 1737–1743. [Google Scholar] [CrossRef]
- Grzyb, A.; Wolna-Maruwka, A.; Niewiadomska, A. Environmental factors affecting the mineralization of crop residues. Agronomy 2020, 10, 1951. [Google Scholar] [CrossRef]
- Grzebisz, W. Plant Production: A Textbook for School Students Educating in the Profession of a Farmer Technician: Collective Work; Environment and Basics of Agrotechnics; Hortpress: Warsaw, Poland, 2008. [Google Scholar]
- Chen, B.; Liu, E.; Tian, Q.; Yan, C.; Zhang, Y. Soil nitrogen dynamics and crop residues. A review. Agron. Sustain. Dev. 2014, 34, 429–442. [Google Scholar] [CrossRef]
- Zandt, D.; Fritz, C.; Wichern, F. In the Land of Plenty: Catch crops trigger nitrogen uptake by soil microorganisms. Plant Soil 2018, 423, 549–562. [Google Scholar] [CrossRef]
- Montaño, N.M.; García-Oliva, F.; Jaramillo, V.J. Dissolved organic carbon affects soil microbial activity and nitrogen dynamics in a mexican tropical deciduous Forest. Plant Soil 2007, 295, 265–277. [Google Scholar] [CrossRef]
- Dail, D.B.; Davidson, E.A.; Chorover, J. Rapid abiotic transformation of nitrate in an acid forest soil. Biogeochemistry 2001, 54, 131–146. [Google Scholar] [CrossRef]
- Myrold, D.D.; Bottomley, P.J. Nitrogen mineralization and immobilization. In Agronomy Monographs; Schepers, J.S., Raun, W.R., Eds.; American Society of Agronomy, Crop Science Society of America, Soil Science Society of America: Madison, WI, USA, 2015; pp. 157–172. [Google Scholar]
- Olk, D.C.; Cassman, K.G.; Schmidt-Rohr, K.; Anders, M.M.; Mao, J.-D.; Deenik, J.L. Chemical stabilization of soil organic nitrogen by phenolic lignin residues in anaerobic agroecosystems. Soil Biol. Biochem. 2006, 38, 3303–3312. [Google Scholar] [CrossRef]
- Coskun, D.; Britto, D.T.; Shi, W.; Kronzucker, H.J. Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 2017, 3, 17074. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.M.; Alzerreca, J.J.; Suwa, Y.; Klotz, M.G. Diversity of ammonia monooxygenase operon in autotrophic ammonia-oxidizing bacteria. Arch. Microbiol. 2002, 177, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Phillips, C.J.; Harris, D.; Dollhopf, S.L.; Gross, K.L.; Prosser, J.I.; Paul, E.A. Effects of agronomic treatments on structure and function of ammonia-oxidizing communities. Appl. Environ. Microbiol. 2000, 66, 5410–5418. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Könneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.; Eldyasti, A. Ammonia-oxidizing bacteria (AOB) opportunities and applications: A review. Rev Environ. Sci. Biotechnol. 2018, 17, 285–321. [Google Scholar] [CrossRef]
- Avrahami, S.; Liesack, W.; Conrad, R. Effects of temperature and fertilizer on activity and community structure of soil ammonia oxidizers. Environ. Microbiol. 2003, 5, 691–705. [Google Scholar] [CrossRef]
- Kowalchuk, G.A.; Stephen, J.R. Ammonia-Oxidizing Bacteria: A model for molecular microbial ecology. Annu. Rev. Microbiol. 2001, 55, 485–529. [Google Scholar] [CrossRef]
- De Boer, W.; Kowalchuk, G.A. Nitrification in acid soils: Microorganisms and mechanisms. Soil Biol. Biochem. 2001, 33, 853–866. [Google Scholar] [CrossRef]
- Nicol, G.W.; Leininger, S.; Schleper, C.; Prosser, J.I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 2008, 10, 2966–2978. [Google Scholar] [CrossRef]
- Morimoto, S.; Hayatsu, M.; Takada Hoshino, Y.; Nagaoka, K.; Yamazaki, M.; Karasawa, T.; Takenaka, M.; Akiyama, H. Quantitative analyses of ammonia-oxidizing archaea (aoa) and ammonia-oxidizing bacteria (AOB) in fields with different soil types. Microb. Environ. 2011, 26, 248–253. [Google Scholar] [CrossRef]
- Enwall, K.; Philippot, L.; Hallin, S. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 2005, 71, 8335–8343. [Google Scholar] [CrossRef]
- Bernhard, A. The nitrogen cycle: Processes, players, and human impact. Nat. Educ. Knowl. 2010, 3, 25. [Google Scholar]
- Lücker, S.; Nowka, B.; Rattei, T.; Spieck, E.; Daims, H. The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front. Microbiol. 2013, 4, 27. [Google Scholar] [CrossRef]
- Sorokin, D.Y.; Lücker, S.; Vejmelkova, D.; Kostrikina, N.A.; Kleerebezem, R.; Rijpstra, W.I.C.; Damsté, J.S.S.; Le Paslier, D.; Muyzer, G.; Wagner, M. Nitrification expanded: Discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. ISME J. 2012, 6, 2245–2256. [Google Scholar] [CrossRef]
- Daims, H.; Lücker, S.; Wagner, M. A new perspective on microbes formerly known as nitrite-oxidizing bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef]
- Ferguson, S.J.; Richardson, D.J.; van Spanning, R.J.M. Biochemistry and molecular biology of nitrification. In Biology of the Nitrogen Cycle; Bothe, H., Ferguson, S., Newton, W.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 209–222. [Google Scholar]
- Fiencke, C.; Spieck, E.; Bock, E. Nitrifying bacteria. In Nitrogen Fixation in Agriculture, Forestry, Ecology, and the Environment; Werner, D., Newton, W.E., Eds.; Springer: Heidelberg, Germany, 2005; pp. 255–276. [Google Scholar]
- Ward, B.B. Nitrification. In Encyclopedia of Ecology; Jørgensen, S.E., Fath, B.D., Eds.; Elsevier: Amsterdam, The Netherlands, 2013; pp. 351–358. [Google Scholar]
- Millar, N.; Robertson, G.P. Nitrogen transfers and transformations in row-crop ecosystems. In The Ecology of Agricultural Ecosystems: Long-Term Research on the Path to Sustainability; Hampilton, S., Doll, J.E., Robertson, P., Eds.; Oxford University Press: New York, NY, USA, 2015; pp. 213–225. [Google Scholar]
- Zhu, X.; Burger, M.; Doane, T.A.; Horwath, W.R. Ammonia oxidation pathways and nitrifier denitrification are significant sources of N2O and NO under low oxygen availability. Proc. Natl. Acad. Sci. USA 2013, 110, 6328–6333. [Google Scholar] [CrossRef]
- Schulze, E.-D.; Mooney, H.A. Biodiversity and Ecosystem Function; Springer: Berlin/Heidelberg, Germany, 2002. [Google Scholar]
- Beeckman, F.; Motte, H.; Beeckman, T. Nitrification in agricultural soils: Impact, actors and mitigation. Curr. Opin. Biotechnol. 2018, 50, 166–173. [Google Scholar] [CrossRef]
- Akhtar, M.; Hussain, F.; Ashraf, M.Y.; Qureshi, T.M.; Akhter, J.; Awan, A.R. Influence of Salinity on Nitrogen Transformations in Soil. Commun. Soil Sci. Plant Anal. 2012, 43, 1674–1683. [Google Scholar] [CrossRef]
- Podlaska, B.; Russel, S. Characteristics of nitrifying bacteria and their role in the nitrogen cycle. In Interdisciplinary Issues in Engineering and Environmental Protection; Traczewska, T.M., Kaźmierczak, B., Eds.; Publishing House of the Wrocław University of Technology: Wrocław, Poland, 2003; pp. 263–279. [Google Scholar]
- Viotti, P.; Collivignarelli, M.C.; Martorelli, E.; Raboni, M. Oxygen control and improved denitrification efficiency by dosing ferrous ions in the anoxic reactor. Desalin. Water Treat. 2016, 57, 18240–18247. [Google Scholar] [CrossRef]
- Butterbach-Bahl, K.; Baggs, E.M.; Dannenmann, M.; Kiese, R.; Zechmeister-Boltenstern, S. Nitrous Oxide emissions from soils: How well do we understand the processes and their controls? Philos. Trans. R. Soc. B. 2013, 368, 20130122. [Google Scholar] [CrossRef] [PubMed]
- Kloos, K.; Mergel, A.; Rösch, C.; Bothe, H. Denitrification within the genus Azospirillum and other associative bacteria. Funct. Plant Biol. 2001, 28, 991–998. [Google Scholar] [CrossRef]
- Su, F.; Takaya, N.; Shoun, H. Nitrous oxide-forming codenitrification catalyzed by cytochrome P450nor. Biosci. Biotechnol. Biochem. 2004, 68, 473–475. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, E.; Nomura, N.; Maseda, H.; Otagaki, H.; Nakajima-Kambe, T.; Nakahara, T.; Uchiyama, H. Participation of nitrite reductase in conversion of NO2− to NO3− in a heterotrophic nitrifier, Burkholderia cepacia NH-17, with denitrification activity. Microb. Environ. 2003, 18, 203–209. [Google Scholar] [CrossRef]
- Okada, N.; Nomura, N.; Nakajima-Kambe, T.; Uchiyama, H. Characterization of the aerobic denitrification in Mesorhizobium sp. strain NH-14 in comparison with that in related Rhizobia. Microb. Environ. 2005, 20, 208–215. [Google Scholar] [CrossRef]
- Dilfuza, E. Role of microorganisms in nitrogen cycling in soils. In Soil Nutrients; Minarsari, M., Ed.; Nova Science Publishers Inc.: Hauppauge, NJ, USA, 2011; pp. 159–176. [Google Scholar]
- Fuka, M.M.; Braker, S.H.G.; Philippot, L. Molecular tools to assess the diversity and density of denitrifying bacteria in their habitats. In Biology of the Nitrogen Cycle; Bothe, H., Ferguson, S., Newton, W.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 313–330. [Google Scholar]
- Rana, A.; Pandey, R.K.; Ramakrishnan, B. Enzymology of the nitrogen cycle and bioremediation of toxic nitrogenous compounds. In Smart Bioremediation Technologies; Bhatt, P., Ed.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 45–61. [Google Scholar]
- Laughlin, R.J.; Stevens, R.J. Evidence for fungal dominance of denitrification and codenitrification in a grassland soil. Soil Sci. Soc. Am. J. 2002, 66, 1540–1548. [Google Scholar] [CrossRef]
- McLain, J.E.; Martens, D.A. N2O production by heterotrophic n transformations in a semiarid soil. Appl. Soil Ecol. 2006, 32, 253–263. [Google Scholar] [CrossRef]
- Shoun, H. Denitrification and anaerobic energy producing mechanisms by fungi. Tanpakushitsu Kakusan Koso 2006, 51, 419–429. [Google Scholar]
- Cabello, P.; Roldan, M.D.; Moreno-Vivian, C. Nitrate reduction and the nitrogen cycle in archaea. Microbiology 2004, 150, 3527–3546. [Google Scholar] [CrossRef]
- Philippot, L. Denitrifying genes in bacterial and archaeal genomes. BBA Gene Struct. Expr. 2002, 1577, 355–376. [Google Scholar] [CrossRef]
- Stremińska, M.A.; Błaszczyk, M. The biogeochemical cycle of nitrogen in soils of coniferous forest ecosystems. Post. Mikrobiol. 2004, 43, 235–250. [Google Scholar]
- Gaillard, R.; Duval, B.D.; Osterholz, W.R.; Kucharik, C.J. Simulated effects of soil texture on nitrous oxide emission factors from corn and soybean agroecosystems in Wisconsin. J. Environ. Qual. 2016, 45, 1540–1548. [Google Scholar] [CrossRef] [PubMed]
- Payne, E.G.; Fletcher, T.D.; Cook, P.L.; Deletic, A.; Hatt, B.E. Processes and drivers of nitrogen removal in stormwater biofiltration. Crit. Rev. Environ. Sci. Technol. 2014, 44, 796–846. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, F.; Xiao, R.; Li, Y.; He, Y.; Wu, J. Effects of vegetation on ammonium removal and nitrous oxide emissions from pilot-scale drainage ditches. Aquat. Bot. 2016, 130, 37–44. [Google Scholar] [CrossRef]
- Clough, T.J.; Lanigan, G.J.; de Klein, C.A.M.; Samad, M.S.; Morales, S.E.; Rex, D.; Bakken, L.R.; Johns, C.; Condron, L.M.; Grant, J. Influence of soil moisture on codenitrification fluxes from a urea-affected pasture soil. Sci Rep 2017, 7, 2185. [Google Scholar] [CrossRef]
- Spott, O.; Russow, R.; Stange, C.F. Formation of hybrid N2O and hybrid N2 due to codenitrification: First review of a barely considered process of microbially mediated N-nitrosation. Soil Biol. Biochem. 2011, 43, 1995–2011. [Google Scholar] [CrossRef]
- Long, A.; Heitman, J.; Tobias, C.; Philips, R.; Song, B. Co-occurring anammox, denitrification, and codenitrification in agricultural soils. Appl. Environ. Microbiol. 2013, 79, 168–176. [Google Scholar] [CrossRef]
- Strous, M.; Fuerst, J.A.; Kramer, E.H.M.; Logemann, S.; Muyzer, G.; van de Pas-Schoonen, K.T.; Webb, R.; Kuenen, J.G.; Jetten, M.S.M. Missing lithotroph identified as new Planctomycete. Nature 1999, 400, 446–449. [Google Scholar] [CrossRef]
- Kartal, B.; Kuypers, M.M.M.; Lavik, G.; Schalk, J.; Op den Camp, H.J.; Jetten, M.S.M.; Strous, M. Anammox bacteria disguised as denitrifiers: Nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 2007, 9, 635–642. [Google Scholar] [CrossRef]
- Ali, M.; Oshiki, M.; Awata, T.; Isobe, K.; Kimura, Z.; Yoshikawa, H.; Hira, D.; Kindaichi, T.; Satoh, H.; Fujii, T. Physiological characterization of anaerobic ammonium oxidizing bacterium ‘Candidatus J Ettenia Caeni’. Environ. Microbiol. 2015, 17, 2172–2189. [Google Scholar] [CrossRef]
- Oshiki, M.; Shinyako-Hata, K.; Satoh, H.; Okabe, S. Draft genome sequence of an anaerobic ammonium-oxidizing bacterium ‘Candidatus Brocadia Sinica’. Genome Announc. 2015, 3, e00267-15. [Google Scholar] [CrossRef]
- Strous, M.; Pelletier, E.; Mangenot, S.; Rattei, T.; Lehner, A.; Taylor, M.W.; Horn, M.; Daims, H.; Bartol-Mavel, D.; Wincker, P. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 2006, 440, 790–794. [Google Scholar] [CrossRef]
- Kuenen, J.G. Anammox bacteria: From discovery to application. Nat. Rev. Microbiol. 2008, 6, 320–326. [Google Scholar] [CrossRef]
- van Niftrik, L.; Geerts, W.J.; van Donselaar, E.G.; Humbel, B.M.; Webb, R.I.; Fuerst, J.A.; Verkleij, A.J.; Jetten, M.S.; Strous, M. Linking ultrastructure and function in four genera of anaerobic ammonium-oxidizing bacteria: Cell plan, glycogen storage, and localization of cytochrome c proteins. J. Bacteriol. 2008, 190, 708–717. [Google Scholar] [CrossRef] [PubMed]
- Han, P.; Gu, J.-D. More refined diversity of anammox bacteria recovered and distribution in different ecosystems. Appl. Microbiol. Biotechnol. 2013, 97, 3653–3663. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.C.; Maas, B.; Dapena, A.; van de Pas-Schoonen, K.; van de Vossenberg, J.; Kartal, B.; van Niftrik, L.; Schmidt, I.; Cirpus, I.; Kuenen, J.G. biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 2005, 71, 1677–1684. [Google Scholar] [CrossRef] [PubMed]
- van Teeseling, M.C.F.; Mesman, R.J.; Kuru, E.; Espaillat, A.; Cava, F.; Brun, Y.V.; Van Nieuwenhze, M.S.; Kartal, B.; van Niftrik, L. Anammox planctomycetes have a peptidoglycan cell wall. Nat Commun. 2015, 6, 6878. [Google Scholar] [CrossRef]
- Jetten, M.S.; van Niftrik, L.; Strous, M.; Kartal, B.; Keltjens, J.T.; Op den Camp, H.J. Biochemistry and molecular biology of anammox bacteria. Crit. Rev. Biochem. Mol. Biol. 2009, 44, 65–84. [Google Scholar] [CrossRef]
- Byrne, N.; Strous, M.; Crépeau, V.; Kartal, B.; Birrien, J.-L.; Schmid, M.; Lesongeur, F.; Schouten, S.; Jaeschke, A.; Jetten, M. Presence and activity of anaerobic ammonium-oxidizing bacteria at deep-sea hydrothermal vents. ISME J. 2009, 3, 117–123. [Google Scholar] [CrossRef]
- Jaeschke, A.; Op den Camp, H.J.; Harhangi, H.; Klimiuk, A.; Hopmans, E.C.; Jetten, M.S.; Schouten, S.; Sinninghe Damsté, J.S. 16S rRNA gene and lipid biomarker evidence for anaerobic ammonium-oxidizing bacteria (anammox) in California and Nevada Hot Springs. FEMS Microbiol. Ecol. 2009, 67, 343–350. [Google Scholar] [CrossRef]
- Jin, R.-C.; Yang, G.-F.; Yu, J.-J.; Zheng, P. The Inhibition of the anammox process: A review. Chem. Eng. 2012, 197, 67–79. [Google Scholar] [CrossRef]
- Tang, C.; Zheng, P.; Mahmood, Q.; Chen, J. Effect of substrate concentration on stability of anammox biofilm reactors. J. Cent. South Univ. Technol. 2010, 17, 79–84. [Google Scholar] [CrossRef]
- Zhu, G.; Xia, C.; Shanyun, W.; Zhou, L.; Liu, L.; Zhao, S. Occurrence, activity and contribution of anammox in some freshwater extreme environments. Environ. Microbiol. Rep. 2015, 7, 961–969. [Google Scholar] [CrossRef]
- Gao, D.-W.; Tao, Y. Versatility and application of anaerobic ammonium-oxidizing bacteria. Appl. Microbiol. Biotechnol. 2011, 91, 887–894. [Google Scholar] [CrossRef]
- Pereira, A.D.; Cabezas, A.; Etchebehere, C.; de Chernicharo, C.A.L.; de Araújo, J.C. Microbial communities in anammox reactors: A review. Environ. Technol. Rev. 2017, 6, 74–93. [Google Scholar] [CrossRef]
- Tomaszewski, M.; Cema, G.; Ziembińska-Buczyńska, A. Influence of temperature and pH on the anammox process: A review and meta-analysis. Chemosphere 2017, 182, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Jaroszynski, L.W.; Cicek, N.; Sparling, R.; Oleszkiewicz, J.A. Impact of free ammonia on anammox rates (anoxic ammonium oxidation) in a moving bed biofilm reactor. Chemosphere 2012, 88, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Kang, S.H.; Chung, Y.C.; Ahn, D.H. Factors affecting the activity of anammox bacteria during start up in the continuous culture reactor. Water Sci. Technol. 2007, 55, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Jaroszynski, L.W.; Cicek, N.; Sparling, R.; Oleszkiewicz, J.A. Importance of the operating pH in maintaining the stability of anoxic ammonium oxidation (anammox) activity in moving bed biofilm reactors. Bioresour. Technol. 2011, 102, 7051–7056. [Google Scholar] [CrossRef]
- Dapena-Mora, A.; Fernández, I.; Campos, J.L.; Mosquera-Corral, A.; Méndez, R.; Jetten, M.S.M. Evaluation of activity and inhibition effects on anammox process by batch tests based on the nitrogen gas production. Enzym. Microb. Technol. 2007, 40, 859–865. [Google Scholar] [CrossRef]
- Hu, B.; Rush, D.; van der Biezen, E.; Zheng, P.; van Mullekom, M.; Schouten, S.; Sinninghe Damsté, J.S.; Smolders, A.J.P.; Jetten, M.S.M.; Kartal, B. New Anaerobic, ammonium-oxidizing community enriched from peat soil. Appl. Environ. Microbiol. 2011, 77, 966–971. [Google Scholar] [CrossRef] [PubMed]
- Coolen, M.J.; Abbas, B.; Van Bleijswijk, J.; Hopmans, E.C.; Kuypers, M.M.; Wakeham, S.G.; Sinninghe Damsté, J.S. Putative ammonia-oxidizing crenarchaeota in suboxic waters of the Black Sea: A basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ. Microbiol. 2007, 9, 1001–1016. [Google Scholar] [CrossRef]
- Penton, C.R.; Devol, A.H.; Tiedje, J.M. Molecular evidence for the broad distribution of anaerobic ammonium-oxidizing bacteria in freshwater and marine sediments. Appl. Environ. Microbiol. 2006, 72, 6829–6832. [Google Scholar] [CrossRef] [PubMed]
- Schubert, C.J.; Durisch-Kaiser, E.; Wehrli, B.; Thamdrup, B.; Lam, P.; Kuypers, M.M. Anaerobic ammonium oxidation in a tropical freshwater system (Lake Tanganyika). Environ. Microbiol. 2006, 8, 1857–1863. [Google Scholar] [CrossRef]
- Humbert, S.; Tarnawski, S.; Fromin, N.; Mallet, M.-P.; Aragno, M.; Zopfi, J. Molecular detection of anammox bacteria in terrestrial ecosystems: Distribution and diversity. ISME J. 2010, 4, 450–454. [Google Scholar] [CrossRef] [PubMed]
- Raymond, J.; Siefert, J.L.; Staples, C.R.; Blankenship, R.E. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004, 21, 541–554. [Google Scholar] [CrossRef]
- Herridge, D.F.; Peoples, M.B.; Boddey, R.M. Global inputs of biological nitrogen fixation in agricultural systems. Plant Soil 2008, 311, 1–18. [Google Scholar] [CrossRef]
- Łyszcz, M.; Gałązka, A. The process of biological fixation of atmospheric nitrogen. In Habitat and Agrotechnical Conditions of Plant Production in Poland; Studies and Reports IUNG-PIB; Institute of Soil Science and Plant Cultivation, National Research Institute: Puławy, Poland, 2016; pp. 59–70. [Google Scholar]
- Cheng, Q. Perspectives in biological nitrogen fixation research. J. Integr. Plant Biol. 2008, 50, 786–798. [Google Scholar] [CrossRef]
- Król, M.J. A Azospirillum—Associative Free Nitrogen-Fixing Bacteria; Institute of Soil Science and Plant Cultivation, National Research Institute: Puławy, Poland, 2006. [Google Scholar]
- Natywa, M.; Selwet, M.; Ambroży, K.; Pociejowska, M. Effect of nitrogen fertilization and irrigation on the number of Azotobacter bacteria in the soil under maize cultivation at various stages of plant development. Pol. J. Agric. 2013, 14, 53–58. [Google Scholar]
- Peters, J.W.; Boyd, E.S.; Hamilton, T.L.; Rubio, L.M. Biochemistry of Mo-nitrogenase. In Nitrogen Cycling in Bacteria: Molecular Analysis; Moir, J.W.B., Ed.; Caister Academic Press: Norfolk, UK, 2011; pp. 59–99. [Google Scholar]
- Rees, D.C.; Akif Tezcan, F.; Haynes, C.A.; Walton, M.Y.; Andrade, S.; Einsle, O.; Howard, J.B. Structural basis of biological nitrogen fixation. Philos. Trans. R. Soc. A 2005, 363, 971–984. [Google Scholar] [CrossRef] [PubMed]
- Siemann, S.; Schneider, K.; Dröttboom, M.; Müller, A. The Fe-only nitrogenase and the Mo nitrogenase from Rhodobacter capsulatus. Eur. J. Biochem. 2002, 269, 1650–1661. [Google Scholar] [CrossRef]
- Wielbo, J.; Skorupska, A. Evolution of the symbiotic system of Rhizobium-legumes. Post. Mikrobiol. 2003, 42, 263–283. [Google Scholar]
- Mus, F.; Alleman, A.B.; Pence, N.; Seefeldt, L.C.; Peters, J.W. Exploring the alternatives of biological nitrogen fixation. Metallomics 2018, 10, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Nordult, S.; Ureta, A. Evidence for conformational protection of nitrogenase in Gluconacetobacter diazotrophicus. J. Bacteriol. 2002, 184, 5805–5809. [Google Scholar]
- Egamberdieva, D.; Kucharova, Z. Cropping effects on microbial population and nitrogenase activity in saline arid soil. Turk. J. Biol. 2008, 32, 85–90. [Google Scholar]
- Vitousek, P.M.; Cassman, K.; Cleveland, C.; Crews, T.; Field, C.B.; Grimm, N.B.; Howarth, R.W.; Marino, R.; Martinelli, L.; Rastetter, E.B. Towards an ecological understanding of biological nitrogen fixation. In The Nitrogen Cycle at Regional to Global Scales; Boyer, E.W., Howarth, R.W., Eds.; Springer: Dordrecht, The Netherlands, 2002; pp. 1–45. [Google Scholar]
- Niewiadomska, A.; Sulewska, H.; Wolna-Maruwka, A.; Ratajczak, K.; Głuchowska, K.; Waraczewska, Z.; Budka, A. An assessment of the influence of co-inoculation with endophytic bacteria and rhizobia, and the influence of PRP SOL and PRP EBV fertilisers on the microbial parameters of soil and nitrogenase activity in yellow lupine (Lupinus luteus L.) cultivation. Pol. J. Environ. Stud. 2018, 27, 2687–2702. [Google Scholar] [CrossRef]
- Liu, Y.; Wu, L.; Baddeley, J.A.; Watson, C.A. Models of biological nitrogen fixation of legumes. A review. Agron. Sust. Dev. 2011, 31, 155–172. [Google Scholar] [CrossRef]
- Long, S.R. Genes and signals in the Rhizobium-legume symbiosis. Plant Physiol. 2001, 125, 69–72. [Google Scholar] [CrossRef]
- Salvagiotti, F.; Cassman, K.G.; Specht, J.E.; Walters, D.T.; Weiss, A.; Dobermann, A. Nitrogen uptake, fixation and response to fertilizer n in soybeans: A review. Field Crop. Res. 2008, 108, 1–13. [Google Scholar] [CrossRef]
- Sellstedt, A.; Richau, K.H. Aspects of nitrogen-fixing Actinobacteria in particular free-living and symbiotic Frankia. FEMS Microbiol. Lett. 2013, 342, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, T.; Pawlowski, K. Frankia and actinorhizal plants: Symbiotic nitrogen fixation. In Rhizotrophs: Plant Growth Promotion to Bioremediation; Mehanz, S., Ed.; Springer: Singapore, 2017; pp. 237–261. [Google Scholar]
- Fauvart, M.; Michiels, J. Rhizobial secreted proteins as determinants of host specificity in the Rhizobium–legume symbiosis. FEMS Microbiol. Lett. 2008, 285, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Mus, F.; Crook, M.B.; Garcia, K.; Garcia Costas, A.; Geddes, B.A.; Kouri, E.D.; Paramasivan, P.; Ryu, M.-H.; Oldroyd, G.E.D.; Poole, P.S. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl. Environ. Microbiol. 2016, 82, 3698–3710. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Jurkiewicz, A.; Fukai, E.; Quistgaard, E.M.; Albrektsen, A.S.; James, E.K.; Thirup, S.; Stougaard, J. LysM domains mediate lipochitin–oligosaccharide recognition and nfr genes extend the symbiotic host range. EMBO J. 2007, 26, 3923–3935. [Google Scholar] [CrossRef] [PubMed]
- Sprent, J.I.; James, E.K. Legume evolution: Where do nodules and mycorrhizas fit in? Plant Physiol. 2007, 144, 575–581. [Google Scholar] [CrossRef]
- Boivin, S.; Fonouni-Farde, C.; Frugier, F. How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 2016, 7, 1240. [Google Scholar] [CrossRef]
- Verma, R.; Annapragada, H.; Katiyar, N.; Shrutika, N.; Das, K.; Murugesan, S. Rhizobium. In Beneficial Microbes in Agro-Ecology; Amaresan, N., Annapurna, K., Kumar, K., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 37–54. [Google Scholar]
- Chandra, S.; Choure, K.; Dubey, R.C.; Maheshwari, D.K. Rhizosphere competent Mesorhizobium loti MP6 induces root hair curling, inhibits Sclerotinia sclerotiorum and enhances growth of indian mustard (Brassica campestris). Braz. J. Microbiol. 2007, 38, 124–130. [Google Scholar] [CrossRef]
- Tamiru, G.; Muleta, D. The effect of rhizobia isolates against black root rot disease of faba bean (Vicia faba L.) caused by Fusarium solani. Open Agric. J. 2018, 12, 131–147. [Google Scholar] [CrossRef]
- Shaukat, S.S.; Siddiqui, I.A. The influence of mineral and carbon sources on biological control of charcoal rot fungus, Macrophomina phaseolina by fluorescent Pseudomonas in tomato. Lett. Appl. Microbiol. 2003, 36, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Król, M.J.; Zielewicz-Dukowska, J. Genetic aspects of N2 binding of Azospirillum bacteria. Post. Mikrobiol 2005, 44, 47–56. [Google Scholar]
- Martyniuk, S. The importance of the biological process of atmospheric nitrogen fixation in ecological agriculture. J. Res. Appl. Agr. Eng. 2008, 53, 9–14. [Google Scholar]
- Svistoonoff, S.; Hocher, V.; Gherbi, H. Actinorhizal root nodule symbioses: What is signalling telling on the origins of nodulation? Curr. Opin. Plant Biol. 2014, 20, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Abrol, Y.P.; Raghuram, N.; Sachdev, M.S. Agricultural Nitrogen Use and Its Environmental Implications; IK International Pvt Ltd: New Dehli, India, 2007. [Google Scholar]
- Meisinger, J.J.; Jokela, W.E. Mmonia Volatilization from Dairy and Poultry Manure: Managing Nutrients and Pathogens from Animal Agriculture; NRAES-130; Natural Resource, Agriculture, and Engineering Service: Ithaca, NY, USA, 2000; pp. 334–354. [Google Scholar]
- Jones, K.M.; Kobayashi, H.; Davies, B.W.; Taga, M.E.; Walker, G.C. How rhizobial symbionts invade plants: The Sinorhizobium–Medicago model. Nat. Rev. Microbiol. 2007, 5, 619–633. [Google Scholar] [CrossRef]
- Sommer, S.G.; Hutchings, N.J. Ammonia emission from field applied manure and its reduction—Invited paper. Eur. J. Agron. 2001, 15, 1–15. [Google Scholar] [CrossRef]
- Yang, J.; Jiao, Y.; Yang, W.Z.; Gu, P.; Bai, S.G.; Liu, L.J. Review of methods for determination of ammonia volatilization in farmland. In Proceedings of the IOP Conference Series: Earth and Environmental Science; IOP Publishing: Bristol, UK, 2018; Volume 113. [Google Scholar]
- Zhou, W.; Tian, Y.H.; Cao, Y.S.; Yin, B. A comparative study on two methods for determination of ammonia volatilization. Acta Pedofil. Sin. 2011, 48, 1090–1095. [Google Scholar]
- Ghaly, A.E.; Ramakrishnan, V.V. Nitrogen sources and cycling in the ecosystem and its role in air, water and soil pollution: A critical review. J. Pollut. Eff. Control 2015, 3, 1–26. [Google Scholar]
- Randall, G.W.; Vetsch, J.A.; Huffman, J.R. Corn production on a subsurface-drained mollisol as affected by time of nitrogen application and nitrapyrin. Agron. J. 2003, 95, 1213–1219. [Google Scholar] [CrossRef]
- Bolan, N.S.; Hedley, M.J. Role of Carbon, Nitrogen, and Sulfur Cycles in Soil Acidification. In Handbook of Soil Acidity; Rengel, Z., Ed.; Marcel Dekker Inc.: New York, NY, USA, 2003; pp. 29–56. [Google Scholar]
- Davis, R.L.; Patton, J.J.; Teal, R.K.; Tang, Y.; Humphreys, M.T.; Mosali, J.; Girma, K.; Lawles, J.W.; Moges, S.M.; Malapati, A. Nitrogen balance in the magruder plots following 109 years in continuous winter wheat. J. Plant Nutr. 2003, 26, 1561–1580. [Google Scholar] [CrossRef]
- Ascott, M.J.; Gooddy, D.C.; Wang, L.; Stuart, M.E.; Lewis, M.A.; Ward, R.S.; Binley, A.M. Global patterns of nitrate storage in the vadose zone. Nat. Commun. 2017, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, L.M.; Cuttle, S.P.; Crabtree, B.; Hopkins, A.; Shepherd, A.; Scholefield, D.; Del Prado, A. Cost effectiveness of nitrate leaching mitigation measures for grassland livestock systems at locations in England and Wales. Sci. Total Environ. 2011, 409, 1104–1115. [Google Scholar] [CrossRef]
- Delgado, J.A.; Shaffer, M.J.; Lal, H.; McKinney, S.P.; Gross, C.M.; Cover, H. Assessment of nitrogen losses to the environment with a nitrogen trading tool (NTT). Comput. Electron. Agric. 2008, 63, 193–206. [Google Scholar] [CrossRef]
- Wang, M.; Pendall, E.; Fang, C.; Li, B.; Nie, M. A global perspective on agroecosystem nitrogen cycles after returning crop residue. Agric. Ecosyst. Environ. 2018, 266, 49–54. [Google Scholar] [CrossRef]
- Roberts, T.; Kirkpatrick, W.; County, D.; Slaton, N.; Norman, R. Estimating Nutrient Removal for Row Crops Grown in Arkansas; Agriculture and Natural Resources; University of Arkansas Cooperative Extension Service Printing Services: Fayetteville, AR, USA, 2015. [Google Scholar]
- FAO. World Fertilizer Trends and Outlook to 2020; Summary Report; FAO: Rome, Italy, 2017. [Google Scholar]


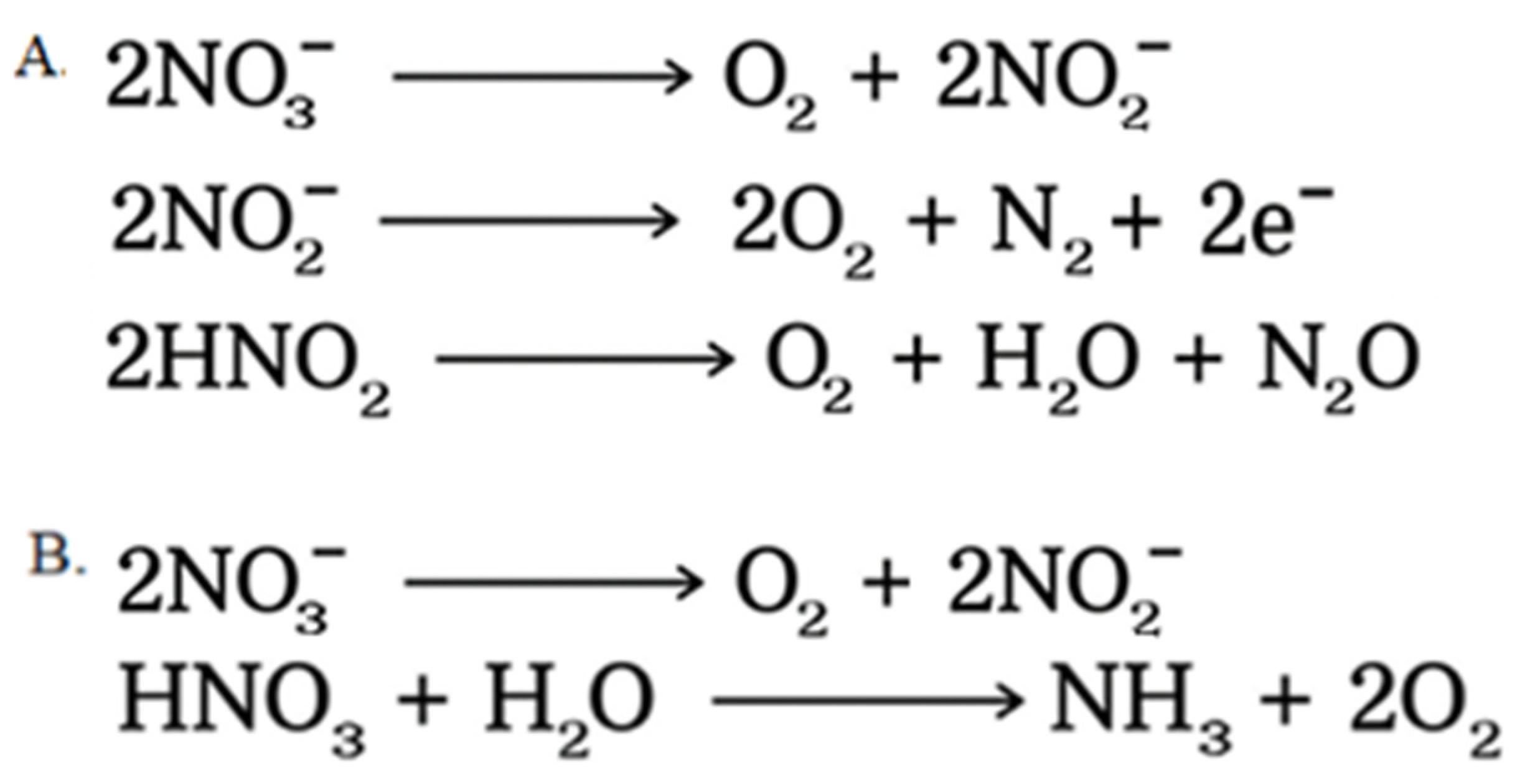


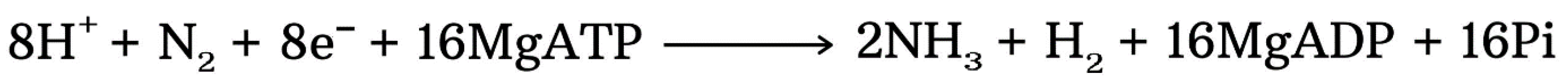

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grzyb, A.; Wolna-Maruwka, A.; Niewiadomska, A. The Significance of Microbial Transformation of Nitrogen Compounds in the Light of Integrated Crop Management. Agronomy 2021, 11, 1415. https://doi.org/10.3390/agronomy11071415
Grzyb A, Wolna-Maruwka A, Niewiadomska A. The Significance of Microbial Transformation of Nitrogen Compounds in the Light of Integrated Crop Management. Agronomy. 2021; 11(7):1415. https://doi.org/10.3390/agronomy11071415
Chicago/Turabian StyleGrzyb, Aleksandra, Agnieszka Wolna-Maruwka, and Alicja Niewiadomska. 2021. "The Significance of Microbial Transformation of Nitrogen Compounds in the Light of Integrated Crop Management" Agronomy 11, no. 7: 1415. https://doi.org/10.3390/agronomy11071415
APA StyleGrzyb, A., Wolna-Maruwka, A., & Niewiadomska, A. (2021). The Significance of Microbial Transformation of Nitrogen Compounds in the Light of Integrated Crop Management. Agronomy, 11(7), 1415. https://doi.org/10.3390/agronomy11071415






