Transcriptomic Analysis of L. japonicus Symbiosis Reveals New Candidate Genes for Local and Systemic Regulation of Nodule Function
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Experimental Setup
2.2. RNA Extraction and Real Time PCR
2.3. Microarray Design
2.4. Microarray Hybridization and Data Analysis
2.5. Protein–Protein Interaction (PPI) Network Analysis
2.6. Clustering Analyses
3. Results
3.1. Transcriptomic Analysis of Lotus japonicus Symbiosis
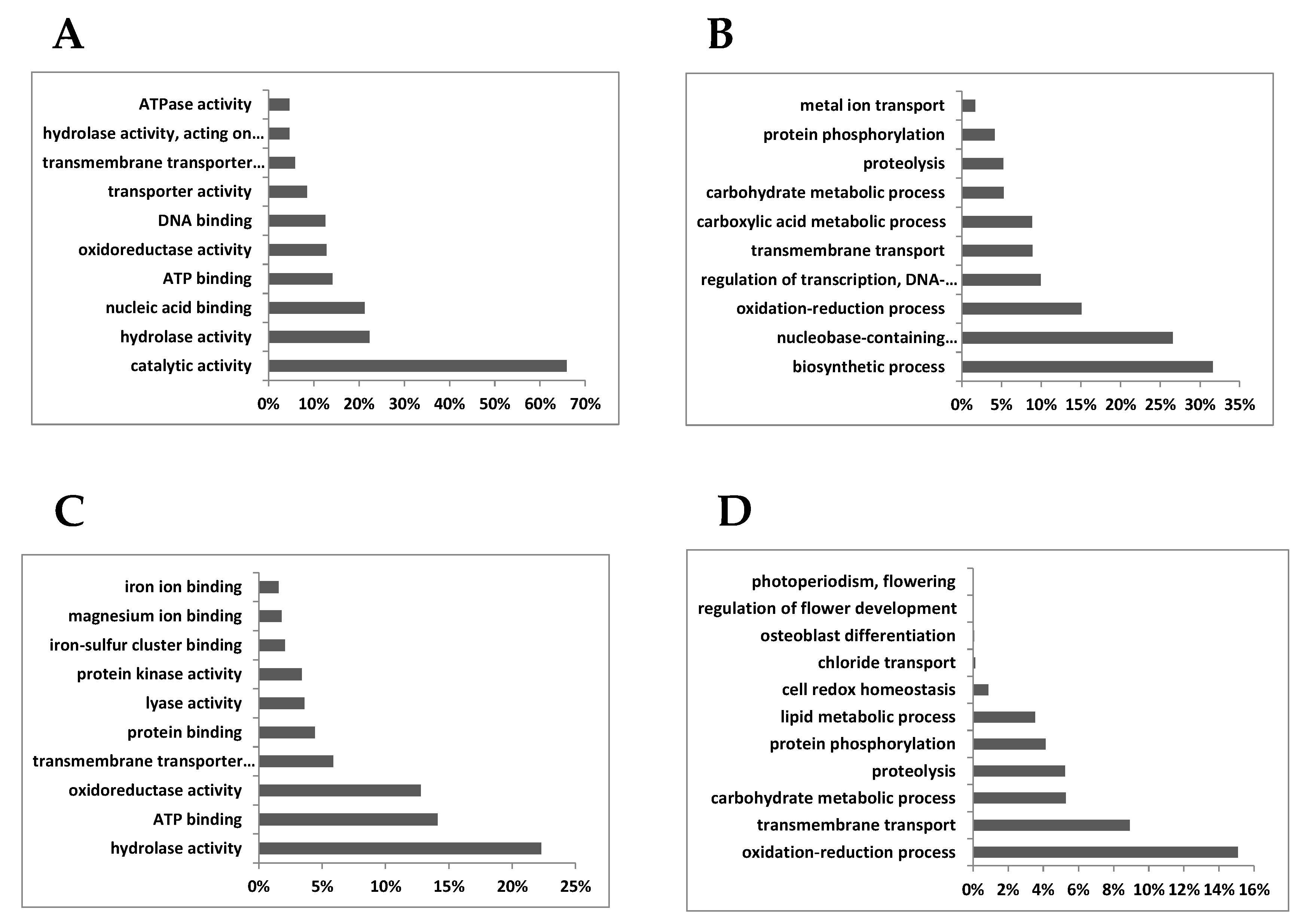
3.2. DEG in Roots
3.2.1. Metabolism
3.2.2. Transport
3.2.3. Signaling and Regulation of Transcription
3.2.4. Redox
3.2.5. Other Categories
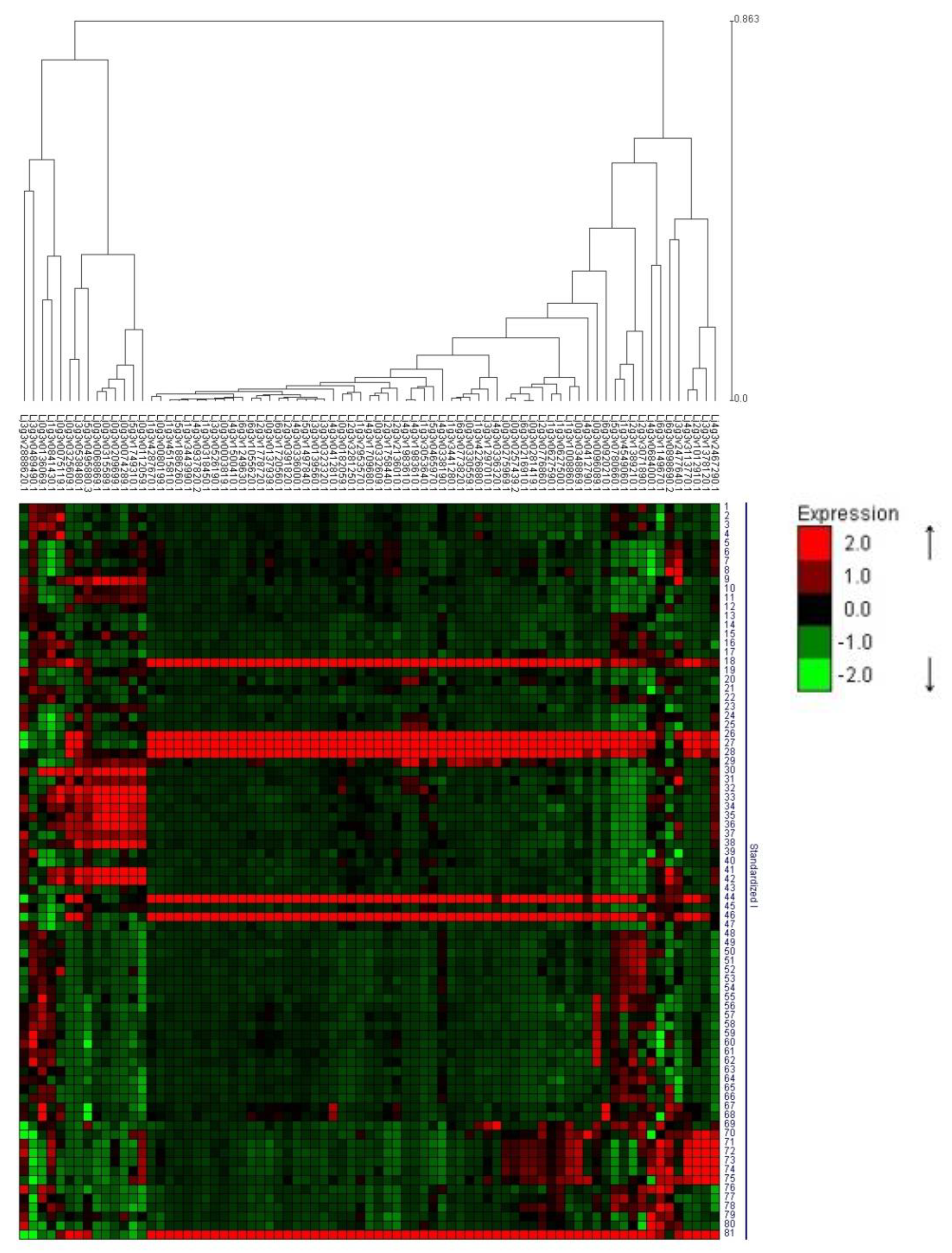
3.2.6. Clustering Analyses of Root DEGs
3.3. DEG in Shoots
3.4. Validation of the Array Data and PPI Network Analysis
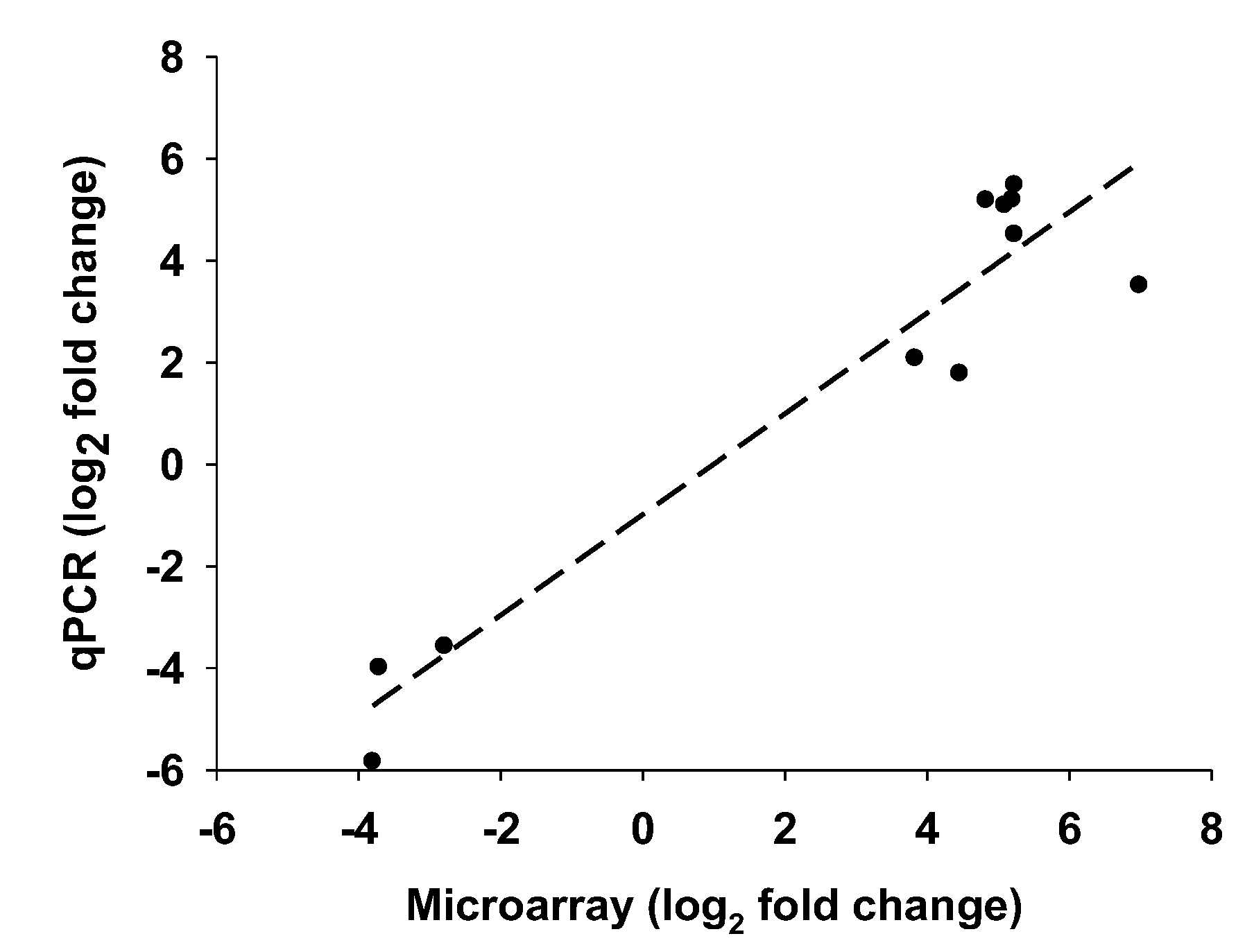
3.4.1. Validation of the Array Data
3.4.2. Protein–Protein Interaction (PPI) Network Analysis
3.5. Hunt for New Genes with Possible Role in Systemic Signaling
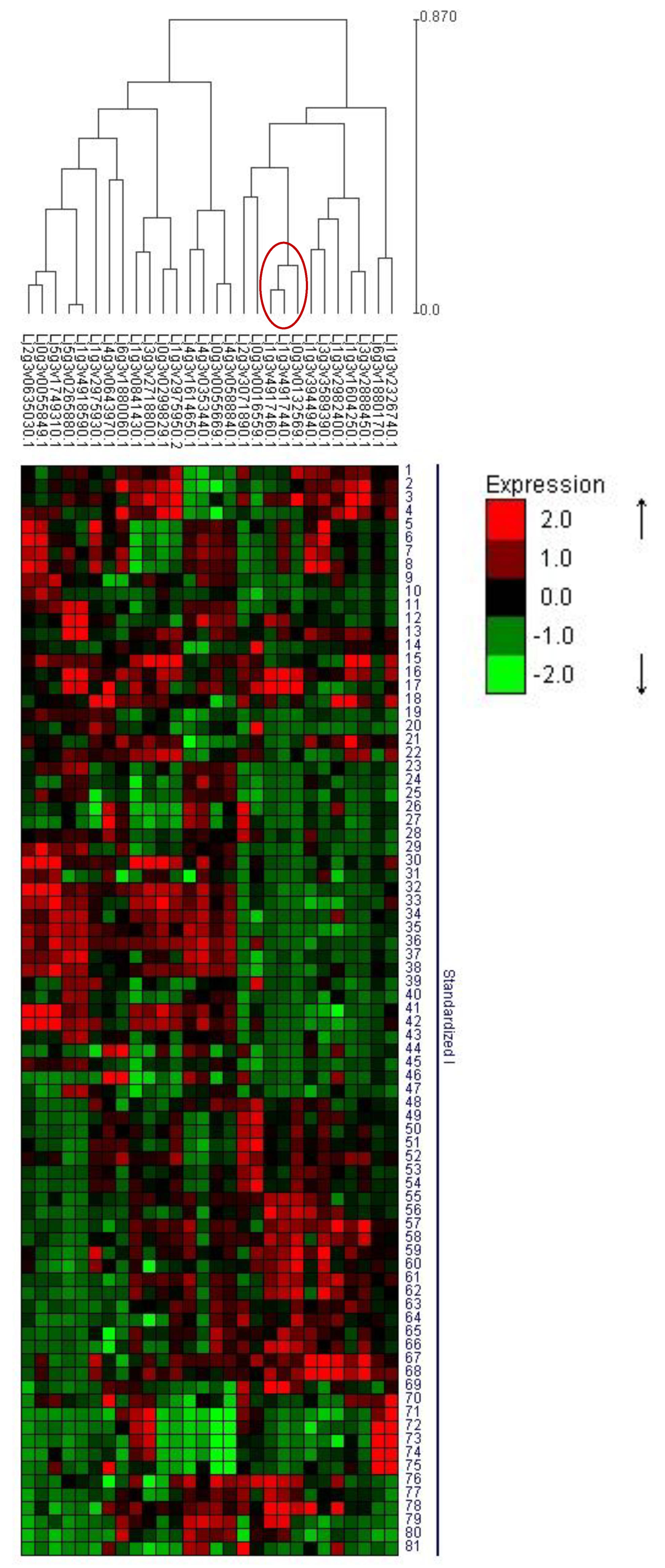
3.6. Clustering with AM-Responsive Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Udvardi, M.; Poole, P.S. Transport and Metabolism in Legume-Rhizobia symbioses. Annu. Rev. Plant Biol. 2013, 64, 781–805. [Google Scholar] [CrossRef]
- Foyer, C.H.; Lam, H.-M.; Nguyen, H.T.; Siddique, K.H.M.; Varshney, R.K.; Colmer, T.D.; Cowling, W.; Bramley, H.; Mori, T.A.; Hodgson, J.M.; et al. Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2016, 2, 16112. [Google Scholar] [CrossRef] [PubMed]
- Madsen, L.H.; Tirichine, L.; Jurkiewicz, A.; Sullivan, J.T.; Heckmann, A.B.; Bek, A.S.; Ronson, C.W.; James, E.K.; Stougaard, J. The molecular network governing nodule organogenesis in the model legume Lotus japonicus. Nat. Commun. 2010, 1, 1–12. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Sekhar Nandety, R.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.A.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2019. [Google Scholar] [CrossRef]
- Colebatch, G.; Desbrosses, G.; Ott, T.; Krusell, L.; Montanari, O.; Kloska, S.; Kopka, J.; Udvardi, M.K. Global changes in transcriptomic orchestrate metabolic differentiation during symbiotic nitrogen fixation in Lotus japonicus. Plant J. 2004, 39, 487–512. [Google Scholar] [CrossRef] [PubMed]
- Manthey, K.; Krajinski, F.; Hohnjec, N.; Firnhaber, C.; Pühler, A.; Perlik, A.M.; Küster, H. Transcriptome profiling in root nodules and arbuscular mycorrhiza identifies a collection of novel genes induced during Medicago truncatula root endosymbiosis. Mol. Plant-Microbe Interact. 2004, 10, 1063–1077. [Google Scholar] [CrossRef] [PubMed]
- Soyano, T.; Hayashi, M. Transcriptional networks leading to symbiotic nodule organogenesis. Curr. Opin. Plant Biol. 2014, 20, 146–154. [Google Scholar] [CrossRef]
- Gutjahr, C.; Parniske, M. Cell and developmental biology of arbuscular micorrhiza symbiosis. Annu. Rev. Cell Dev. Biol. 2013, 29, 593–617. [Google Scholar] [CrossRef]
- Genre, A.; Russo, G. Does a common pathway transduce symbiotic signals in plant-microbe interactions? Front. Plant Sci. 2016, 7, 96. [Google Scholar] [CrossRef]
- Clarke, V.C.; Loughlin, P.C.; Day, D.A.; Smith, P.M.C. Transport processes of the legume symbiosome membrane. Front. Plant Sci. 2014, 5, 699. [Google Scholar] [CrossRef]
- De Bang, T.C.; Lay, K.S.; Scheible, W.-R.; Takahashi, H. Small peptide signaling pathways modulating macronutrients utilization in plants. Curr. Opin. Plant Biol. 2017, 39, 31–39. [Google Scholar] [CrossRef]
- Matamoros, M.A.; Baird, L.M.; Escuredo, P.R.; Dalton, D.A. Stress-induced legume root nodule senescence. Physiological, biochemical and structural alterations. Plant Physiol. 1999, 121, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Cabeza, R.; Koester, B.; Liese, R.; Linger, A. An RNA sequencing transcriptome analysis reveals novel insights into molecular aspects of the nitrate impact on the nodule activity of Medicago truncatula. Plant Physiol. 2014, 164, 400–411. [Google Scholar] [CrossRef]
- Barbulova, A.; Rogato, A.; D’Apuzzo, E.; Omrane, S.; Chiurazzi, M. Differential effects of combined N sources on early steps of the nod factor-dependent transduction pathway in Lotus japonicus. Mol. Plant-Microbe Interact. 2007, 8, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Nishida, H.; Tanaka, S.; Handa, Y.; Ito, M.; Sakamoto, Y.; Matsunaga, S.; Betsuyaku, S.; Miura, K.; Soyano, T.; Kawaguchi, M.; et al. A NIN-LIKE protein mediates nitrate-induced control of root nodule symbiosis in Lotus japonicus. Nat. Commun. 2018, 9, 499. [Google Scholar] [CrossRef]
- Burghardt, R.T.; Gujlin, J.; Chun, C.L.; Liu, J.; Sadowsky, M.J.; Stupar, R.M.; Young, N.D.; Tiffin, P. Transcriptomics basis of genome by genome variation in a legume-rhizobia mutualism. Mol. Ecol. 2017, 26, 6122–6135. [Google Scholar] [CrossRef]
- Shimoda, Y.; Nishigaya, Y.; Yamaya-Ito, H.; Inagaki, N.; Umehara, Y.; Hirakawa, H.; Sato, S.; Yamazaki, T.; Hayashi, M. The Rhizobial autotransporter determines the symbiotic nitrogen fixation activity of Lotus japonicus in a host-specific manner. Proc. Natl. Acad. Sci. USA 2020, 117, 1806–1815. [Google Scholar] [CrossRef]
- Høgslund, N.; Radutoiu, S.; Krusell, L.; Voroshilova, V.; Hannah, M.A.; Goffard, N.; Sánchez, D.H.; Lippold, F.; Ott, T.; Sato, S.; et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of Lotus japonicus mutant and wild-type plants. PLoS ONE 2009, 4, e6556. [Google Scholar] [CrossRef]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-García, N.; Yu, H.-J.; Hwang, H.-J.; Oh, M.; Kim, G.B.; Surendrarao, A.K.; Chasman, D.; et al. Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef]
- Sato, S.; Andersen, S.U. Genome sequencing. In The Lotus japonicus Genome; Tabata, S., Stougaard, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 35–40. [Google Scholar]
- García-Calderón, M.; Chiurazzi, M.; Espuny, M.R.; Márquez, A.J. Photorespiratory metabolism and nodule function: Behavior of Lotus japonicus mutants deficient in plastid glutamine synthetase. Mol. Plant-Microbe Interact. 2012, 25, 211–219. [Google Scholar] [CrossRef]
- Handberg, K.; Stougaard, J. Lotus japonicus, an autonomous, diploid legume species for classical and molecular genetics. Plant J. 1992, 2, 487–496. [Google Scholar] [CrossRef]
- Kistner, C.N.; Matamoros, M. RNA isolation using phase extraction and LiCl precipitation. In The Lotus japonicus Handbook; Márquez, A.J., Ed.; Springer: Dodrecht, Germany, 2005; pp. 123–124. [Google Scholar]
- Verdier, J.; Bandyopadhyay, K.; Udvardi, M.K. A tutorial on Lotus japonicus transcriptomic tools. In The Lotus japonicus Genome; Tabata, S., Stougaard, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 183–199. [Google Scholar]
- Agilent User Manual. Available online: https://www.agilent.com/cs/library/usermanuals/Public/G4140-90040_GeneExpression_OneColor_6.9.pdf (accessed on 28 April 2020).
- Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Dai, X.; Liu, T.; Zhao, P.X. LegumeIP: An integrative database for comparative genomics and transcriptomics of model legumes. Nucl. Ac. Res. 2012, 40, D1221–D1229. [Google Scholar] [CrossRef] [PubMed]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene onthology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, D.H.; Lippold, F.; Redestig, H.; Hannah, M.A.; Erban, A.; Krämer, U.; Kopka, J.; Udvardi, M.K. Integrative functional genomics of salt acclimatization in the model legume Lotus japonicus. Plant J. 2008, 53, 973–987. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, D.H.; Pieckenstain, F.L.; Szymanski, J.; Erban, A.; Bromke, M.; Hannah, M.A.; Kramer, U.; Kopka, J.; Udvardi, M.K. Comparative functional genomics of salt stress in related model and cultivated plants identifies and overcomes limitations to translational genomics. PLoS ONE 2011, 6, e17094. [Google Scholar] [CrossRef]
- Guether, M.; Balestrini, R.; Hannah, M.; He, J.; Udvardi, M.K.; Bonfante, P. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009, 182, 200–212. [Google Scholar] [CrossRef]
- Díaz, P.; Betti, M.; Sánchez, D.H.; Udvardi, M.K.; Monza, J.; Márquez, A.J. Deficiency in plastidic glutamine synthetase alters proline metabolism and transcriptomic response in Lotus japonicus under drought stress. New Phytol. 2010, 188, 1001–1013. [Google Scholar] [CrossRef]
- Verdier, J.; Torrez-Jerez, I.; Wang, M.; Adriankaja, A.; Allen, S.N.; Tang, Y.; Murray, J.D.; Udvardi, M.K. Establishment of the Lotus japonicus Gene Expression Atlas (LjGEA) and its use to explore legume seed maturation. Plant J. 2013, 74, 351–362. [Google Scholar] [CrossRef]
- Shamir, R.; Maron-Katz, A.; Tanay, A.; Linhart, C.; Steinfeld, I.; Sharan, R.; Shiloh, Y.; Elkon, R. EXPANDER and integrative program suite for microarray data analysis. BMC Bioinf. 2005, 6, 232. [Google Scholar] [CrossRef]
- Giovannetti, M.; Goschi, C.; Dietzen, C.; Andersen, S.U.; Kopriva, S.; Busch, W. Identification of novel genes involved in phosphate accumulation in Lotus japonicus through genome wide association mapping of root system architecture and anion content. PLoS Genet. 2019, 15, e1008126. [Google Scholar] [CrossRef] [PubMed]
- Sańko-Sawczenko, I.; Lotocka, B.; Mielecki, J.; Rekosz-Burlaga, H.; Czarnocka, W. Transcriptomic changes in Medicago truncatula and Lotus japonicus during drought stress. Int. J. Mol. Sci. 2019, 20, 1204. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, Y.; Saito, K. Comparative transcriptome analysis between Solanum lycopersicum L. and Lotus japonicus during arbuscular mychorrizal development. Soil Biol. 2017, 63, 127–136. [Google Scholar]
- Kwon, Y.; Yu, S.-I.; Lee, H.; Han Yim, J.; Zhu, J.K.; Lee, B.-H. Arabidopsis serine decarboxylase mutant implicates the role of ethanolamine in plant growth and development. Int. J. Mol. Sci. 2012, 13, 3176–3188. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A. Hormone regulation of root nodule formation in Lotus. In The Lotus japonicus Genome; Tabata, S., Stougaard, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 85–93. [Google Scholar]
- Akashi, T.; Koshimizu, S.; Aoki, T.; Ayabe, S.-H. Identification of cDNAs encoding pterocarpan reductase involved in isoflavan phytoalexin biosynthesis in Lotus japonicus by EST mining. FEBS Lett. 2006, 580, 5666–5670. [Google Scholar] [CrossRef]
- Desbrosses, G.; Kopka, C.; Ott, T.; Udvardi, M.K. Lotus japonicus LjKUP is induced during late nodule development and encodes a potassium transporter of the plasma membrane. Mol. Plant Microbe Interact. 2004, 17, 789–797. [Google Scholar] [CrossRef]
- Krusell, L.; Krause, K.; Ott, T.; Desbrosses, G.; Krämer, U.; Sato, S.; Nakamura, Y.; Tabata, S.; James, E.K.; Sandal, N.; et al. The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 2005, 17, 1625–1636. [Google Scholar] [CrossRef]
- Vincill, E.D.; Szczyglowski, K.; Roberts, D.M. GmN70 and LjN70. Anion transporter of the symbiosome membrane of nodules with a transport preference for nitrate. Plant Physiol. 2016, 137, 1435–1444. [Google Scholar] [CrossRef]
- Clarke, V.C.; Loughlin, P.C.; Gavrin, A.; Chen, C.; Brear, E.M.; Day, D.A.; Smith, P.M.C. Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteom. 2015, 14, 1301–1322. [Google Scholar] [CrossRef]
- Soyano, T.; Shimoda, Y.; Hayashi, M. NODULE INCEPTION antagonistically regulates gene expression with nitrate in Lotus japonicus. Plant Cell Physiol. 2015, 56, 368–376. [Google Scholar] [CrossRef]
- D’Apuzzo, E.; Rogato, A.; Simon-Rosin, U.; El Alaoui, H.; Barbulova, A.; Betti, M.; Imou, M.; Katinakis, P.; Márquez, A.; Marini, A.-M.; et al. Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with different transcriptional regulation and spatial expression. Plant Physiol. 2004, 134, 1763–1774. [Google Scholar]
- Feng, Z.; Sun, X.; Wang, G.; Liu, H.; Zhu, J. LBD29 regulates the cell cycle progression in response to auxin during lateral root formation in Arabidopsis thaliana. Ann. Bot. 2012, 110, 1–10. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Balestrini, R.; Ott, T.; Güther, M.; Bonfante, P.; Udvardi, M.K.; De Tullio, M.C. Ascorbate oxidase: The unexpected involvement of a “wasteful enzyme” in the symbioses with nitrogen-fixing bacteria and arbuscular mycorrhizal fungi. Plant Physiol. Biochem. 2012, 59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Becana, M.; Wienkoop, S.; Matamoros, M.A. Sulfur transport and metabolism in legume root nodules. Front. Plant Sci. 2018, 9, 1434. [Google Scholar] [CrossRef]
- Cao, Y.; Halane, M.K.; Gassmann, W.; Stacey, G. The role of plant innate immunity in the legume-rhiziobium symbiosis. Annu. Rev. Plant Biol. 2017, 68, 535–561. [Google Scholar] [CrossRef]
- Takahara, M.; Magori, S.; Soyano, T.; Okamoto, S.; Yoshida, C.; Yano, K.; Sato, S.; Tabata, S.; Yamaguchi, K.; Shigenobu, S.; et al. TOO MUCH LOVE, a novel kelch repeat-containing F-box protein, functions in the long-distance regulation of the legume-Rhizobium symbiosis. Plant Cell Physiol. 2013, 54, 433–447. [Google Scholar] [CrossRef]
- Yano, K.; Yoshida, S.; Müller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S.; et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Vandereyken, K.; Van Leene, J.; De Coninck, B.; Cammue, B.P.A. Hub protein controversy: Tacking a closer look to at plant stress response hubs. Front. Plant Sci. 2018, 9, 694. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chen, W.; Li, X.; Jiang, H.; Wu, P.; Xia, K.; Yang, Y.; Wu, G. Knockdown of LjIPT3 influences nodule development in Lotus japonicus. Plant Cell Physiol. 2014, 55, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, A.; Saida, Y.; Yoshimizu, M.; Takanashi, K.; Sosso, D.; Frommer, W.B.; Yazaki, K. Characterization of LjSWEET3, a sugar transporter in nodules of Lotus japonicus. Plant Cell Physiol. 2017, 58, 298–306. [Google Scholar] [PubMed]
- Kryvoruchko, I.S.; Sinharoy, S.; Torres-Jerez, I.; Sosso, D.; Pislariu, C.I.; Guan, D.; Murray, J.; Benedito, V.A.; Frommer, W.B.; Udvardi, M.K. MtSWEET11, a nodule-specific sucrose transporter of Medicago truncatula. Plant Physiol. 2016, 171, 554–565. [Google Scholar] [CrossRef]
- Nishimura, R.; Hayashi, M.; Wu, G.-J.; Kouchi, H.; Imaizumi-Anraku, H.; Murakami, Y.; Kawasaki, S.; Akao, S.; Ohmori, M.; Nagasawa, M.; et al. HAR1 mediates systemic regulation of symbiotic organ development. Nature 2002, 420, 426–429. [Google Scholar] [CrossRef]
- Miyazawa, H.; Oka-Kira, E.; Sato, N.; Takahashi, H.; Wu, G.-J.; Sato, S.; Hayashi, M.; Betsuyaku, S.; Nakazono, M.; Tabata, S.; et al. The receptor-like kinase KLAVIER mediates systemic regulation of nodulation and non-symbiotic shoot development in Lotus japonicus. Development 2010, 137, 4317–4325. [Google Scholar] [CrossRef]
- Krusell, L.; Sato, N.; Fukuhara, I.; Koch, B.E.V.; Grossmann, C.; Okamoto, S.; Oka-Kira, E.; Otsubo, Y.; Aubert, G.; Nakagawa, T.; et al. The Clavata2 genes of pea and Lotus japonicus affect autoregulation of nodulation. Plant J. 2011, 65, 861–871. [Google Scholar] [CrossRef]
- Degli Esposti, M.; Martínez Romero, E. A survey of the energy metabolism of nodudlating symbionts reveals a new form of respiratory complex I. FEMS Microbiol. Ecol. 2016, 92, fiw084. [Google Scholar] [CrossRef][Green Version]
- Denancé, N.; Szurek, B.; Noel, L.D. Emerging function of nodulin-like proteins in non-nodulating plant species. Plant Cell. Physiol. 2014, 55, 469–474. [Google Scholar] [CrossRef]
- Jasinski, M.; Banasiak, J.; Radom, M.; Kalitkiewicz, A.; Figlerowicz, M. Full-size ABC transporters from the ABCG subfaminly in Medicago truncatula. Mol. Plant-Microbe Interact. 2009, 22, 921–931. [Google Scholar] [CrossRef]
- Lerán, S.; Varala, K.; Boyer, J.-C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernández, E.; Forde, B.; et al. A unified nomenclature of NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Valkov, V.T.; Chiurazzi, M. Nitrate transport and signalling. In The Lotus japonicus Genome; Tabata, S., Stougaard, J., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 125–136. [Google Scholar]
- Valkov, V.T.; Rogato, A.; Martins Alves, L.; Sol, S.; Noguero, M.; Léran, S.; Lacombe, B.; Chiurazzi, M. The nitrate transporter family protein LjNPF8.6 controls the N-fixing nodule activity. Plant Physiol. 2016, 175, 1269–1282. [Google Scholar] [CrossRef]
- Pal’ove-Balang, P.; García-Calderón, M.; Pérez-Delgado, C.M.; Pavlovkin, J.; Betti, M.; Márquez, A.J. A Lotus japonicus mutant defective in nitrate uptake is also affected in the nitrate response to nodulation. Plant Biol. 2015, 17, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Zdyb, A.; Demchenko, K.; Heumann, J.; Mrosk, C.; Grzeganek, P.; Göbel, C.; Feussner, I.; Pawlowski, K.; Hause, B. Jasmonate biosynthesis in legume and actinorhizal nodules. New Phytol. 2011, 189, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Kohlen, W.; Pin Ng, J.L.; Deinum, E.E.; Mathesius, U. Auxin transport, metabolism, and signaling during nodule initiation: Indeterminate and determinate nodules. J. Exp. Bot. 2016, 69, 229–244. [Google Scholar] [CrossRef]
- Pin Ng, J.L.; Perrine-Walker, F.; Wasson, A.P.; Mathesius, U. The control of auxin transport in parasitic and symbiotic root-microbe interactions. Plants 2015, 4, 606–643. [Google Scholar]
- Sasaki, T.; Suzaki, T.; Soyano, T.; Kojima, N.; Sakakibara, H.; Kawaguchi, M. Shoot-derived cytokinins systemically regulate root nodulation. Nat. Comm. 2014, 5, 4983. [Google Scholar] [CrossRef]
- Suzaki, T.; Yano, K.; Ito, M.; Umehara, Y.; Suganuma, N.; Kawaguchi, M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development 2012, 139, 3997–4006. [Google Scholar] [CrossRef]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ohnishi-Ogawa, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef]
- Suzaki, T.; Nishida, H. Autoregulation of legume nodulation by sophisticated transcriptional regulatory networks. Mol. Plant 2019, 12, 1179–1181. [Google Scholar] [CrossRef]
- Barrabah, F.; Bourcy, M.; Eschstruth, A.; Cayrel, A.; Guefrachi, I.; Mergaert, P.; Wen, J.; Mysore, K.S.; Gourion, B.; Ratet, P. A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol. 2016, 203, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Asamizu, E.; Shimoda, Y.; Kouchi, H.; Tabata, S.; Sato, S. A positive role for LjERF1 in the nodulation process is revealed by systematic analysis of nodule-associated transcription factors of Lotus japonicus. Plant Physiol. 2008, 147, 2030–2040. [Google Scholar] [CrossRef] [PubMed]
- Armijo, G.; Gutiérrez, R.A. Emerging players in the nitrogen signaling pathway. Mol. Plant 2017, 10, 1019–1022. [Google Scholar] [CrossRef]
- Soyano, T.; Hirakawa, H.; Sato, S.; Hayashi, M.; Kawaguchi, M. NODULE INCEPTION creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc. Natl. Acad. Sci. USA 2014, 40, 14607–14612. [Google Scholar] [CrossRef] [PubMed]
- Kawaharada, Y.; James, E.K.; Kelly, S.; Sandal, N.; Stougaard, J. The ethylene responsive factor required for nodulation1 (ERN1) transcription factor is required for infection-thread formation in Lotus japonicus. Mol. Plant-Microbe Interact. 2017, 30, 194–204. [Google Scholar] [CrossRef] [PubMed]


| Gene/Category | Description | log2 FC |
|---|---|---|
| Carbon metabolism | ||
| Lj1g3v4226880.1 | Carbonic anhydrase LjCAA1 | 5.15 |
| Lj5g3v0780660.1 | Carbonic anhydrase LjCAA2 | 5.10 |
| Lj2g3v1389210.1 ** | Beta-glucosidase | 4.17 |
| Amino acid metabolism | ||
| Lj4g3v0338190.1/Lj6g3v2218710.1 | L,L-diaminopimelate aminotransferase | 5.69 |
| Lj0g3v0209699.1 ** | Serine decarboxylase | 4.59 |
| Lj5g3v0495880.3 ** | Chorismate mutase | 4.28 |
| Lj4g3v0412790.1 | BCAA aminotransferase | 4.01 |
| Lj0g3v0315589.1 ** | Serine decarboxylase | 4.00 |
| Hormone metabolism | ||
| Lj0g3v0281419.1 | Putative gibberellin 2-beta-dioxygenase | 7.03 |
| Lj6g3v0216910.1/Lj4g3v0254300.1 | Putative gibberellin 2-beta-dioxygenase | 6.98 |
| Lj5g3v1749310.1 | Tify 10a repressor of JA response | 5.74 |
| Lj6g3v1249630.1 | Cytokinin dehydrogenase | 5.59 |
| Lj3g3v0526190.1 | PIN-like auxin transporter | 4.69 |
| Lj1g3v0841430.1 ** | Iaa-amino acid hydrolase | 4.12 |
| Lj0g3v0075119.1 ** | 12-oxophytodienoate reductase | 3.74 |
| Secondary metabolism | ||
| Lj6g3v0898690.2 | CYP83B1 glucosinolate biosynthesis | 5.49 |
| Lj5g3v1886260.1 | Laccase | 5.31 |
| Lj1g3v3053640.1 ** | Hyoscyamine 6-dioxygenase | 5.03 |
| Lj0g3v0074289.1 ** | Benzyl alcohol o-benzoyltransferase | 4.39 |
| Protein modification and degradation | ||
| Lj0g3v0257439.2 ** | 2-aminoethanethiol dioxygenase | 4.94 |
| Lj1g3v0318450.1 ** | Casein kinase-like | 4.70 |
| Lj1g3v2953570.1 ** | 2-aminoethanethiol dioxygenase | 4.51 |
| Lj3g3v0323320.1 | Calmodulin binding protein | 4.50 |
| Cell division and development | ||
| Lj4g3v1983610.1 ** | Early nodulin-like protein | 5.09 |
| Lj0g3v0330559.1 ** | Casp-like protein | 4.98 |
| Lj2g3v3281250.1 | Pectinesterase | 4.48 |
| Lj1g3v3441280.1 ** | Similar to early nodulin enod18 | 4.24 |
| Lj0g3v0080199.1 ** | Similar to nodule-specific protein LjNOD70 | 3.89 |
| Signaling | ||
| Lj6g3v1055620.1 | Cysteine-rich receptor-kinase-like | 4.19 |
| Lj4g3v3113770.1 ** | Transmembrane kinase receptor | 4.18 |
| Lj0g3v0068869.1 ** | F-box protein | 4.13 |
| Stress and defense | ||
| Lj0g3v0096089.1 ** | Pathogen-related protein | 5.36 |
| Lj5g3v1497840.1 | Respiratory burst oxidase-like protein | 5.01 |
| Lj4g3v1709680.1 ** | Dehydration-responsive protein rd22 | 4.21 |
| Transcription and RNA processing | ||
| Lj2g3v0391820.1 ** | MADS-A18 | 5.59 |
| Lj3g3v0538480.1 ** | Ethylene-responsive transcription factor | 4.83 |
| Lj0g3v0136069.1 ** | MYC2-like transcription factor | 4.78 |
| Lj0g3v0275969.1 | C3H family transcription factor | 4.52 |
| Lj1g3v3443990.1 ** | Transcriptional regulator tac1-like | 4.18 |
| Lj6g3v1149670.1 ** | AT-rich interactive domain protein | 3.59 |
| Transport | ||
| Lj2g3v1758440.1 | Polyol transporter LjPLT4 | 6.99 |
| Lj3g3v0489490.1 ** | Mate efflux family protein | 6.78 |
| Lj0g3v0348869.1 | Potassium transporter LjKUP | 6.05 |
| Lj3g3v0139560.1 ** | Nodulin MtN21-like transporter | 5.46 |
| Lj4g3v1500410.1 | NRAMP metal transporter | 5.33 |
| Lj1g3v4515810.1/810.2 | LjNPF5.7/LjNPF5.5 | 5.23 |
| Lj2g3v0776860.1 | LjSST1 sulfate transporter | 5.23 |
| Lj4g3v0336000.1 ** | Nodulin MtN21-like transporter | 5.20 |
| Lj5g3v0465970.1 | Leghemoglobin Lb3 | 5.09 |
| Lj0g3v0003819.1 | Similar to LjN70 | 5.09 |
| Lj0g3v0332609.1 | Polyol transporter LjPLT14 | 4.75 |
| Lj0g3v0035419.1 | SWEET sugar transporter | 4.49 |
| Lj1g3v4287670.1 ** | Oligopeptide transporter | 4.49 |
| Lj1g3v1008860.1/ ** Lj1g3v1008910.1 | Oligopeptide transporter 6-like | 4.24 |
| Lj6g3v0773820.1 ** | Purine permease 9 | 4.13 |
| Lj4g3v0937820.2 | Polyol transporter LjPLT11 | 4.06 |
| Lj2g3v1778720.1/Lj2g3v1758430.1 | Polyol transporter LjPLT3 | 3.96 |
| Lj4g3v2467290.1 ** | Nodulin MtN21-like transporter | 3.86 |
| Lj3g3v2477640.1 | ABC family transporter | 3.83 |
| Redox | ||
| Lj4g3v0412810.1 ** | Cytochrome p450 71D11 | 4.52 |
| Lj2g3v2136010.1 | L-ascorbate oxidase LjAO1 | 3.87 |
| Other | ||
| Lj0g3v0182059.1 ** | Putative exonuclease | 5.80 |
| Lj2g3v1012910.1 ** | Esterase/lipase | 5.78 |
| Lj0g3v0112059.1 ** | Butyrate-CoA ligase peroxisomal | 5.28 |
| Lj6g3v0920210.1 ** | Fatty acid desaturase | 4.75 |
| Not assigned | ||
| Lj1g3v4549060.1 ** | Unknown function | 6.06 |
| Lj3g3v2888620.1 ** | Unknown function | 5.86 |
| Lj6g3v1720560.1 ** | Proline-rich protein | 4.83 |
| Lj4g3v0336320.1 ** | Similar to GmEKN | 4.67 |
| Lj3g3v1378120.1 ** | Uncharacterized protein | 4.63 |
| Lj0g3v0325609.1 ** | NADH-ubiquinone reductase complex subunit | 4.59 |
| Lj1g3v0627590.1 ** | Plant cadmium resistance 2-like | 4.51 |
| Lj4g3v0684000.1 ** | Uncharacterized protein | 4.35 |
| Lj1g3v0627500.1 ** | Plant cadmium resistance 2-like | 4.00 |
| Lj2g3v3071890.1 ** | Uncharacterized protein | 3.87 |
| Lj3g3v1297010.1 ** | Uncharacterized protein | 3.81 |
| Gene/Category | Description | log2 FC |
|---|---|---|
| Amino acid metabolism | ||
| Lj0g3v0006719.1 | nitrate reductase | −3.96 |
| Lj4g3v0588830.1 | Fd-nitrite reductase | −3.72 |
| Lj2g3v2291670.1 | LjASN1 asparagine synthetase | −3.02 |
| Hormone metabolism | ||
| Lj6g3v0996850.1 | Auxin-induced protein | −2.99 |
| Secondary metabolism | ||
| Lj5g3v0962910.1 ** | Secoisolariciresinol dehydrogenase | −2.92 |
| Protein modification and degradation | ||
| Lj1g3v3368880.1 ** | Serpin-like protein | −3.72 |
| Lj5g3v2258350.1 ** | ACT-domain protein kinase | −3.42 |
| Lj0g3v0119929.1 ** | Ubiquitin-nedd8-like protein rub2 | −3.17 |
| Lj0g3v0038179.1 ** | Ring u-box protein | −2.71 |
| Lj6g3v1880290.1 ** | Basic 7s globulin-like | −2.66 |
| Cell division and development | ||
| Lj0g3v0278459.1 ** | β-1,3-glucanase LjGlu1 | −3.18 |
| Lj4g3v1287310.1/Lj2g3v2002970.1 ** | Expansin a7 like | −2.64 |
| Stress and defense | ||
| Lj0g3v0348489.1 | Pathogenesis-related protein | −4.27 |
| Lj0g3v0105499.1 ** | DMR6-like oxygenase | −3.27 |
| Lj0g3v0134279.1 | Lectin precursor | −2.67 |
| Lj2g3v2017460.1 | Thaumatin-like protein | −2.64 |
| Transcription and RNA processing | ||
| Lj5g3v2099760.1 | Similar to LBD38 | −3.71 |
| Lj5g3v1050280.1 | Similar to LBD29 | −2.79 |
| Lj1g3v0051030.1 | Similar to LBD38 | −2.70 |
| Transport | ||
| Lj3g3v3069030.1 | LjNRT2.1 | −5.42 |
| Lj3g3v3069050.1 | LjNRT2.2 | −5.35 |
| Lj4g3v0135670.1 ** | Putative s-type anion channel | −5.12 |
| Lj4g3v0343320.1 | ABC transporter family protein | −4.98 |
| Lj4g3v1415270.1 | High-affinity nitrate transporter | −4.97 |
| Lj4g3v0353440.1 | Non-symbiotic hemoglobin LjGlb1-1 | −4.66 |
| Lj5g3v1314550.1 | LjAMT1;1 ammonium transporter | −3.21 |
| Lj0g3v0055669.1 ** | Chloride channel protein clc-b | −2.74 |
| Redox | ||
| Lj0g3v0330859.1 ** | Cytochrome p450 family 71 protein | −3.46 |
| Lj1g3v4897990.1 ** | Ferredoxin | −2.73 |
| Lj1g3v4917460.1 ** | Glutaredoxin | −2.72 |
| Lj1g3v4917440.1 ** | Glutaredoxin | −2.67 |
| Lj6g3v1643270.1 ** | Ferredoxin-NADP reductase | −2.67 |
| Other | ||
| Lj1g3v4918590.1 | F-box protein pp2-b15-like | −3.24 |
| Not assigned | ||
| Lj3g3v0740450.1 ** | Uncharacterized protein | −5.41 |
| Lj6g3v0497280.1 ** | Uncharacterized protein | −3.11 |
| Lj3g3v0669220.1 ** | Uncharacterized protein | −3.05 |
| Lj0g3v0216409.1 ** | Uncharacterized protein | −2.99 |
| Lj0g3v0081509.1 ** | Uncharacterized protein | −2.94 |
| Lj3g3v0826140.1 ** | Uncharacterized protein | −2.62 |
| Lj6g3v2017410.1 ** | Uncharacterized protein | −2.55 |
| Gene/Category | Description | log2 FC |
|---|---|---|
| Carbon metabolism | ||
| Lj1g3v2975950.2 | Starch synthase Iib | 4.11 |
| Hormone metabolism | ||
| Lj1g3v1604250.1 | Allene oxide synthase Ljaos2 | 5.09 |
| Lj5g3v1749310.1 | Tify 10a repressor of JA response | 4.44 |
| Lj3g3v2718800.1 | IAA-amino acid conjugate hydrolases | 4.09 |
| Lj1g3v0841430.1 | IAA-amino acid conjugate hydrolases | 3.99 |
| Secondary metabolism | ||
| Lj3g3v2888450.1 ** | Tricyclene synthase | 4.23 |
| Lj0g3v0299829.1 ** | probable 2-oxoglutarate-dependent dioxygenase | 3.72 |
| Cell division and development | ||
| Lj1g3v3944940.1 ** | Protein flowering locus t-like | 5.02 |
| Signaling | ||
| Lj1g3v2982400.1 ** | DMP9 membrane protein | 4.46 |
| Lj1g3v2975930.1 ** | DMP9 membrane protein | 4.18 |
| Stress and defense | ||
| Lj2g3v0635030.1 | Rj2 homolog protein | 4.40 |
| Lj3g3v3589390.1 | Glucan endo-1,3-beta-glucosidase | 3.74 |
| Transport | ||
| Lj6g3v1880060.1 | Glucose-6-phosphate phosphate transporter | 3.99 |
| Other | ||
| Lj0g3v0055849.1 | Phospholipase a1-chloroplastic-like | 3.69 |
| Not assigned | ||
| Lj0g3v0089179.1 ** | Uncharacterized protein | 3.96 |
| Lj0g3v0016559.1 ** | Uncharacterized protein | 3.95 |
| Lj4g3v0643970.1 ** | Uncharacterized protein | 3.93 |
| Gene/Category | Description | log2 FC |
|---|---|---|
| Amino acid metabolism | ||
| Lj4g3v0588840.1 | Fd nitrite reductase | −3.98 |
| Protein modification and degradation | ||
| Lj5g3v0265880.1 | Prolyl endopeptidase-like | −2.87 |
| Transcription and RNA processing | ||
| Lj6g3v1880170.1 | Nuclear transcription factor | −3.34 |
| Transport | ||
| Lj4g3v0353440.1 ** | Non-symbiotic hemoglobin LjGlb1-1 | −5.93 |
| Lj0g3v0055669.1 ** | Chloride channel protein clc-b | −3.77 |
| Redox | ||
| Lj1g3v4917460.1 ** | Glutaredoxin | −3.80 |
| Lj0g3v0132569.1 ** | Glutaredoxin | −3.78 |
| Lj1g3v4917440.1 ** | Glutaredoxin | −3.32 |
| Not assigned | ||
| Lj2g3v3071890.1 ** | Uncharacterized protein | −4.25 |
| Lj1g3v4918590.1 ** | Uncharacterized protein | −3.41 |
| Lj1g3v0488870.1 ** | Uncharacterized protein | −3.16 |
| Lj4g3v1614650.1 ** | Uncharacterized protein | −3.14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Delgado, C.M.; García-Calderón, M.; Monje-Rueda, M.D.; Márquez, A.J.; Betti, M. Transcriptomic Analysis of L. japonicus Symbiosis Reveals New Candidate Genes for Local and Systemic Regulation of Nodule Function. Agronomy 2020, 10, 819. https://doi.org/10.3390/agronomy10060819
Pérez-Delgado CM, García-Calderón M, Monje-Rueda MD, Márquez AJ, Betti M. Transcriptomic Analysis of L. japonicus Symbiosis Reveals New Candidate Genes for Local and Systemic Regulation of Nodule Function. Agronomy. 2020; 10(6):819. https://doi.org/10.3390/agronomy10060819
Chicago/Turabian StylePérez-Delgado, Carmen M., Margarita García-Calderón, María Dolores Monje-Rueda, Antonio J. Márquez, and Marco Betti. 2020. "Transcriptomic Analysis of L. japonicus Symbiosis Reveals New Candidate Genes for Local and Systemic Regulation of Nodule Function" Agronomy 10, no. 6: 819. https://doi.org/10.3390/agronomy10060819
APA StylePérez-Delgado, C. M., García-Calderón, M., Monje-Rueda, M. D., Márquez, A. J., & Betti, M. (2020). Transcriptomic Analysis of L. japonicus Symbiosis Reveals New Candidate Genes for Local and Systemic Regulation of Nodule Function. Agronomy, 10(6), 819. https://doi.org/10.3390/agronomy10060819






