Genetic Variation and Phylogeny of Wabisuke Camellias by Amplified Fragment Length Polymorphism (AFLP) Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. AFLP Analysis
3. Results
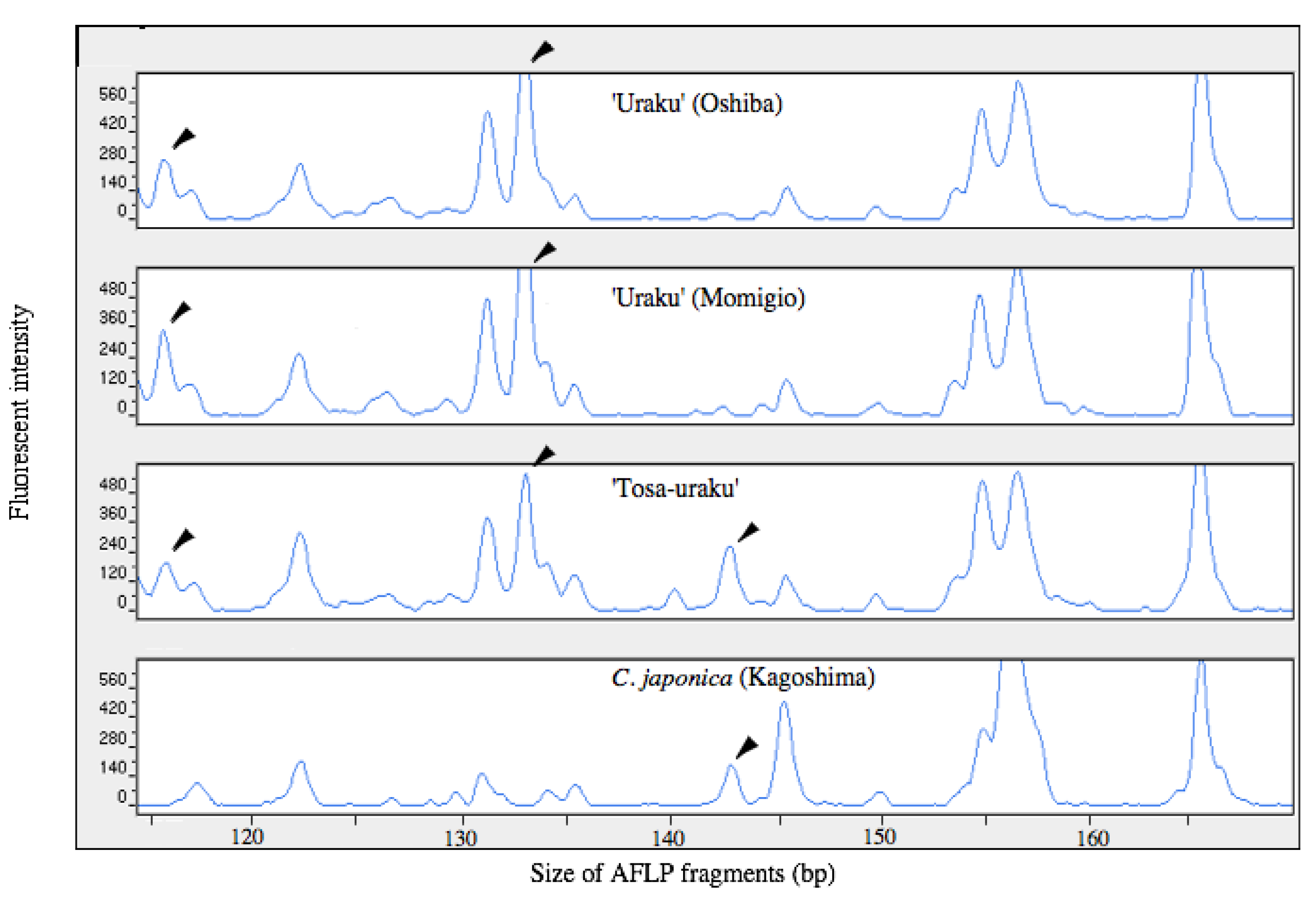
3.1. Genetic Identity of Two Old ‘Uraku’ Accessions
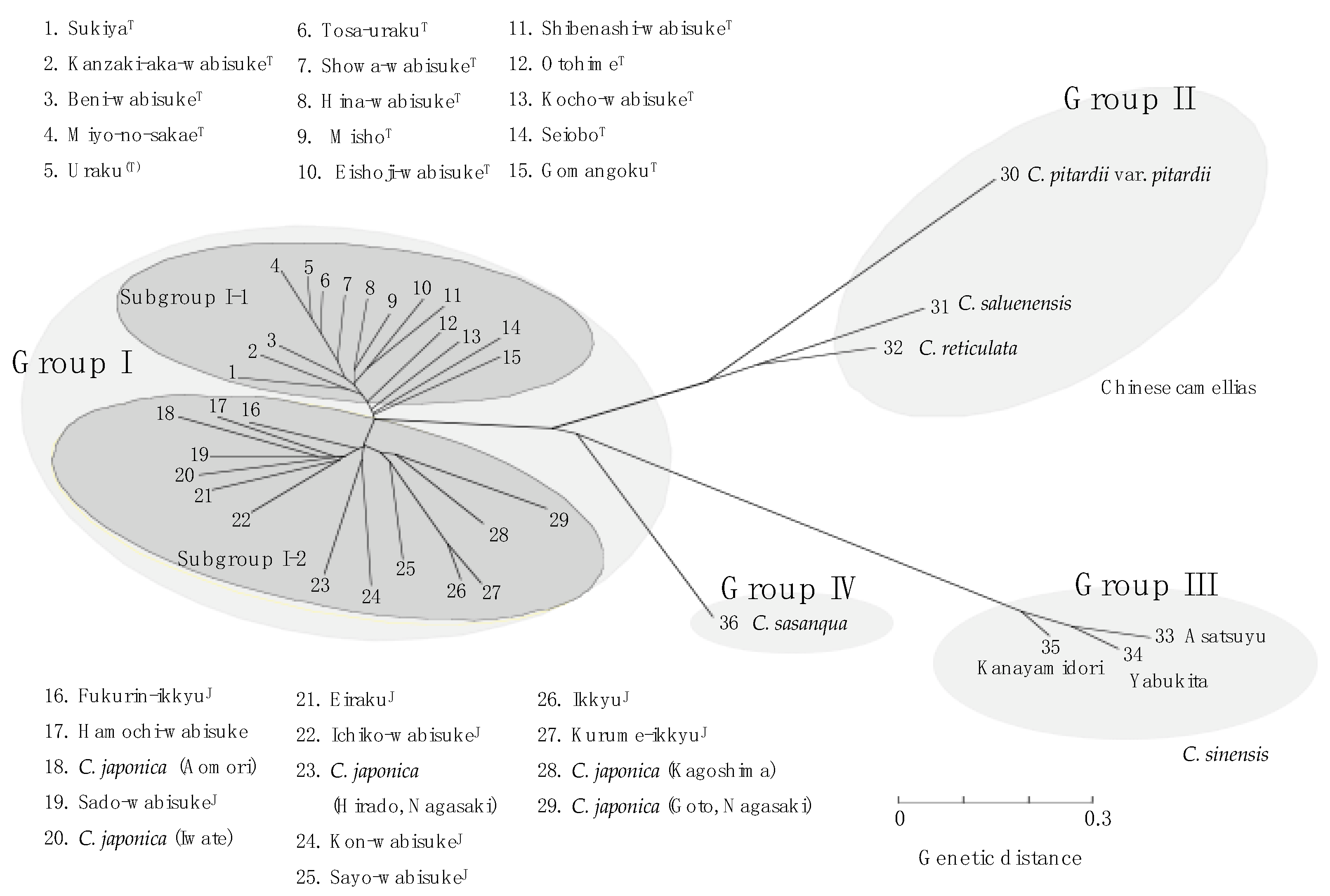
3.2. Genetic Variation in Wabisuke Camellias
4. Discussion
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kirino, S. Chaseki no Hana. Tsubaki to Wabisuke; Bunka-Shuppan-Kyoku: Tokyo, Japan, 1986; pp. 36–43. (In Japanese) [Google Scholar]
- Savige, T.J. Uraku. In The International Camellia Register; The International Camellia Society: Wirlinga, NSW, Australia, 1993; Volume 2, p. 1904. [Google Scholar]
- Miyajima, I.; Ozaki, Y.; Itateyama, M.; Ikematsu, R.; Okubo, H.; Goi, M. Genetic identity of old Camellia ‘Uraku’ trees existing in Japan. J. Jpn. Soc. Hortic. Sci. 2001, 70, 366–371. [Google Scholar] [CrossRef]
- Tanaka, T.; Kirino, S.; Hakoda, N.; Fujieda, K.; Mizutani, T. Studies on the origin of Camellia wabiske. Proc. Sch. Agric. Kyushu Tokai Univ. 2001, 20, 1–7. [Google Scholar]
- Tamura, T.; Hagiya, K.; Hakoda, N. Tsubaki-zoku. In Engei-Shokubutsu-Daijiten; Aiga, T., Ed.; Shogakukan: Tokyo, Japan, 1989; pp. 267–275. (In Japanese) [Google Scholar]
- Sakata, Y. Studies on the flower colours in the genus Camellia, with special reference to the phylogenies of the genus. Bull. Fac. Agr. Kagoshima Univ. 1988, 38, 9–62. [Google Scholar]
- Kato, M.; Simura, T. Cytological studies on Camellia species II. The karyotype analysis in C. sinensis and C. wabiske. Jpn. J. Breed. 1971, 21, 265–268. [Google Scholar] [CrossRef][Green Version]
- Tanikawa, N.; Onozaki, T.; Nakayama, M.; Shibata, M. Maternal origin of ‘Tarokaja’ and other wabisuke camellia cultivars indicated by chloroplast DNA variation. J. Jpn. Soc. Hortic. Sci. 2010, 79, 77–83. [Google Scholar] [CrossRef][Green Version]
- Tanikawa, N.; Yoshida, K.; Kondo, T.; Mizuno, T.; Iwashina, T.; Nakayama, M. Urakunoside, a new tetraglycosyl kaempferol from petals of the wabisuke camellia cv. Tarokaja. Biosci. Biotechnol. Biochem. 2011, 75, 2046–2048. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Perera, L.; Russell, J.R.; Provan, J.; McNicol, J.W.; Powell, W. Exaluating genetic relationships between indigenous coconut (Cocos nucifera L.) accessions from Sri Lanka by means of AFLP profiling. Theor. Appl. Genet. 1998, 96, 545–550. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, S.; Yonemori, K.; Sato, A.; Yamada, M.; Sugiura, A. Analysis of the genetic relationships among pollination-constant and non-astringent (PCNA) cultivars of persimmon (Diospyros kaki Thunb.) from Japan and China using amplified fragment length polymorphism (AFLP). J. Jpn. Soc. Hortic. Sci. 2000, 69, 665–670. [Google Scholar] [CrossRef]
- Ellis, R.P.; McNicol, J.W.; Baird, E.; Booth, A.; Lawrence, P.; Thomas, B.; Powell, W. The use of AFLPs to examine genetic relatedness in barley. Mol. Breed. 1997, 3, 359–369. [Google Scholar] [CrossRef]
- Rabey, H.A.; Al-Malki, A.L.; Abulnaja, K.O.; Ebrahim, M.K.; Kumosani, T.; Khan, J.A. Phylogeny of ten species of the genus Hordeum L. as revealed by AFLP markers and seed storage protein electrophoresis. Mol. Biol. Rep. 2013, 41, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Yildizdogan, Z.; Ikten, C.; Mutlu, N.; Toker, C. Phylogeny of Lens taxa based on morphological and AFLP markers. J. Biotechnol. 2015, 208, S30. [Google Scholar] [CrossRef]
- Drabkova, L.Z.; Pospiskova, M.; Businsky, R. Phylogeny and infrageneric delimitation in Spiraea (Rosaceae) inferred from AFLP markers and a comparison with morphology. Bot. J. Linn. Soc. 2017, 185, 525–541. [Google Scholar] [CrossRef]
- Kobayashi, N.; Horikoshi, T.; Katsuyama, H.; Handa, T.; Tankayanagi, K. A simple and efficient DNA extraction method for plants, especially woody plants. Plant Tiss. Cult. Biotechnol. 1998, 4, 76–80. [Google Scholar]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felesenstein, J. PHYLIP (Phylogeny Inference Package) Version 3.5c; Distributed by author; Department of Genetics, University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Japan Camellia Society (Ed.) Nihon-Tsubaki-Sazanka-Meikan; Seibundo-Shinkosha: Tokyo, Japan, 1998; p. 359. (In Japanese) [Google Scholar]
- Yokoyama, Y.; Kirino, S. Nippon-no-Chinka; Tanko-Sha: Kyoto, Japan, 2005; p. 421. (In Japanese) [Google Scholar]
- Kirino, S. Tsubaki; Gakken: Tokyo, Japan, 2005; p. 192. (In Japanese) [Google Scholar]
- Kondo, K. Cytological studies in cultivated species of Camellia. Ph.D. Thesis, University of North Carolina, Chapel Hill, NC, USA, 1975. [Google Scholar]
| Species/Cultivars | Information on Accession Origin/Collection |
|---|---|
| Wabisuke camellias | |
| Ohae-uraku | ‘Uraku’ from Saito City, Miyazaki Prefecture, Japan |
| Momigio-uraku | ‘Uraku’ from Saito City, Miyazaki Prefecture, Japan |
| ‘Beni-wabisuke’ | Synonym. Kuro-wabisuke, Kyushu University (KU) collection |
| ‘Eiraku’ | KU collection |
| ‘Eishiji-wabisuke’ | KU collection |
| ‘Fukurin-ikkyu’ | KU collection |
| ‘Goman-Goku’ | KU collection |
| ‘Hamochi-wabisuke’ | KU collection |
| ‘Hina-wabisuke’ | KU collection |
| ‘Ichiko-wabisuke’ | KU collection |
| ‘Ikkyu’ | KU collection |
| ‘Kanzaki-aka-wabisuke’ | KU collection |
| ‘Kocho-wabisuke’ | KU collection |
| ‘Kon-wabisuke’ | KU collection |
| ‘Kurume-ikkyu’ | KU collection |
| ‘Misho’ | KU collection |
| ‘Miyo-no-sakae’ | KU collection |
| ‘Otohime’ | KU collection |
| ‘Sado-wabisuke’ | KU collection |
| ‘Sayo-wabisuke’ | KU collection |
| ‘Seiobo’ | KU collection |
| ‘Shibenashi-wabisuke’ | KU collection |
| ‘Showa-wabisuke’ | KU collection |
| ‘Sukiya’ | KU collection |
| ‘Tosa-uraku’ | KU collection |
| C. japonica | |
| Aomori | Natural accession collected from Aomori Prefecture, Japan |
| Iwate | Natural accession collected from Iwate Prefecture, Japan |
| Goto | Natural accession collected from Goto Islands, Nagasaki Prefecture, Japan |
| Hirado | Natural accession collected from Hirado, Nagasaki Prefecture, Japan |
| Kagoshima | Natural accession collected from Kagoshima Prefecture, Japan |
| Chinese relative species | |
| C. pitardii var. pitardii | Collected from Sichuan, China |
| C. reticulata | Collected from Yunnan, China |
| C. saluenensis | Collected from Yunnan, China |
| Japanese tea cultivars | |
| ‘Asatsuyu’ | Saga Tea Research Laboratory (STRL) collection |
| ‘Kanayamidori’ | STRL collection |
| ‘Yabukita’ | STRL collection |
| Outgroup | |
| C. sasanqua | Natural accession collected from Saga Prefecture, Japan |
| Primer Pairs | No. of Fragments | ||
|---|---|---|---|
| Total | Monomorphic (%) | Polymorphic (%) | |
| E-ACT/M-CAC | 63 | 9 (14) | 54 (86) |
| E-AAC/M-CAA | 51 | 4 (8) | 47 (92) |
| E-AAC/M-CAC | 41 | 8 (20) | 33 (80) |
| E-AAG/M-CAA | 41 | 14 (34) | 27 (66) |
| E-AAG/M-CAC | 57 | 7 (12) | 50 (88) |
| E-ACA/M-CTC | 32 | 7 (22) | 25 (78) |
| E-AGG/M-CTC | 48 | 5 (10) | 43 (90) |
| Total | 333 | 54 (16) | 279 (84) |
| Within and between Groups | Genetic Distance | |
|---|---|---|
| Mean | Range | |
| Within groups | ||
| Wabisuke camellias | 0.3380 | 0.1083–0.5495 |
| C. japonica | 0.3682 | 0.3138–0.4391 |
| Chinese camellias | 0.5337 | 0.3313–0.7113 |
| C. sinensis | 0.1660 | 0.1231–0.2274 |
| Between groups | ||
| Wabisuke—C. japonica | 0.3902 | 0.2788–0.5150 |
| Wabisuke—Chinese camellias | 0.8395 | 0.6683–1.1582 |
| Wabisuke—C. sinensis | 1.0576 | 0.8705–1.2339 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H.; Koike, I.; Nakashima, T.; Hiramatsu, M.; Miyajima, I.; Mizunoe, Y.; Okubo, H.; Ozaki, Y. Genetic Variation and Phylogeny of Wabisuke Camellias by Amplified Fragment Length Polymorphism (AFLP) Analysis. Agronomy 2021, 11, 1974. https://doi.org/10.3390/agronomy11101974
Kim J-H, Koike I, Nakashima T, Hiramatsu M, Miyajima I, Mizunoe Y, Okubo H, Ozaki Y. Genetic Variation and Phylogeny of Wabisuke Camellias by Amplified Fragment Length Polymorphism (AFLP) Analysis. Agronomy. 2021; 11(10):1974. https://doi.org/10.3390/agronomy11101974
Chicago/Turabian StyleKim, Jung-Hee, Itsumi Koike, Toshiki Nakashima, Michikazu Hiramatsu, Ikuo Miyajima, Yuki Mizunoe, Hiroshi Okubo, and Yukio Ozaki. 2021. "Genetic Variation and Phylogeny of Wabisuke Camellias by Amplified Fragment Length Polymorphism (AFLP) Analysis" Agronomy 11, no. 10: 1974. https://doi.org/10.3390/agronomy11101974
APA StyleKim, J.-H., Koike, I., Nakashima, T., Hiramatsu, M., Miyajima, I., Mizunoe, Y., Okubo, H., & Ozaki, Y. (2021). Genetic Variation and Phylogeny of Wabisuke Camellias by Amplified Fragment Length Polymorphism (AFLP) Analysis. Agronomy, 11(10), 1974. https://doi.org/10.3390/agronomy11101974






