Marginal Grapevine Germplasm from Apulia (Southern Italy) Represents an Unexplored Source of Genetic Diversity
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. SSR Genotyping
2.3. Genotyping by Sequencing and SNP Calling
2.4. Genetic Diversity and Population Structure
2.5. Divergent Loci
3. Results
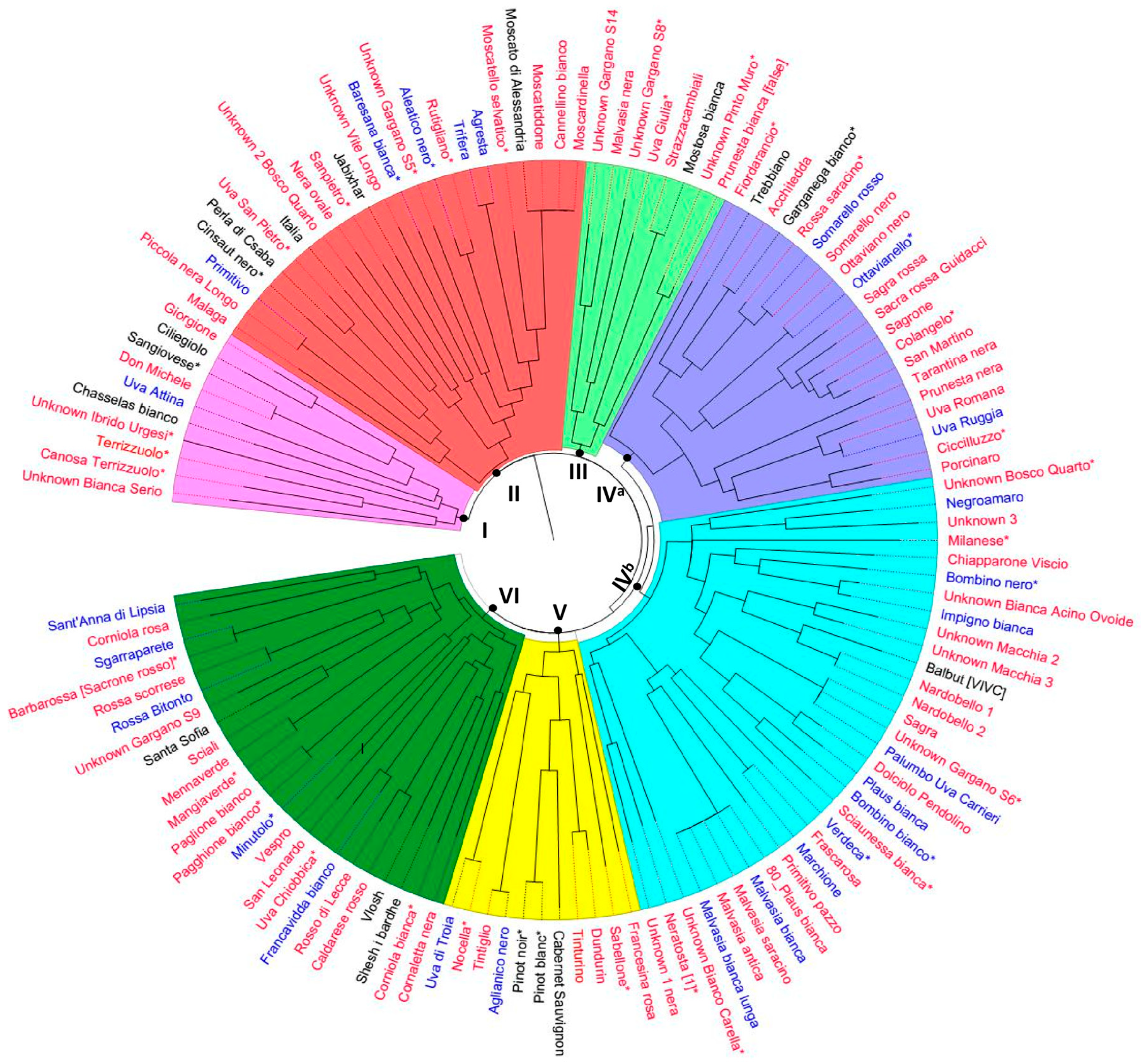
3.1. SSR-Based Analysis of Grapevine Genotypes
3.2. Genetic Relationships between Grapevine Genotypes Based on SSR Markers
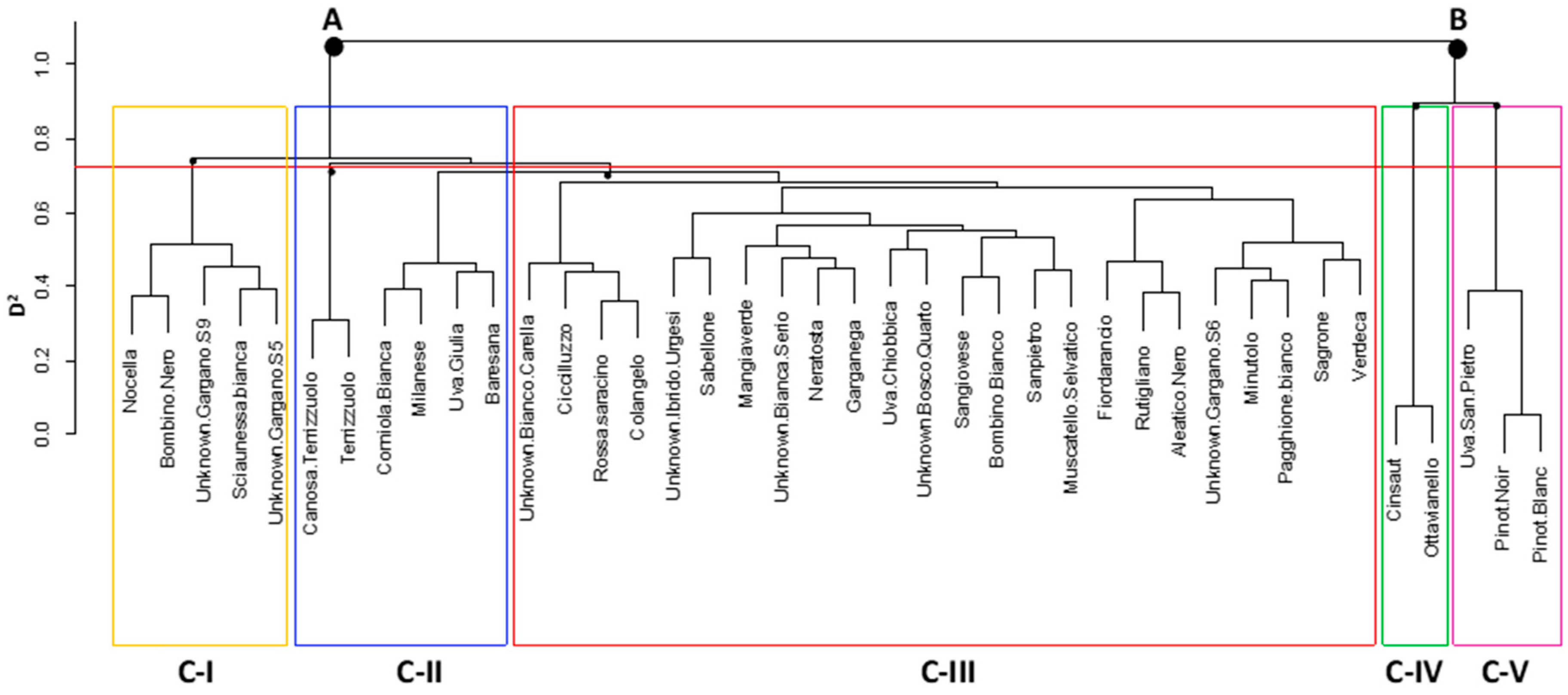
3.3. GBS-Derived SNP Analysis
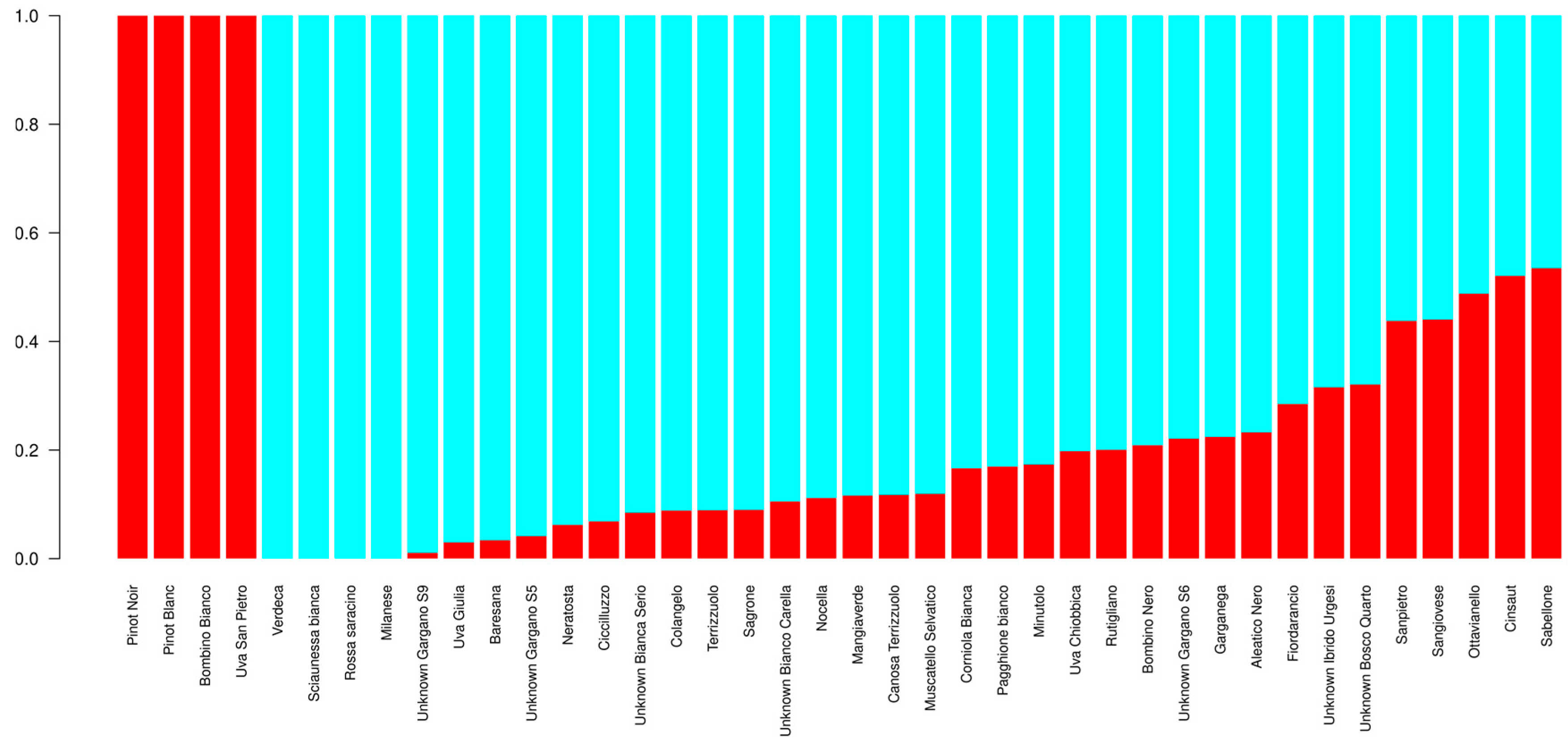
3.4. Genetic Diversity Assessed by SNPs
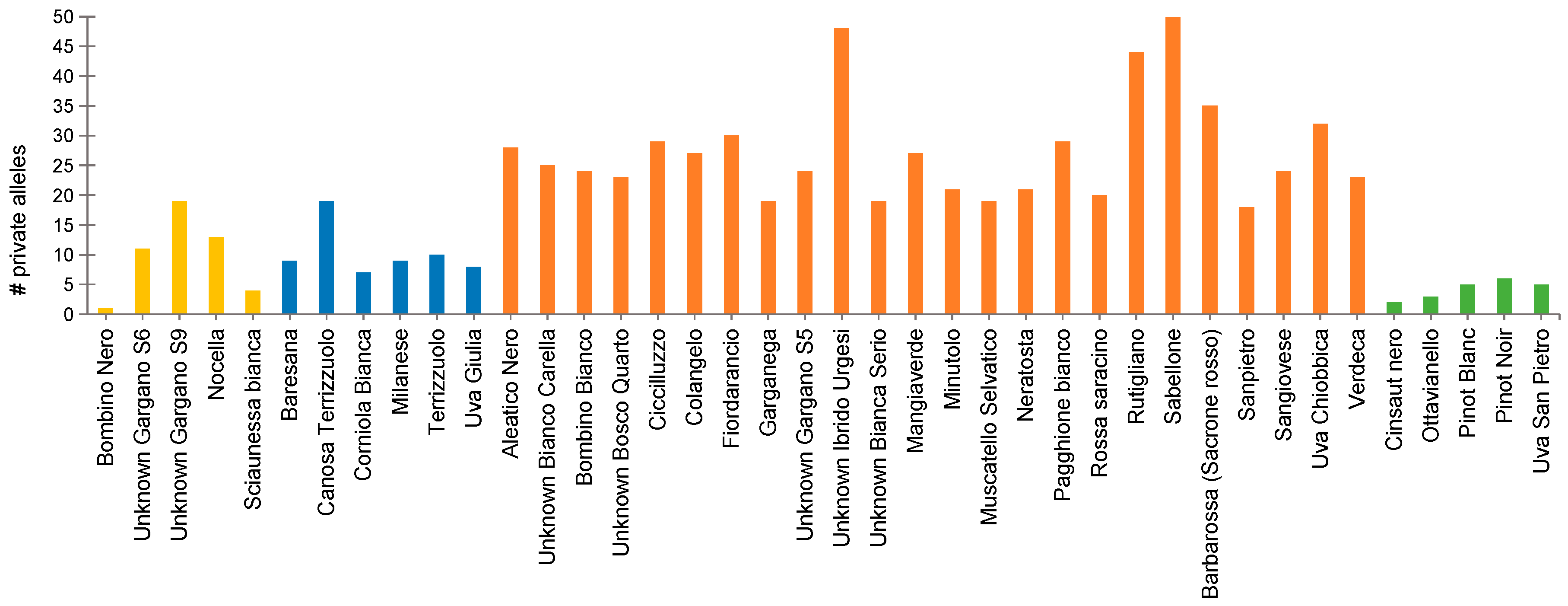
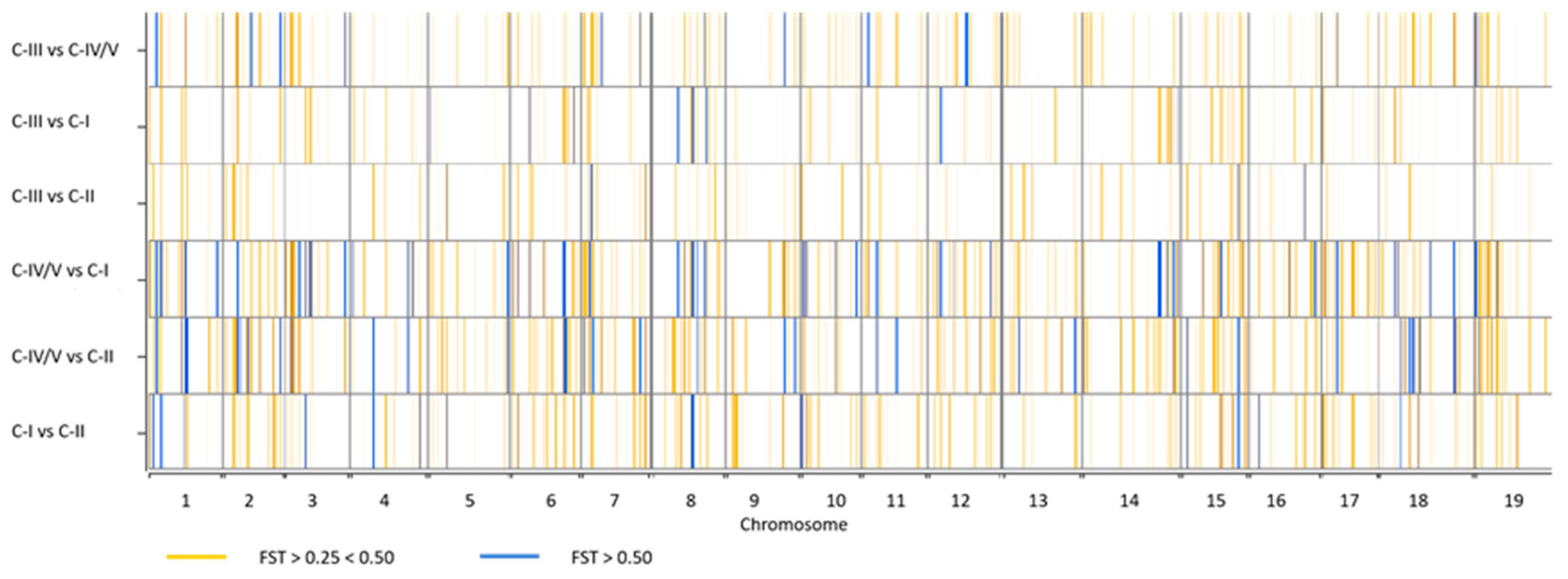
3.5. Detection of Private SNP Alleles and Divergent Loci
4. Discussion
4.1. Cultivar Identification and Genetic Diversity by Means of SSRs
4.2. High Throughput SNP Genotyping Reveals Patterns of Genetic Divergence in Apulian Minor/Neglected Varieties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- McGovern, P.E.; Glusker, D.L.; Exner, L.J.; Voigt, M.M. Neolithic resinated wine. Nature 1996, 381, 480–481. [Google Scholar] [CrossRef]
- Arroyo-García, R.; Ruíz-García, L.; Bolling, L.; Ocete, R.; López, M.A.; Arnold, C.; Ergul, A.; Söylemezoğlu, G.; Uzun, H.I.; Cabello, F.; et al. Multiple origins of cultivated grapevine (Vitis vinifera L. ssp. sativa) based on chloroplast DNA polymorphisms. Mol. Ecol. 2006, 15, 3707–3714. [Google Scholar] [CrossRef] [PubMed]
- This, P.; Lacombe, T.; Thomas, M.R. Historical origins and genetic diversity of wine grapes. Trends Genet. 2006, 22, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Maul, E.; Toepfer, R. Vitis International Variety Catalogue (VIVC): A Cultivar Database Referenced by Genetic Profiles and Morphology. In Proceedings of the BIO Web of Conferences, Mainz, Germany, 5–10 July 2015. [Google Scholar] [CrossRef]
- Registro Nazionale Delle Varietà Di Vite, Ministero Delle Politiche Agricole, Alimentari e Forestali. Available online: http://catalogoviti.politicheagricole.it/catalogo.php (accessed on 14 April 2020).
- Lacombe, T.; Audeguin, L.; Boselli, M.; Bucchetti, B.; Cabello, F.; Chatelet, P.; Crespan, M.; D’Onofrio, C.; Dias, J.E.; Ercisli, S.; et al. Grapevine European catalogue: Towards a comprehensive list. Vitis 2011, 50, 65–68. [Google Scholar]
- Sardaro, R.; Bozzo, F.; Petrillo, F.; Fucilli, V. Measuring the financial sustainability of vine landraces for better conservation programs of Mediterranean agro-biodiversity. Land Use Policy 2017, 68, 160–167. [Google Scholar] [CrossRef]
- La Notte, P.; Civita, F.; Raimondi, S.; Schneider, A. Atlante Dei Vitigni Tradizionali Di Puglia 2018; Caramia, B., Ed.; CRSFA: Locorotondo (BR), Italy, 2018. [Google Scholar]
- Schneider, A.; Raimondi, S.; Pirolo, C.S.; Marinoni, D.; Ruffa, P.; Venerito, P.; La Notte, P. Genetic characterization of grape cultivars from Apulia (Southern Italy) and synonymies in other Mediterranean regions. Am. J. Enol. Vitic. 2014, 65, 244–249. [Google Scholar] [CrossRef]
- Van Leeuwen, C.; Schultz, H.R.; de Cortazar-Atauri, I.G.; Duchene, E.; Ollat, N.; Pieri, P.; Bois, B.; Goutouly, J.P.; Quénol, H.; Touzard, J.M.; et al. Why climate change will not dramatically decrease viticultural suitability in main wine-producing areas by 2050. Proc. Natl. Acad. Sci. USA 2013, 110, E3051–E3052. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, M.; Lux, R.; Schultz, H.R. Constructing a framework for risk analyses of climate change effects on the water budget of differently sloped vineyards with a numeric simulation using the Monte Carlo method coupled to a water balance model. Front. Plant Sci. 2014, 5, 645. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Montemurro, C.; Strippoli, G.; Fanelli, V.; de Giovanni, C.; Antonacci, D.; Vivaldi, G.A.; Pellegrini, G.; Blanco, A.; Camposeo, S.; et al. Ecophysiological response to water stress and regulation of gene expression for a 9-cis-epoxycarotenoid dioxygenase in Vitis vinifera L. ‘Italia’. Acta Hortic. 2015, 1082, 277–284. [Google Scholar] [CrossRef]
- De Giovanni, C.; Di Rienzo, V.; Miazzi, M.; Fanelli, V.; Blanco, A.; Montemurro, C. A DNA methylation survey of NCED genes in Vitis vinifera L. under stress conditions. Acta Hortic. 2015, 1082, 277–284. [Google Scholar] [CrossRef]
- Hajjeh, H.; Miazzi, M.; Faretra, F. Overwintering of Erysiphe necator Schw. in southern Italy. J. Plant Pathol. 2009, 90, 323–330. [Google Scholar]
- Miazzi, M.; Hajjeh, H.R. Differential sensitivity to triadimenol of Erysiphe necator isolates belonging to different genetic groups. J. Plant Pathol. 2011, 93, 729–735. [Google Scholar]
- Sanzani, S.M.; Miazzi, M.M.; Di Rienzo, V.; Fanelli, V.; Gambacorta, G.; Taurino, M.R.; Montemurro, C. A rapid assay to detect toxigenic Penicillium spp. contamination in wine and musts. Toxins 2016, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Marrano, L.; Grzeskowiak, P.; Sanz, M.; Lorenzi, S.; Prazzoli, M.L.; Arzumano, A.; Amanova, M.; Failla, O.; Maghradze, D.; Grando, M.S. Genetic diversity and relationships in the grapevine germplasm collection from Central Asia. Vitis 2015, 54, 233–237. [Google Scholar]
- Gristina, A.S.; De Michele, R.; Garfı, G.; La Mantia, T.; Fontana, I.; Spinelli, P.; Motisi, A.; Carimi, F. Urgent need for preservation of grapevine (Vitis vinifera L. subsp. vinifera) germplasm from small circum-Sicilian islands as revealed by SSR markers and traditional use investigations. Genet. Resour. Crop Evol. 2016. [Google Scholar] [CrossRef]
- Drori, E.; Rahimi, O.; Marrano, A.; Henig, Y.; Brauner, H.; Salmon-Divon, M.; Netzer, Y.; Prazzoli, M.L.; Stanevsky, M.; Failla, O.; et al. Collection and characterization of grapevine genetic resources (Vitis vinifera) in the Holy Land, towards the renewal of ancient winemaking practices. Sci. Rep. 2017, 7, 4463. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, V.; Savoia, M.A.; Gadaleta, S.; Piarulli, L.; Montemurro, C.; La Notte, F.; Miazzi, M.M.; Bruno, M.; Falbo, M.; Petrillo, F.; et al. Molecular characterization of wine grape cultivars from Calabria. Acta Hortic. 2019, 1248. [Google Scholar] [CrossRef]
- Halász, J.; Kodad, O.; Galiba, G.M.; Skola, I.; Ercisli, S.; Ledbetter, C.A. Genetic variability is preserved among strongly differentiated and geographically diverse almond germplasm: An assessment by simple sequence repeat markers. Tree Gen. Genom. 2019, 15, 12. [Google Scholar] [CrossRef]
- This, P.; Jung, A.; Boccacci, P.; Borrego, J.; Costantini, L.; Crespan, M.; Eisenheld, C.; Grando, M.S.; Lacombe, T.; Lacou, V.; et al. Development of a common set of standard varieties and standardized method of scoring microsatellites markers for the analysis of grapevine genetic resources. Theor. Appl. Genet. 2004, 109, 1448–1458. [Google Scholar] [CrossRef]
- Boucheffa, S.; Tamendjari, A.; Sanchez-Gimeno, A.C.; Rovellini, P.; Venturini, S.; di Rienzo, V.; Miazzi, M.M.; Montemurro, C. Diversity assessment of Algerian wild and cultivated olives (Olea europaea L.) by molecular; morphological and chemical traits. Eur. J. Lipid Sci. Technol. 2019, 1800302. [Google Scholar] [CrossRef]
- Pasqualone, A.; Di Rienzo, V.; Nasti, R.; Blanco, A.; Gomes, T.; Montemurro, C. Traceability of Italian Protected Designation of Origin (PDO) table olives by means of microsatellite molecular markers. J. Agric. Food Chem. 2013, 61, 3068–3073. [Google Scholar] [CrossRef] [PubMed]
- Sabetta, W.; Miazzi, M.M.; di Rienzo, V.; Fanelli, V.; Pasqualone, A.; Montemurro, C. Development and application of protocols to certify the authenticity and the traceability of Apulian typical products in olive sector. Rivista Italiana Delle Sostanze Grasse 2017, 94, 37–43. [Google Scholar]
- Pasqualone, A.; Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Caponio, F.; Montemurro, C. High resolution melting analysis of DNA microsatellites in olive pastes and virgin olive oils obtained by talc addition. Eur. J. Lipid Sci. Technol. 2015, 117, 2000–2010. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Fanelli, V.; Miazzi, M.M.; Sabetta, W.; Montemurro, C. A reliable analytical procedure to discover table grape DNA adulteration in industrial wines and musts. Acta Hortic. 2017, 1188, 365–370. [Google Scholar] [CrossRef]
- Emanuelli, F.; Lorenzi, S.; Grzeskowiak, L.; Catalano, V.; Stefanini, M.; Troggio, M.; Grando, M.S.; Myles, S.; Martinez-Zapater, J.M.; Zyprian, E.; et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013, 13, 39. [Google Scholar] [CrossRef] [PubMed]
- Jaillon, O.; Aury, J.-M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Troggio, M.; Cartwright, D.A.; Cestaro, A.; Pruss, D.; Pindo, M.; FitzGerald, L.M.; Vezzulli, S.; Reid, J.; et al. A high-quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2007, 2, e1326. [Google Scholar] [CrossRef]
- Adam-Blondon, A.F.; Jaillon, O.; Vezzulli, S.; Zharkikh, A.; Troggio, M.; Velasco, R. Genome sequence initiatives. In Genetics Genomics and Breeding of Grapes; Adam-Blondon, A.-F., Martinez-Zapater, J.M., Chittaranjan, K., Eds.; Science Publishers, CRC Press: Boca Raton, FL, USA, 2011; pp. 211–234. [Google Scholar]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom. Data 2017, 14, 56–62. [Google Scholar] [CrossRef]
- Roach, M.J.; Johnson, D.L.; Bohlmann, J.; van Vuuren, H.J.J.; Jones, S.J.M.; Pretorius, S.I.; Schmidt, S.A.; Borneman, A.R. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018, 14, e1007807. [Google Scholar] [CrossRef]
- Liang, Z.; Duan, S.; Sheng, J.; Zhu, S.; Ni, X.; Shao, J.; Mao, R.; Zhu, Y.; Deng, W.; Yang, M.; et al. Whole-genome resequencing of 472 Vitis accessions for grapevine diversity and demographic history analyses. Nat. Commun. 2019, 10, 1190. [Google Scholar] [CrossRef]
- Laucou, V.; Launay, A.; Bacilieri, R.; Lacombe, T.; Adam-Blondon, A.F.; Berard, A.; Le Paslier, M.C. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10 K genome-wide SNPs. PLoS ONE 2018, 13, e0192540. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.L.; Zhao, H.L.; Li, Q.; Zhang, G.H.; Jiang, J.F.; Liu, C.H.; Yu, Y.H. Genome-wide association study of berry-related traits in grape [Vitis vinifera L.] based on genotyping-by-sequencing markers. Hortic. Res. 2019, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Lijavetzky, D.; Cabezas, J.A.; Ibáñez, A.; Rodriguez, V.; Martínez-Zapater, J.M. High throughput SNP discovery and genotyping in grapevine (Vitis vinifera L.) by combining a re-sequencing approach and SNPlex technology. BMC Genom. 2007, 8, 424. [Google Scholar] [CrossRef] [PubMed]
- Miazzi, M.M.; D’Agostino, N.; Gadaleta, S.; di Rienzo, V.; Fanelli, V.; Sabetta, W.; Montemurro, C.; Taranto, F. GBS-derived SNP catalogue from a wide grape (V. vinifera L.) germplasm collection that includes the most representative Apulian autochthonous varieties. Acta Hortic. 2019, 22, 1248. [Google Scholar] [CrossRef]
- De Lorenzis, G.; Mercati, F.; Bergamini, C.; Cardone, M.F.; Lupini, A.; Mauceri, A.; Caputo, A.F.; Abbate, L.; Barbagallo, M.G.; Antonacci, D.; et al. SNP genotyping elucidates the genetic diversity of Magna Graecia grapevine germplasm and its historical origin and dissemination. BMC Plant Biol. 2019, 19, 5–15. [Google Scholar] [CrossRef]
- Cipriani, G.; Spadotto, A.; Jurman, I.; Di Gaspero, G.; Crespan, M.; Meneghetti, S.; Frare, E.; Vignani, R.; Cresti, M.; Morgante, M.; et al. The SSR-based molecular profile of 1005 grapevine (Vitis vinifera L.) accessions uncovers new synonymy and parentages and reveals a large admixture amongst varieties of different geographic origin. Theor. Appl. Genet. 2010, 121, 1569–1585. [Google Scholar] [CrossRef]
- Lacombe, T.; Boursiquot, J.M.; Laucou, V.; Di Vecchi-Staraz, M.; Péros, J.-P.; This, P. Large-scale parentage analysis in an extended set of grapevine cultivars (Vitis vinifera L.). Theor. Appl. Genet. 2013, 126, 401–414. [Google Scholar] [CrossRef]
- Marrano, A.; Micheletti, D.; Lorenzi, S.; Neale, D.; Grando, M.S. Genomic signatures of different adaptations to environmental stimuli between wild and cultivated Vitis vinifera L. Hortic. Res. 2018, 5, 34. [Google Scholar] [CrossRef]
- Corrado, G.; Piffanelli, P.; Caramante, M.; Coppola, M.; Rao, R. SNP genotyping reveals genetic diversity between cultivated landraces and contemporary varieties of tomato. BMC Genom. 2013, 14, 835. [Google Scholar] [CrossRef]
- Akagi, T.; Hanada, T.; Yaegaki, H.; Gradziel, T.M.; Tao, R. Genome-wide view of genetic diversity reveals paths of selection and cultivar differentiation in peach domestication. DNA Res. 2016, 23, 271–282. [Google Scholar] [CrossRef]
- De Palma, L.; Poli, G.; Lopriore, G.; Tarantino, A.; Tarricone, L.; Soleti, F. Individuazione, Studio e Caratterizzazione Di Vitigni Di Antica Coltivazione Nell’Alto Tavoliere Pugliese; Centro Grafico Francescano: Foggia, Italy, 2008. [Google Scholar]
- Spadoni, A.; Sion, S.; Gadaleta, S.; Savoia, M.; Piarulli, L.; Fanelli, V.; di Rienzo, V.; Taranto, D.; Miazzi, M.; Montemurro, C.; et al. A simple and rapid method for genomic DNA extraction and microsatellite analysis in tree plants. J. Agric. Sci. Technol. 2019, 21, 1215–1226. [Google Scholar]
- OIV Statistical Report on World Vitiviniculture. Available online: http://www.oiv.int/en/ (accessed on 14 April 2020).
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Sefc, K.M.; Regner, F.; Turetschek, E.; Glossl, J.; Steinkellner, H. Identification of microsatellite sequences in Vitis riparia and their applicability for genotyping of different Vitis species. Genome 1999, 42, 367–373. [Google Scholar] [CrossRef]
- Di Rienzo, V.; Miazzi, M.M.; Fanelli, V.; Savino, V.; Pollastro, S.; Colucci, F.; Miccolupo, A.; Blanco, A.; Pasqualone, A.; Montemurro, C. An enhanced analytical procedure to discover table grape DNA adulteration in industrial musts. Food Control 2016, 60, 124–130. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A high capacity genotyping by sequencing analysis pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Hang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. GigaScience 2015, 4, 7. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Wright, S. The genetical structure of populations. Ann. Eugen. 1949, 15, 323–354. [Google Scholar] [CrossRef]
- Kalinowski, S.T. Counting alleles with rarefaction: Private alleles and hierarchical sampling designs. Conserv. Genet. 2004, 5, 539–543. [Google Scholar] [CrossRef]
- Lynch, M.; Ritland, K. Estimation of pairwise relatedness with molecular markers. Genetics 1999, 152, 1753–1766. [Google Scholar] [PubMed]
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Gao, X.; Starmer, J. AWclust: Point-and-click software for non-parametric population structure analysis. BMC Bioinform. 2008, 9, 77. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Calò, A.; Scienza, A.; Costacurta, A.; Molina, M. Vitigni d’Italia; Edagricole, Ed.; Agricole Calderini S.R.L.: Bologna, Italy, 2001. [Google Scholar]
- Lentini, A. Archaeobotany brewing and winemaking in Mediterranean Basin and Transcaucasus area. In Cyprus in the Prehistory of Wine; Flourentzos, P., Belgiorno, M.R., Micosia, L.A., Eds.; de Strobel Publ: Nicosia, Cyprus, 2005; pp. 6–11. [Google Scholar]
- Istituto Di Servizi Per Il Mercato Agricolo Alimentare. Available online: www.ismea.it (accessed on 14 April 2020).
- Voluntary Consortia National Confederation for the Protection of the Italian Wines. Available online: www.federdoc.com (accessed on 14 April 2020).
- Zulini, L.; Russo, M.; Peterlunger, E. Genotyping wine and table grape cultivars from Apulia (Southern Italy) using microsatellite markers. Vitis Geilweilerhof 2002, 41, 183–188. [Google Scholar]
- Di Vecchi Staraz, M.; Bandinelli, R.; Boselli, M.; This, P.; Boursiquot, J.M.; Laucou, V.; Lacombe, T.; Varès, D. Genetic structuring and parentage analysis for evolutionary studies in grapevine: Kin group and origin of the cultivar Sangiovese revealed. J. Am. Soc. Hortic. Sci. 2007, 132, 514–524. [Google Scholar] [CrossRef]
- Bacilieri, R.; Lacombe, T.; Le Cunff, L.; Di Vecchi-Staraz, M.; Laucou, V.; Genna, B.; Péros, J.-P.; This, P.; Boursiquot, J.M. Genetic structure in cultivated grapevines is linked to geography and human selection. BMC Plant Biol. 2013, 13, 25. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Ganopoulos, I.; Tani, E.; Tsaftaris, A. Epigenetics, epigenomics and crop improvement. Adv. Bot. Res. 2018, 86, 287–324. [Google Scholar]
- Biscotti, N.; Delviscio, G.; Bonsanto, D.; Casavecchia, S.; Biondi, E. Indagini su popolazioni selvatiche di Vitis vinifera L. rinvenute nel Parco Nazionale del Gargano (Foggia), in Puglia. Inf. Bot. Ital. 2015, 4, 179–186. [Google Scholar]
- Zdunić, G.; Preece, J.E.; Aradhya, M.; Velasco, D.; Koehmstedt, A. Genetic diversity and differentiation within and between cultivated (Vitis vinifera L. ssp. sativa) and wild (Vitis vinifera L. ssp. sylvestris) grapes. Vitis 2013, 52, 29–32. [Google Scholar]
- Migliaro, D.; De Nardi, B.; Vezzulli, S.; Crespan, M. An upgraded core set of 11 SSR markers for grapevine cultivar identification: The case of berry color mutants. Am. J. Enol. Vitic. 2017, 68, 496–498. [Google Scholar] [CrossRef]
- Xin, H.; Zhang, J.; Zhu, W.; Wang, N.; Fang, P.; Han, Y.; Li, S. The effects of artificial selection on sugar metabolism and transporter genes in grape. Tree Gen. Genom. 2013, 9, 1343–1349. [Google Scholar] [CrossRef]
- Yasuda, T.; Konishi, N.; Kojima, S. Ammonium uptake capacity and response of cytosolic glutamine synthetase 1; 2 to ammonium supply are key factors for the adaptation of ammonium nutrition in Arabidopsis thaliana. Soil Sci. Plant Nutr. 2017, 63. [Google Scholar] [CrossRef]
- Taranto, F.; D’Agostino, N.; Rodriguez, M.; Pavan, S.; Minervini, A.P.; Pecchioni, N.; Papa, R.; De Vita, P. Whole genome scan reveals molecular signatures of divergence and selection related to important traits in durum wheat germplasm. Front. Genet. (accepted).
- Taranto, F.; Nicolia, A.; Pavan, S.; De Vita, P.; D’Agostino, N. Biotechnological and digital revolution for climate-smart plant breeding. Agronomy 2018, 8, 277. [Google Scholar] [CrossRef]
| SSR Locus Name | Na | Ne | I | Ho | He | F | PIC |
|---|---|---|---|---|---|---|---|
| VrZAG21 | 11 | 5.10 | 1.87 | 0.855 | 0.806 | −0.061 | 0.780 |
| VrZAG62 | 11 | 7.40 | 2.11 | 0.914 | 0.865 | −0.057 | 0.850 |
| VrZAG64 | 7 | 5.20 | 1.73 | 0.889 | 0.806 | −0.102 | 0.778 |
| VvMD25 | 7 | 4.10 | 1.56 | 0.748 | 0.758 | 0.013 | 0.721 |
| VvMD27 | 9 | 5.40 | 1.85 | 0.827 | 0.814 | −0.015 | 0.791 |
| VvMD28 | 15 | 6.80 | 2.13 | 0.849 | 0.853 | 0.004 | 0.836 |
| VvMD32 | 12 | 6.40 | 2.04 | 0.789 | 0.845 | 0.066 | 0.827 |
| VvMD7 | 11 | 5.40 | 1.88 | 0.844 | 0.815 | −0.036 | 0.792 |
| VvS2 | 14 | 5.50 | 2.00 | 0.891 | 0.82 | −0.086 | 0.799 |
| VrZAG79 | 13 | 7.40 | 2.22 | 0.891 | 0.866 | −0.028 | 0.853 |
| Total | 110 | - | - | - | - | - | |
| Min | 7 | 4.10 | 1.56 | 0.748 | 0.758 | −0.102 | 0.721 |
| Max | 15 | 7.40 | 2.22 | 0.914 | 0.866 | 0.066 | 0.853 |
| Mean | 11 | 5.90 | 1.94 | 0.849 | 0.825 | −0.03 | 0.802 |
| Genotypes | Type | No. Private Alleles | Markers |
|---|---|---|---|
| Malaga | Minor | 2 | VVMD28/ZG79 |
| Unknown Gargano S8 | Minor | 1 | VVS2 |
| Canosa Terrizzuolo | Minor | 1 | ZG79 |
| Sabellone | Minor | 1 | VVS2 |
| Primitivo pazzo | Minor | 1 | VVS2 |
| Unknown Bianca Serio | Minor | 2 | VVS2/ZG79 |
| Unknown Bianca Acino Ovoide | Minor | 1 | VVMD28 |
| Neratosta (1) | Minor | 1 | VVMD28 |
| Unknown Vite Longo | Minor | 3 | VVMD7/VVMD28/VVMD32 |
| Aleatico nero | Apulian | 1 | ZG21 |
| Unknown Ibrido Urgesi | Minor | 4 | VVMD7/VVS2/ZG62/ZG79 |
| Cornaletta nera | Minor | 1 | VVMD32 |
| Nera ovale | Minor | 1 | VVS2 |
| Unknown Gargano S5 | Minor | 1 | VVMD7 |
| Cabernet- Sauvignon | International | 1 | VVS2 |
| Genotype | Cases of Synonymy | |||
|---|---|---|---|---|
| Acchitedda | Garganega bianco | |||
| Cannellino bianco | Moscardinella | Moscatiddone | Moscato di Alessandria | |
| Caldarese rosso | Rosso di Lecce | |||
| Dolciolo Pendolino | Plaus bianca | |||
| Don Michele | Sangiovese | |||
| Frascarosa | Marchione | |||
| unknown Gargano S6 | Palumbo Uva Carrieri | Sagra | ||
| Malvasia antica | Malvasia bianca lunga | Malvasia saracino | Malvasia bianca | 80-Plaus bianca |
| Nardobello 2 | Nardobello 1 | balbut | ||
| Nocella | Uva di Troia | |||
| ottavianello | Ottaviano nero | |||
| Piccola nera Longo | Primitivo | |||
| Pinot blanc | Pinot noir | |||
| Rossa Bitonto | Rossa scorrese | |||
| Sciali | Santa Sofia | |||
| Sgarraparete | Barbarossa | |||
| Somarello rosso | Somarello nero | |||
| Strazzacambiali | Mostosa bianca | |||
| Tintiglio | Aglianico nero | |||
| Trifera | Agresta | |||
| Uva Ruggia | Prunesta nera | Uva Romana | ||
| Pagghione bianco | Paglione bianco | |||
| Mangiaverde | Mennaverde | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miazzi, M.M.; D’Agostino, N.; di Rienzo, V.; Venerito, P.; Savino, V.N.; Fucilli, V.; Ruffa, P.; Roseti, V.; Pirolo, C.; La Notte, P.; et al. Marginal Grapevine Germplasm from Apulia (Southern Italy) Represents an Unexplored Source of Genetic Diversity. Agronomy 2020, 10, 563. https://doi.org/10.3390/agronomy10040563
Miazzi MM, D’Agostino N, di Rienzo V, Venerito P, Savino VN, Fucilli V, Ruffa P, Roseti V, Pirolo C, La Notte P, et al. Marginal Grapevine Germplasm from Apulia (Southern Italy) Represents an Unexplored Source of Genetic Diversity. Agronomy. 2020; 10(4):563. https://doi.org/10.3390/agronomy10040563
Chicago/Turabian StyleMiazzi, Monica Marilena, Nunzio D’Agostino, Valentina di Rienzo, Pasquale Venerito, Vito Nicola Savino, Vincenzo Fucilli, Paola Ruffa, Vincenzo Roseti, Costantino Pirolo, Pierfederico La Notte, and et al. 2020. "Marginal Grapevine Germplasm from Apulia (Southern Italy) Represents an Unexplored Source of Genetic Diversity" Agronomy 10, no. 4: 563. https://doi.org/10.3390/agronomy10040563
APA StyleMiazzi, M. M., D’Agostino, N., di Rienzo, V., Venerito, P., Savino, V. N., Fucilli, V., Ruffa, P., Roseti, V., Pirolo, C., La Notte, P., Montemurro, C., & Taranto, F. (2020). Marginal Grapevine Germplasm from Apulia (Southern Italy) Represents an Unexplored Source of Genetic Diversity. Agronomy, 10(4), 563. https://doi.org/10.3390/agronomy10040563









