Determination of Genetic Distance, Genome Size and Chromosome Numbers to Support Breeding in Ornamental Lavandula Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genome Size
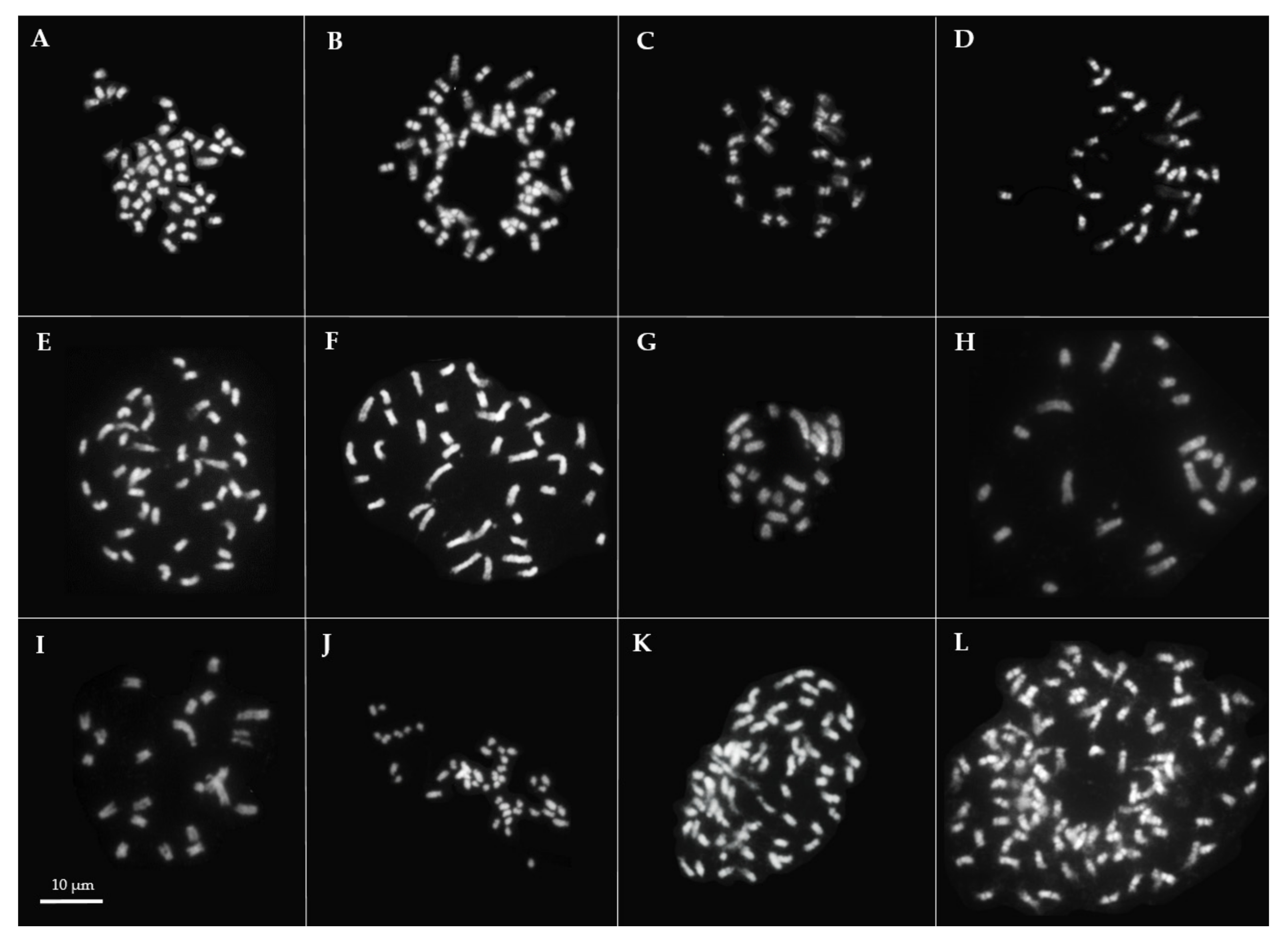
2.3. Chromosome Number
2.4. AFLP
2.5. Statistical Analysis of AFLP Data
3. Results
3.1. Genetic Distances, Genome Sizes and Chromosome Numbers in the Lavandula Progenitor Collection
3.2. Genetic Distances, Genome Sizes and Chromosome Numbers in Lavandula Hybrids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moja, S.; Guitton, Y.; Nicolè, F.; Legendre, L.; Pasquier, B.; Upson, T.; Jullien, F. Genome size and plastid trnK-matK markers give new insights into the evolutionary history of the genus Lavandula L. Plant Biosyst. 2016, 150, 1216–1224. [Google Scholar] [CrossRef]
- Lis-Balchin, M. Lavender: The Genus Lavandula; CRC Press: London, UK, 2002; p. 296. [Google Scholar] [CrossRef]
- Upson, T.; Andrews, S. The Genus Lavandula; Royal Botanic Gardens Kew: Richmond, UK, 2004; p. 442. [Google Scholar]
- McNaughton, V. Lavender: The Grower’s Guide; Blooming Books: Caringbah, Australia, 2000; p. 180. [Google Scholar]
- Wunderlich, R. Ein Vorschlag zu einer natürlichen Gliederung der Labiaten auf Grund der Pollenkörner, der Samenentwicklung und des reifen Samens. Oesterreichische Bot. Z. 1967, 114, 383–483. [Google Scholar] [CrossRef]
- Upson, T. Systematics of the Genus Lavandula L. (Lamiaceae). Ph.D. Thesis, University of Reading, Reading, UK, 1997. [Google Scholar]
- Garcia, J. Contribuição para o estudo cário-sistemático do género Lavandula L. Bol. Soc. Broteriana 1942, 13, 183–193. [Google Scholar]
- Buyukli, M. On karyotype and polyploid series in Lavandula L. Citol. Genet. 1970, 4, 268–274. [Google Scholar]
- Suárez-Cervera, M.; Seoane-Camba, J. Taxonomía numérica de algunas especies de Lavandula L., basada en caracteres morfológicos, cariológicos y palinológicos. An. Jard. Bot. Madr. 1986, 42, 395–409. [Google Scholar]
- Paton, A.; Springate, D.; Suddee, S.; Otieno, D.; Grayer, R.; Harley, M.; Willis, F.; Simmonds, M.; Powell, M.; Savolainen, V. Phylogeny and evolution of basils and allies (Ocimeae, Labiatae) based on three plastid DNA regions. Mol. Phylogenet. Evol. 2004, 31, 277–299. [Google Scholar] [CrossRef]
- Ez Zoubi, Y.; Bousta, D.; Farah, A. A Phytopharmacological review of a Mediterranean plant: Lavandula stoechas L. Clin. Phytosci. 2020, 6, 9. [Google Scholar] [CrossRef]
- Darlington, C.; Wylie, A. Chromosome Atlas of Flowering Plants; George Allen and Unwin Ltd.: London, UK, 1955; p. 519. [Google Scholar]
- Rice, A.; Glick, L.; Abadi, S.; Einhorn, M.; Kopelman, N.; Salman-Minkov, A.; Mayzel, J.; Chay, O.; Mayrose, I. The Chromosome Counts Database (CCDB)—A community resource of plant chromosome numbers. New Phytol. 2015, 206, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Stanev, S.; Zagorcheva, T.; Atanassov, I. Lavender cultivation in Bulgaria—21st century developments, breeding challenges and opportunities. Bulg. J. Agric. Sci. 2016, 22, 584–590. [Google Scholar]
- Urwin, N. Lavender Breeding for Commercial Yield©. Comb. Proc. Int. Plant Propagator Soc. 2008, 58, 78–84. [Google Scholar]
- Van Huylenbroeck, J.; Eeckhaut, T.; Leus, L.; Van Laere, K.; Dhooghe, E. Bridging the gap: Tools for interspecific and intergeneric hybridization in ornamentals. In Proceedings of the XXVI International Eucarpia Symposium Section Ornamentals: Editing Novelty, Erfurt, Germany, 1–4 September 2019; Volume 1283, pp. 161–168. [Google Scholar] [CrossRef]
- Akbarzadeh, M.; Van Laere, K.; Leus, L.; De Riek, J.; Van Huylenbroeck, J.; Werbrouck, S.P.O.; Dhooghe, E. Can knowledge of genetic distances, genome sizes and chromosome numbers support breeding programs in hardy geraniums? Genes 2021, 12, 730. [Google Scholar] [CrossRef] [PubMed]
- Denaeghel, H.; Van Laere, K.; Leus, L.; Van Huylenbroeck, J.; Van Labeke, M.C. Interspecific hybridization in Sarcococca supported by analysis of ploidy level, genome size and genetic relationships. Euphytica 2017, 213, 149. [Google Scholar] [CrossRef]
- Tychonievich, J.; Warner, R. Interspecific crossability of selected Salvia species and potential use for crop improvement. J. Am. Soc. Hortic. Sci. 2011, 136, 41–47. [Google Scholar] [CrossRef] [Green Version]
- Kardos, J.; Robacker, C.; Dirr, M.; Rinehart, T. Production and verification of Hydrangea macrophylla × H. angustipetala hybrids. Hortic. Sci. 2009, 44, 1534–1537. [Google Scholar] [CrossRef] [Green Version]
- Handa, T.; Kita, K.; Wongsawad, P.; Kurashige, Y.; Yukawa, T. Molecular phylogeny-assisted breeding of ornamentals. J. Crop Improv. 2006, 17, 51–68. [Google Scholar] [CrossRef]
- Galbraith, D.; Harkins, K.; Maddox, J.; Ayres, N.; Sharma, D.; Firoozabady, E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 1983, 220, 1049–1051. [Google Scholar] [CrossRef]
- Doležel, J.; Sgorbati, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Lysak, M.; Dolezel, J. Estimation of nuclear DNA content in Sesleria (Poaceae). Caryologia 1998, 51, 123–132. [Google Scholar] [CrossRef]
- Greilhuber, J.; Dolezel, J.; Lysák, M.; Bennett, M. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Ann. Bot. 2005, 95, 255–260. [Google Scholar] [CrossRef]
- Kirov, I.; Divashuk, M.; Van Laere, K.; Soloviev, A.; Khrustaleva, L. An easy “SteamDrop” method for high quality plant chromosome preparation. Mol. Cytogenet. 2014, 7, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirov, I.; Khrustaleva, L.; Van Laere, K.; Soloviev, A.; Meeus, S.; Romanov, D.; Fesenko, I. DRAWID: User-friendly java software for chromosome measurements and idiogram drawing. Comp. Cytogenet. 2017, 11, 747–757. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Urwin, N.; Horsnell, J.; Moon, T. Generation and characterisation of colchicine-induced autotetraploid Lavandula angustifolia. Euphytica 2007, 156, 257–266. [Google Scholar] [CrossRef]
- Vinot, M.; Bouscary, A. Etudes sur la Lavande. VI. Les hybrides. Recherches 1971, 18, 29–44. [Google Scholar]
- Sattler, M.; Carvalho, C.; Clarindo, W. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Van Laere, K.; França, S.; Vansteenkiste, H.; Van Huylenbroeck, J.; Steppe, K.; Van Labeke, M.C. Influence of ploidy level on morphology, growth and drought susceptibility in Spathiphyllum wallisii. Acta Physiol. Plant. 2011, 33, 1149–1156. [Google Scholar] [CrossRef]

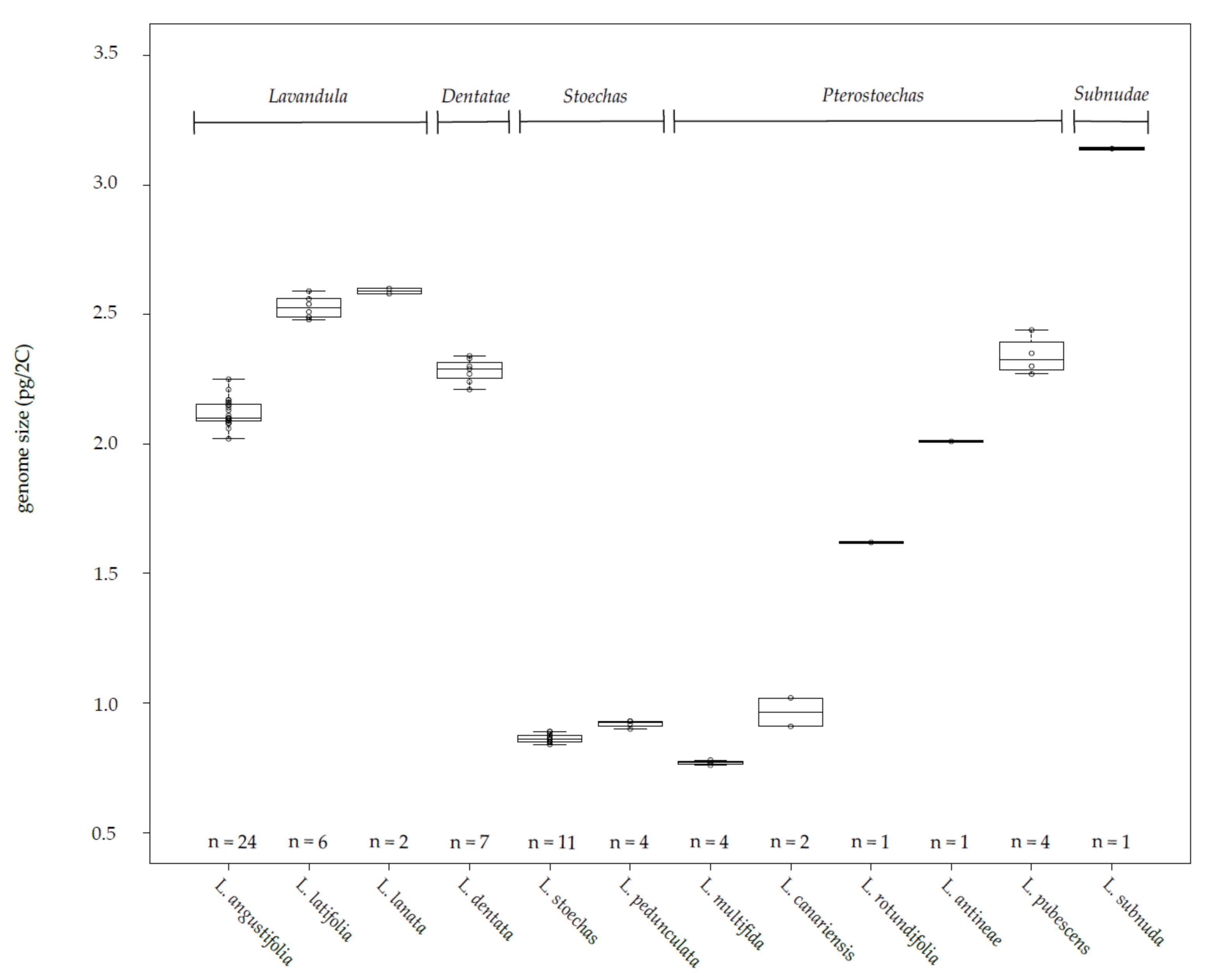

| Progenitors | Progenitor Chromosome Number (2n) |
Progenitor Genome Size (pg 2C−1) | Hybrid Genotype (Code) | Hybrid Chromosome Number (2n) | Hybrid Genome Size (pg 2C−1) |
|---|---|---|---|---|---|
| L. angustifolia × L. latifolia | 42, 48, 50, 54 48, 50, 54 | 2.02 ± 0.09–2.25 ± 0.08 2.48 ± 0.05–2.59 ± 0.07 | L. × intermedia ‘Silver’ (L73) a | 50 | 2.13 ± 0.04 (M) |
| L. × intermedia ‘Old English Group’ (L29) | 50 | 2.36 ± 0.09 (M) | |||
| L. × intermedia ‘Futura’ (L86) | n.a. | 2.28 ± 0.07 (M) | |||
| L. × intermedia ‘Nizza’ (L1) | 50 | 2.32 ± 0.06 (M) | |||
| L. × intermedia ‘Edelweiss’ (L31) | n.a. | 2.37 ± 0.08 (M) | |||
| L. × intermedia ‘Dutch’ (L72) | n.a. | 2.35 ± 0.04 (M) | |||
| L. × intermedia ‘Heavenly Angel’ (L30) | 100 | 4.80 ± 0.06 (T) | |||
| L. angustifolia × L. lanata | 42, 48, 50, 54 50, 54 | 2.02 ± 0.09–2.25 ± 0.08 2.58 ± 0.07–2.60 ± 0.05 | L. × chaytorae ‘Gorgeous’ (L8) | 50 | 2.33 ± 0.09 (M) |
| L. × chaytorae ‘Richard Gray’ (L22) | 50 | 2.30 ± 0.06 (M) | |||
| L. latifolia × L. lanata | 48, 50, 54 50, 54 | 2.48 ± 0.05–2.59 ± 0.07 2.58 ± 0.07–2.60 ± 0.05 | L. × losae (L2) | 50 | 2.55 ± 0.03 (M) |
| L. dentata × L. lanata | 44, 42, 45 50, 54 | 2.21 ± 0.05–2.34 ± 0.05 2.58 ± 0.07–2.60 ± 0.05 | L. × ginginsii ‘Goodwin Creek Grey’ (L9) | 48 | 2.43 ± 0.04 (M) |
| L. dentata × L. latifolia | 44, 42, 45 48, 50, 54 | 2.21 ± 0.05–2.34 ± 0.05 2.48 ± 0.05–2.59 ± 0.07 | L. × allardii ‘African Pride’ (L11) | 48 | 2.49 ± 0.05 (M) |
| L. × heterophylla ‘Devantville Cuche’ (L17) | 48 | 2.42 ± 0.05 (M) | |||
| L. × heterophylla ‘Meerlo’ (L75) | n.a. | 2.43 ± 0.05 (M) | |||
| L. × heterophylla ‘Big Boy James’ (L13) | 66 | 3.50 ± 0.06 (M) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Van Oost, E.; Leus, L.; De Rybel, B.; Van Laere, K. Determination of Genetic Distance, Genome Size and Chromosome Numbers to Support Breeding in Ornamental Lavandula Species. Agronomy 2021, 11, 2173. https://doi.org/10.3390/agronomy11112173
Van Oost E, Leus L, De Rybel B, Van Laere K. Determination of Genetic Distance, Genome Size and Chromosome Numbers to Support Breeding in Ornamental Lavandula Species. Agronomy. 2021; 11(11):2173. https://doi.org/10.3390/agronomy11112173
Chicago/Turabian StyleVan Oost, Ewout, Leen Leus, Bert De Rybel, and Katrijn Van Laere. 2021. "Determination of Genetic Distance, Genome Size and Chromosome Numbers to Support Breeding in Ornamental Lavandula Species" Agronomy 11, no. 11: 2173. https://doi.org/10.3390/agronomy11112173
APA StyleVan Oost, E., Leus, L., De Rybel, B., & Van Laere, K. (2021). Determination of Genetic Distance, Genome Size and Chromosome Numbers to Support Breeding in Ornamental Lavandula Species. Agronomy, 11(11), 2173. https://doi.org/10.3390/agronomy11112173






