Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding
Abstract
1. Introduction
2. Materials and Methods
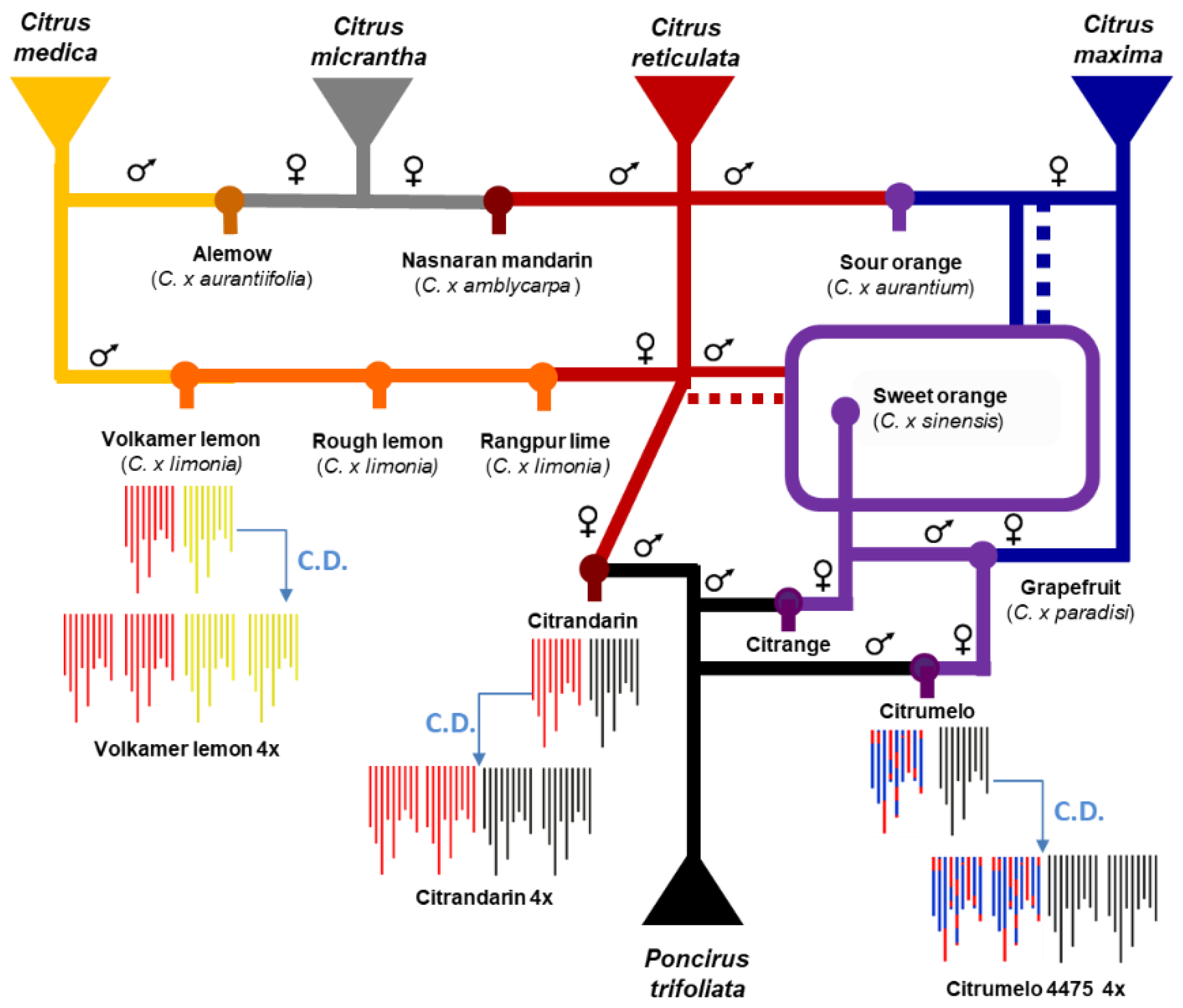
2.1. Plant Material
2.2. GBS Analysis
2.3. Diversity and Genetic Relationship Analysis Based on GBS Data
2.4. Development of P. trifoliata DSNPs Kaspar Markers and Population Genotyping
2.5. Intergeneric Recombination and Mapping Analysis of Tetraploid Citrumelo 4475
2.6. Estimation of Preferential Pairing (PP)
3. Results
3.1. Poncirus DSNP Mining from GBS Data
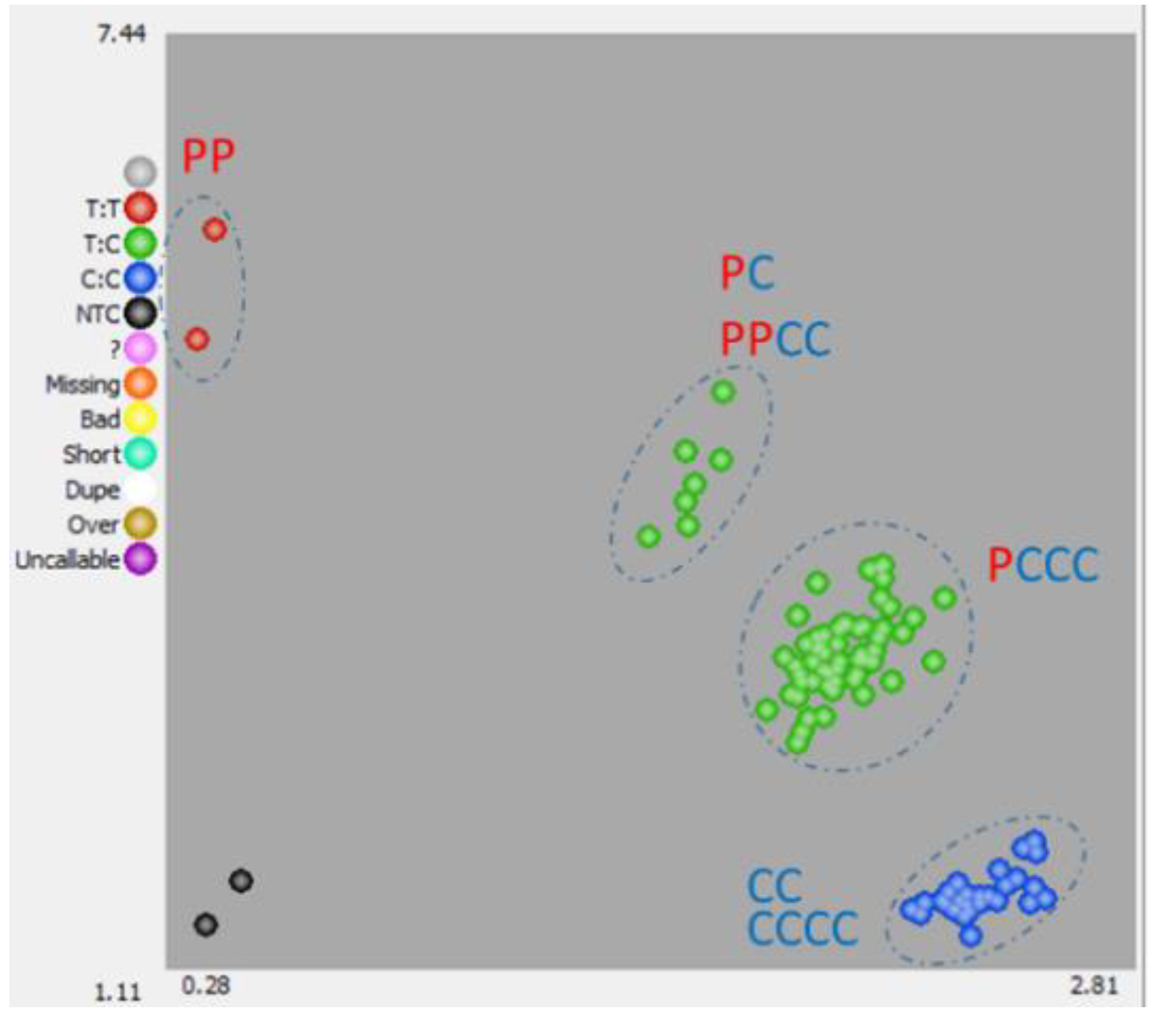
3.2. Development and Validation of Kaspar Markers
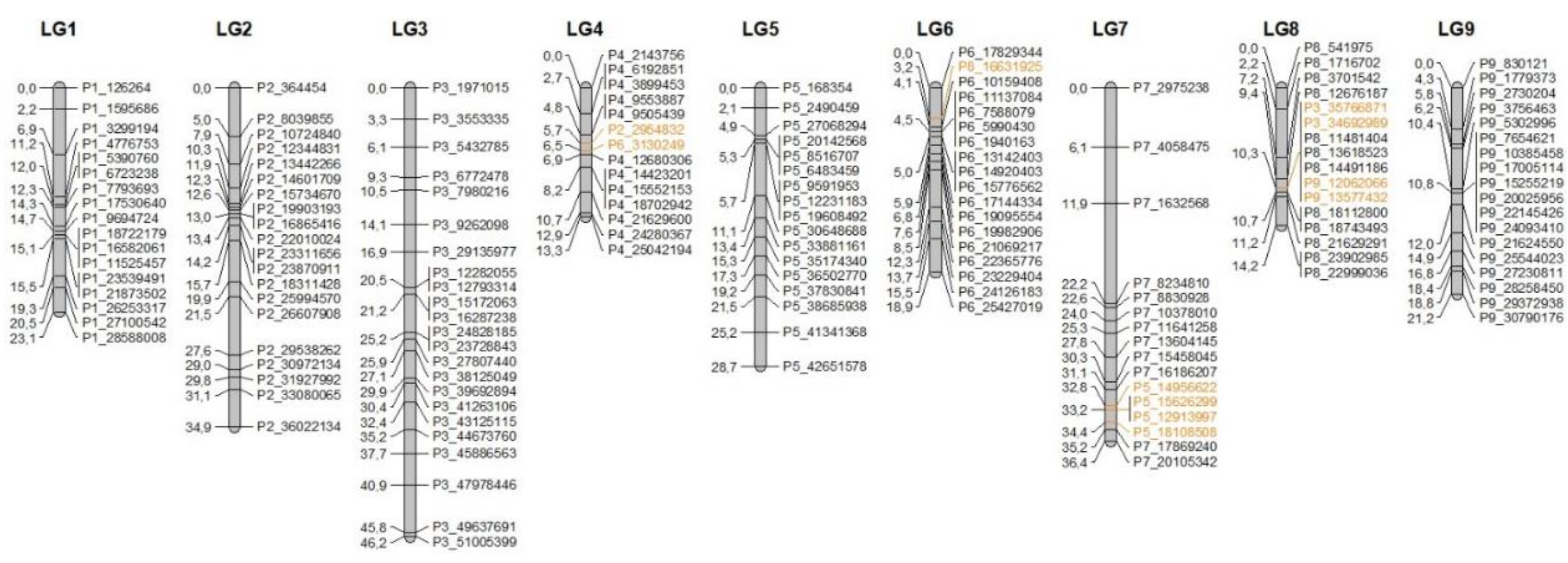
3.3. Genetic Mapping
3.4. Parental Heterozygosity Restitution and Meiotic Inheritance Analysis
3.5. Intergeneric Recombination and Genetic Structure of the Diploid Citrumelo and Citrandarin Gametes
4. Discussion
4.1. A set of 159 KAPar SNP Markers Fully Distinguishing Poncirus from Citrus Species was Validated
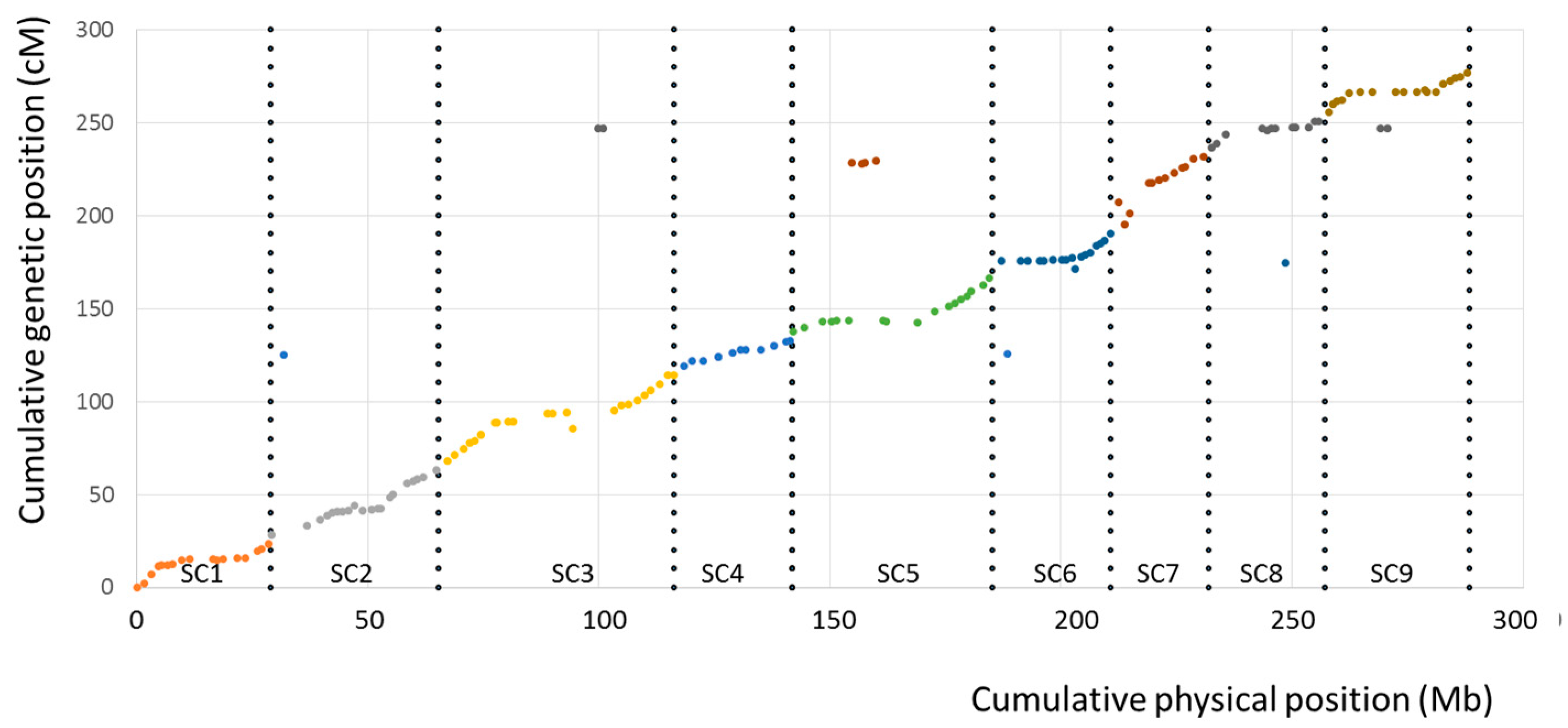
4.2. The Tetraploid Citrumelo Genetic Map Confirms the Large Synteny and Co-Linearity between Citrus and Poncirus Genomes
4.3. The Two Tetraploid Intergeneric Hybrids Display Intermediate Inheritance with a Disomic Tendency
4.4. Intergeneric Recombination Occurs but at Relatively Low Frequency and the Diploid Citrumelo Gametes Transmit a Large Part of the Intergeneric Heterozygosity of the Original Diploid 4475 Citrumelo Rootstock
4.5. Implications for Citrus Rootstock Breeding
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Castle, W.S. A Career Perspective on Citrus Rootstocks, Their Development, and Commercialization. HortScience 2010, 45, 11–15. [Google Scholar] [CrossRef]
- De Ollas, C.; Morillón, R.; Fotopoulos, V.; Puértolas, J.; Ollitrault, P.; Gómez-Cadenas, A.; Arbona, V. Facing Climate Change: Biotechnology of Iconic Mediterranean Woody Crops. Front. Plant Sci. 2019, 10, 427. [Google Scholar] [CrossRef] [PubMed]
- Syvertsen, J.P.; Garcia-Sanchez, F. Multiple abiotic stresses occurring with salinity stress in citrus. Environ. Exp. Bot. 2014, 103, 128–137. [Google Scholar] [CrossRef]
- Román, M.P.; Cambra, M.; Juárez, J.; Moreno, P.; Duran-Vila, N.; Tanaka, F.A.O.; Alves, E.; Kitajima, E.W.; Yamamoto, P.T.; Bassanezi, R.B.; et al. Sudden Death of Citrus in Brazil: A Graft-Transmissible Bud Union Disease. Plant Dis. 2004, 88, 453–467. [Google Scholar] [CrossRef]
- Yang, Z.-N.; Ye, X.-R.; Choi, S.; Molina, J.; Moonan, F.; Wing, R.A.; Roose, M.L.; Mirkov, T.E. Construction of a 1.2-Mb contig including the citrus tristeza virus resistance gene locus using a bacterial artificial chromosome library of Poncirus trifoliata (L.) Raf. Genome 2001, 44, 382–393. [Google Scholar] [CrossRef]
- Spiegel-Roy, P.; Goldschmidt, E.E. The Biology of Citrus. In The Biology of Horticultural Crops; Cambridge University Press: Cambridge, UK, 1996; ISBN 978-0-521-33321-4. [Google Scholar]
- Aritua, V.; Achor, D.; Gmitter, F.G.; Albrigo, G.; Wang, N. Transcriptional and Microscopic Analyses of Citrus Stem and Root Responses to Candidatus Liberibacter asiaticus Infection. PLoS ONE 2013, 8, e73742. [Google Scholar] [CrossRef]
- Johnson, E.G.; Wu, J.; Bright, D.B.; Graham, J.H. Association of ‘Candidatus Liberibacter asiaticus’ root infection, but not phloem plugging with root loss on huanglongbing-affected trees prior to appearance of foliar symptoms. Plant Pathol. 2014, 63, 290–298. [Google Scholar] [CrossRef]
- Da Graça, J.V.; Douhan, G.W.; Halbert, S.E.; Keremane, M.L.; Lee, R.F.; Vidalakis, G.; Zhao, H. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus: Citrus huanglongbing. J. Integr. Plant Biol. 2016, 58, 373–387. [Google Scholar] [CrossRef]
- Ramadugu, C.; Keremane, M.L.; Halbert, S.E.; Duan, Y.P.; Roose, M.L.; Stover, E.; Lee, R.F. Long-Term Field Evaluation Reveals Huanglongbing Resistance in Citrus Relatives. Plant Dis. 2016, 100, 1858–1869. [Google Scholar] [CrossRef]
- Ollitrault, P.; Germanà, M.A.; Froelicher, Y.; Cuenca, J.; Aleza, P.; Morillon, R.; Grosser, J.W.; Guo, W. Ploidy Manipulation for Citrus Breeding, Genetics, and Genomics. In The Citrus Genome; Gentile, A., La Malfa, S., Deng, Z., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 75–105. ISBN 978-3-030-15308-3. [Google Scholar]
- Mouhaya, W.; Allario, T.; Brumos, J.; Andres, F.; Froelicher, Y.; Luro, F.; Talon, M.; Ollitrault, P.; Morillon, R. Sensitivity to high salinity in tetraploid citrus seedlings increases with water availability and correlates with expression of candidate genes. Funct. Plant Biol. 2010, 37, 674–685. [Google Scholar] [CrossRef]
- Podda, A.; Checcucci, G.; Mouhaya, W.; Centeno, D.; Rofidal, V.; Del Carratore, R.; Luro, F.; Morillon, R.; Ollitrault, P.; Maserti, B.E. Salt-stress induced changes in the leaf proteome of diploid and tetraploid mandarins with contrasting Na+ and Cl− accumulation behaviour. J. Plant Physiol. 2013, 170, 1101–1112. [Google Scholar] [CrossRef]
- Ruiz, M.; Quiñones, A.; Martínez-Alcántara, B.; Aleza, P.; Morillon, R.; Navarro, L.; Primo-Millo, E.; Martínez-Cuenca, M.-R. Effects of salinity on diploid (2x) and doubled diploid (4x) Citrus macrophylla genotypes. Sci. Hortic. 2016, 207, 33–40. [Google Scholar] [CrossRef]
- De Souza, J.D.; Rodrigues, V.; Barigah, T.; Ollitrault, P.; Gesteira, A.d.S.; Morillon, R. Whole genome expression and hydraulic architecture of diploid and doubled diploid citrus seedlings in intra and interspecific contexts. Citrus Res. Technol. 2016, 37, 206–210. [Google Scholar] [CrossRef]
- Oliveira, T.M.; Yahmed, J.B.; Dutra, J.; Maserti, B.E.; Talon, M.; Navarro, L.; Ollitraut, P.; Gesteira, A.D.; Morillon, R. Better tolerance to water deficit in doubled diploid ‘Carrizo citrange’ compared to diploid seedlings is associated with more limited water consumption. Acta Physiol. Plant. 2017, 39, 204. [Google Scholar] [CrossRef]
- Khalid, M.F.; Hussain, S.; Anjum, M.A.; Ahmad, S.; Ali, M.A.; Ejaz, S.; Morillon, R. Better salinity tolerance in tetraploid vs diploid volkamer lemon seedlings is associated with robust antioxidant and osmotic adjustment mechanisms. J. Plant Physiol. 2020, 244, 153071. [Google Scholar] [CrossRef] [PubMed]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Iglesias, D.J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P.; Morillon, R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Environ. 2013, 36, 856–868. [Google Scholar] [CrossRef] [PubMed]
- Dutra de Souza, J.; de Andrade Silva, E.M.; Coelho Filho, M.A.; Morillon, R.; Bonatto, D.; Micheli, F.; da Silva Gesteira, A. Different adaptation strategies of two citrus scion/rootstock combinations in response to drought stress. PLoS ONE 2017, 12, e0177993. [Google Scholar] [CrossRef] [PubMed]
- Oustric, J.; Lourkisti, R.; Herbette, S.; Morillon, R.; Paolacci, G.; Gonzalez, N.; Berti, L.; Santini, J. Effect of Propagation Method and Ploidy Level of Various Rootstocks on the Response of the Common Clementine (Citrus clementina Hort. ex Tan) to a Mild Water Deficit. Agriculture 2020, 10, 321. [Google Scholar] [CrossRef]
- Oustric, J.; Morillon, R.; Luro, F.; Herbette, S.; Lourkisti, R.; Giannettini, J.; Berti, L.; Santini, J. Tetraploid Carrizo citrange rootstock (Citrus sinensis Osb. X Poncirus trifoliata L. Raf.) enhances natural chilling stress tolerance of common clementine (Citrus clementina Hort. ex Tan). J. Plant Physiol. 2017, 214, 108–115. [Google Scholar] [CrossRef]
- Oustric, J.; Morillon, R.; Ollitrault, P.; Herbette, S.; Luro, F.; Froelicher, Y.; Tur, I.; Dambier, D.; Giannettini, J.; Berti, L.; et al. Somatic hybridization between diploid Poncirus and Citrus improves natural chilling and light stress tolerances compared with equivalent doubled-diploid genotypes. Trees 2018, 32, 883–895. [Google Scholar] [CrossRef]
- Oustric, J.; Quilichini, Y.; Morillon, R.; Herbette, S.; Luro, F.; Giannettini, J.; Berti, L.; Santini, J. Tetraploid citrus seedlings subjected to long-term nutrient deficiency are less affected at the ultrastructural, physiological and biochemical levels than diploid ones. Plant Physiol. Biochem. 2019, 135, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.; Quiñones, A.; Martínez-Alcántara, B.; Aleza, P.; Morillon, R.; Navarro, L.; Primo-Millo, E.; Martínez-Cuenca, M.-R. Tetraploidy Enhances Boron-Excess Tolerance in Carrizo Citrange (Citrus sinensis L. Osb. × Poncirus trifoliata L. Raf.). Front. Plant Sci. 2016, 7, 701. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.-Q.; Tu, H.; Liang, W.-J.; Long, J.-M.; Wu, X.-M.; Zhang, H.-Y.; Guo, W.-W. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 2015, 15, 89. [Google Scholar] [CrossRef] [PubMed]
- Grosser, J.W.; Chandler, J.L. Somatic hybridization of high yield, cold-hardy and disease resistant parents for citrus rootstock improvement. J. Hortic. Sci. Biotechnol. 2000, 75, 641–644. [Google Scholar] [CrossRef]
- Grosser, J.W.; Ollitrault, P.; Olivares-Fuster, O. Somatic hybridization in citrus: An effective tool to facilitate variety improvement. Vitro Cell. Dev. Biol.-Plant 2000, 36, 434–449. [Google Scholar] [CrossRef]
- Guo, W.W.; Wu, R.C.; Cheng, Y.J.; Deng, X.X. Production and molecular characterization of Citrus intergeneric somatic hybrids between red tangerine and citrange. Plant Breed. 2007, 126, 72–76. [Google Scholar] [CrossRef]
- Dambier, D.; Benyahia, H.; Pensabene-Bellavia, G.; Aka Kaçar, Y.; Froelicher, Y.; Belfalah, Z.; Lhou, B.; Handaji, N.; Printz, B.; Morillon, R.; et al. Somatic hybridization for citrus rootstock breeding: An effective tool to solve some important issues of the Mediterranean citrus industry. Plant Cell Rep. 2011, 30, 883–900. [Google Scholar] [CrossRef]
- Grosser, J.W.; Gmitter, F.G. Protoplast fusion for production of tetraploids and triploids: Applications for scion and rootstock breeding in citrus. Plant Cell Tissue Organ Cult. PCTOC 2011, 104, 343–357. [Google Scholar] [CrossRef]
- Guerra, D.; Schifino-Wittmann, M.T.; Schwarz, S.F.; Weiler, R.L.; Dahmer, N.; Souza, P.V.D. de Tetraploidization in citrus rootstocks: Effect of genetic constitution and environment in chromosome duplication. Crop Breed. Appl. Biotechnol. 2016, 16, 35–41. [Google Scholar] [CrossRef]
- Ollitrault, P.; Froelicher, Y.; Dambier, D.; Seker, M. Rootstock breeding by somatic hybridisation for the mediterranean citrus industry. In Proceedings of the First International Citrus Biotechnology Symposium, 2000, Eliat, Israel, 29 November–3 December 1998; pp. 157–162. [Google Scholar]
- Ollitrault, P.; Guo, W.; Grosser, J.W. Somatic hybridization. In Citrus Genetics, Breeding and Biotechnology; Khan Igar, A., Ed.; CAB International: Wallingford, UK, 2007; pp. 235–260. [Google Scholar]
- Grosser, J.W.; Barthe, G.A.; Castle, B.; Gmitter, F.G., Jr.; Lee, O. The development of improved tetraploid citrus rootstocks to facilitate advanced production systems and sustainable citriculture in Florida. Acta Hortic. 2015, 1065, 319–327. [Google Scholar] [CrossRef]
- Aleza, P.; Froelicher, Y.; Schwarz, S.; Agustí, M.; Hernández, M.; Juárez, J.; Luro, F.; Morillon, R.; Navarro, L.; Ollitrault, P. Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Ann. Bot. 2011, 108, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, M.; Pensabene-Bellavia, G.; Quiñones, A.; García-Lor, A.; Morillon, R.; Ollitrault, P.; Primo-Millo, E.; Navarro, L.; Aleza, P. Molecular Characterization and Stress Tolerance Evaluation of New Allotetraploid Somatic Hybrids Between Carrizo Citrange and Citrus macrophylla W. rootstocks. Front. Plant Sci. 2018, 9, 901. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Comte, A.; Curk, F.; Costantino, G.; Luro, F.; Dereeper, A.; Mournet, P.; Froelicher, Y.; Ollitrault, P. Genotyping by sequencing can reveal the complex mosaic genomes in gene pools resulting from reticulate evolution: A case study in diploid and polyploid citrus. Ann. Bot. 2019, 123, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. The role of genetic and genomic attributes in the success of polyploids. Proc. Natl. Acad. Sci. USA 2000, 97, 7051–7057. [Google Scholar] [CrossRef]
- Stebbins, G.L. Types of Polyploids: Their Classification and Significance. In Advances in Genetics; Demerec, M., Ed.; University of California Press: Berkeley, CA, USA, 1947; Volume 1, pp. 403–429. ISBN 0065-2660. [Google Scholar]
- Udall, J.A.; Wendel, J.F. Polyploidy and Crop Improvement. Crop Sci. 2006, 46, S3–S14. [Google Scholar] [CrossRef]
- Stift, M.; Berenos, C.; Kuperus, P.; van Tienderen, P.H. Segregation Models for Disomic, Tetrasomic and Intermediate Inheritance in Tetraploids: A General Procedure Applied to Rorippa (Yellow Cress) Microsatellite Data. Genetics 2008, 179, 2113–2123. [Google Scholar] [CrossRef]
- Jeridi, M.; Perrier, X.; Rodier-Goud, M.; Ferchichi, A.; D’Hont, A.; Bakry, F. Cytogenetic evidence of mixed disomic and polysomic inheritance in an allotetraploid (AABB) Musa genotype. Ann. Bot. 2012, 110, 1593–1606. [Google Scholar] [CrossRef]
- Kamiri, M.; Stift, M.; Costantino, G.; Dambier, D.; Kabbage, T.; Ollitrault, P.; Froelicher, Y. Preferential Homologous Chromosome Pairing in a Tetraploid Intergeneric Somatic Hybrid (Citrus reticulata + Poncirus trifoliata) Revealed by Molecular Marker Inheritance. Front. Plant Sci. 2018, 9, 1557. [Google Scholar] [CrossRef] [PubMed]
- Rouiss, H.; Bakry, F.; Froelicher, Y.; Navarro, L.; Aleza, P.; Ollitrault, P. Origin of C. latifolia and C. aurantiifolia triploid limes: The preferential disomic inheritance of doubled-diploid ‘Mexican’ lime is consistent with an interploid hybridization hypothesis. Ann. Bot. 2018, 121, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Danzmann, R.G. Secondary tetrasomic segregation of MDH-B and preferential pairing of homeologues in rainbow trout. Genetics 1997, 145, 1083–1092. [Google Scholar] [PubMed]
- Barone, A.; Li, J.; Sebastiano, A.; Cardi, T.; Frusciante, L. Evidence for tetrasomic inheritance in a tetraploid Solanum commersonii (+) S. tuberosum somatic hybrid through the use of molecular markers. Theor. Appl. Genet. 2002, 104, 539–546. [Google Scholar] [CrossRef]
- Jannoo, N.; Grivet, L.; David, J.; D’Hont, A.; Glaszmann, J.-C. Differential chromosome pairing affinities at meiosis in polyploid sugarcane revealed by molecular markers. Heredity 2004, 93, 460–467. [Google Scholar] [CrossRef][Green Version]
- Bousalem, M.; Arnau, G.; Hochu, I.; Arnolin, R.; Viader, V.; Santoni, S.; David, J. Microsatellite segregation analysis and cytogenetic evidence for tetrasomic inheritance in the American yam Dioscorea trifida and a new basic chromosome number in the Dioscoreae. Theor. Appl. Genet. 2006, 113, 439–451. [Google Scholar] [CrossRef]
- Landergott, U.; Naciri, Y.; Schneller, J.J.; Holderegger, R. Allelic configuration and polysomic inheritance of highly variable microsatellites in tetraploid gynodioecious Thymus praecox agg. Theor. Appl. Genet. 2006, 113, 453–465. [Google Scholar] [CrossRef]
- Cuenca, J.; Aleza, P.; Navarro, L.; Ollitrault, P. Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: Application to citrus triploid progeny. Ann. Bot. 2013, 111, 731–742. [Google Scholar] [CrossRef][Green Version]
- Aleza, P.; Cuenca, J.; Hernández, M.; Juárez, J.; Navarro, L.; Ollitrault, P. Genetic mapping of centromeres in the nine Citrus clementina chromosomes using half-tetrad analysis and recombination patterns in unreduced and haploid gametes. BMC Plant Biol. 2015, 15, 80. [Google Scholar] [CrossRef]
- Rouiss, H.; Cuenca, J.; Navarro, L.; Ollitrault, P.; Aleza, P. Tetraploid citrus progenies arising from FDR and SDR unreduced pollen in 4x X 2x hybridizations. Tree Genet. Genomes 2017, 13, 10. [Google Scholar] [CrossRef]
- Rouiss, H.; Cuenca, J.; Navarro, L.; Ollitrault, P.; Aleza, P. Unreduced Megagametophyte Production in Lemon Occurs via Three Meiotic Mechanisms, Predominantly Second-Division Restitution. Front. Plant Sci. 2017, 8, 1211. [Google Scholar] [CrossRef] [PubMed]
- Aleza, P.; Cuenca, J.; Juárez, J.; Navarro, L.; Ollitrault, P. Inheritance in doubled-diploid clementine and comparative study with SDR unreduced gametes of diploid clementine. Plant Cell Rep. 2016, 35, 1573–1586. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, D.; Curk, F.; Evrard, J.C.; Froelicher, Y.; Ollitrault, P. Preferential Disomic Segregation and C. micrantha/C. medica Interspecific Recombination in Tetraploid ‘Giant Key’ Lime; Outlook for Triploid Lime Breeding. Front. Plant Sci. 2020, 11, 939. [Google Scholar] [CrossRef] [PubMed]
- Castle, B.; Stover, E. Rootstock refections: Swingle citrumelo update. Citrus Industry 2000, 81, 18–20. [Google Scholar]
- Roose, M.L. Rootstocks. In Citrus Production Manual; Ferguson, L., Grafton-Cardwell, E.E., Eds.; University of California Agriculture and Natural Resources Communication Services: Oakland, CA, USA, 2014; pp. 95–105. [Google Scholar]
- Oueslati, A.; Salhi-Hannachi, A.; Luro, F.; Vignes, H.; Mournet, P.; Ollitrault, P. Genotyping by sequencing reveals the interspecific C. maxima / C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits. PLoS ONE 2017, 12, e0185618. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A Robust, Simple Genotyping-by-Sequencing (GBS) Approach for High Diversity Species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef] [PubMed]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef]
- Glaubitz, J.C.; Casstevens, T.M.; Lu, F.; Harriman, J.; Elshire, R.J.; Sun, Q.; Buckler, E.S. TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE 2014, 9, e90346. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X. Jacquemoud-Collet DARwin Software. 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 8 December 2020).
- Bruyère, S.; Luro, F.; Morillon, R.; Ollitrault, P. Poncirus phylogenetic diagnostic SNPs markers are useful to analyse zygotic rates in diploid and tetraploid Citrus x Poncirus rootstock seedlings. In Abstract Book Sustainable Citriculture: The role of Applied Knowledge; Mattos, D., Fermino, C.E., Moreira, N.V., Alves de Azevedo, F., Della Coletta, F.H., Vicente Contador, Z.P., Eds.; IAPAR: Londrina, Brazil, 2016; p. 125. [Google Scholar]
- Cuppen, E. Genotyping by Allele-Specific Amplification (KASPar). Cold Spring Harb. Protoc. 2007, 2007, pdb.prot4841. [Google Scholar] [CrossRef] [PubMed]
- Grandke, F.; Ranganathan, S.; van Bers, N.; de Haan, J.R.; Metzler, D. PERGOLA: Fast and deterministic linkage mapping of polyploids. BMC Bioinform. 2017, 18, 12. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Kamiri, M.; Stift, M.; Srairi, I.; Costantino, G.; Moussadik, A.E.; Hmyene, A.; Bakry, F.; Ollitrault, P.; Froelicher, Y. Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic hybrid between C. reticulata and C. limon using SSR markers and cytogenetic analysis. Plant Cell Rep. 2011, 30, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Gmitter, F.G., Jr. Mining of haplotype-based expressed sequence tag single nucleotide polymorphisms in citrus. BMC Genom. 2013, 14, 746. [Google Scholar] [CrossRef]
- Ollitrault, P.; Terol, J.; Garcia-Lor, A.; Bérard, A.; Chauveau, A.; Froelicher, Y.; Belzile, C.; Morillon, R.; Navarro, L.; Brunel, D.; et al. SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genom. 2012, 13, 13. [Google Scholar] [CrossRef]
- Garcia-Lor, A.; Ancillo, G.; Navarro, L.; Ollitrault, P. Citrus (Rutaceae) SNP Markers Based on Competitive Allele-Specific PCR; Transferability Across the Aurantioideae Subfamily. Appl. Plant Sci. 2013, 1, 1200406. [Google Scholar] [CrossRef]
- Cuenca, J.; Aleza, P.; Juárez, J.; García-Lor, A.; Froelicher, Y.; Navarro, L.; Ollitrault, P. Maximum-likelihood method identifies meiotic restitution mechanism from heterozygosity transmission of centromeric loci: Application in citrus. Sci. Rep. 2015, 5, 9897. [Google Scholar] [CrossRef][Green Version]
- Curk, F.; Ancillo, G.; Ollitrault, F.; Perrier, X.; Jacquemoud-Collet, J.-P.; Garcia-Lor, A.; Navarro, L.; Ollitrault, P. Nuclear Species-Diagnostic SNP Markers Mined from 454 Amplicon Sequencing Reveal Admixture Genomic Structure of Modern Citrus Varieties. PLoS ONE 2015, 10, e0125628. [Google Scholar] [CrossRef]
- Bernet, G.P.; Fernandez-Ribacoba, J.; Carbonell, E.A.; Asins, M.J. Comparative genome-wide segregation analysis and map construction using a reciprocal cross design to facilitate citrus germplasm utilization. Mol. Breed. 2010, 25, 659–673. [Google Scholar] [CrossRef]
- Ollitrault, P.; Terol, J.; Chen, C.; Federici, C.T.; Lotfy, S.; Hippolyte, I.; Ollitrault, F.; Bérard, A.; Chauveau, A.; Cuenca, J.; et al. A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genom. 2012, 13, 593. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Fujii, H.; Endo, T.; Ueda, T.; Sugiyama, A.; Nakano, M.; Kita, M.; Yoshioka, T.; Shimizu, T.; Nesumi, H.; et al. Construction of a citrus framework genetic map anchored by 708 gene-based markers. Tree Genet. Genomes 2014, 10, 1001–1013. [Google Scholar] [CrossRef]
- Yu, Y.; Chen, C.; Gmitter, F.G. QTL mapping of mandarin (Citrus reticulata) fruit characters using high-throughput SNP markers. Tree Genet. Genomes 2016, 12, 77. [Google Scholar] [CrossRef]
- Curtolo, M.; Soratto, T.A.T.; Gazaffi, R.; Takita, M.A.; Machado, M.A.; Cristofani-Yaly, M. High-density linkage maps for Citrus sunki and Poncirus trifoliata using DArTseq markers. Tree Genet. Genomes 2018, 14, 5. [Google Scholar] [CrossRef]
- Chen, C.; Bowman, K.D.; Choi, Y.A.; Dang, P.M.; Rao, M.N.; Huang, S.; Soneji, J.R.; McCollum, T.G.; Gmitter, F.G. EST-SSR genetic maps for Citrus sinensis and Poncirus trifoliata. Tree Genet. Genomes 2007, 4, 1–10. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, Y.; Ye, J.; Pan, Z.; Tan, M.; Xie, Z.; Chai, L.; Xu, Q.; Fraser, P.D.; Deng, X. SLAF-Based Construction of a High-Density Genetic Map and Its Application in QTL Mapping of Carotenoids Content in Citrus Fruit. J. Agric. Food Chem. 2019, 67, 994–1002. [Google Scholar] [CrossRef]
- Huang, M.; Roose, M.L.; Yu, Q.; Du, D.; Yu, Y.; Zhang, Y.; Deng, Z.; Stover, E.; Gmitter, F.G. Construction of High-Density Genetic Maps and Detection of QTLs Associated with Huanglongbing Tolerance in Citrus. Front. Plant Sci. 2018, 9, 1694. [Google Scholar] [CrossRef]
- Garavello, M.; Cuenca, J.; Garcia-Lor, A.; Ortega, N.; Navarro, L.; Ollitrault, P.; Aleza, P. Male and female inheritance patterns in tetraploid ‘Moncada’ mandarin. Plant Cell Rep. 2020, 39, 335–349. [Google Scholar] [CrossRef]
- Xie, K.-D.; Xia, Q.-M.; Wang, X.-P.; Liang, W.-J.; Wu, X.-M.; Grosser, J.W.; Guo, W.-W. Cytogenetic and SSR-marker evidence of mixed disomic, tetrasomic, and intermediate inheritance in a citrus allotetraploid somatic hybrid between ‘Nova’ tangelo and ‘HB’ pummelo. Tree Genet. Genomes 2015, 11, 112. [Google Scholar] [CrossRef]
- Sybenga, J. Cytogenetics in Plant Breeding. In Monographs on Theoretical and Applied Genetics; Springer: Berlin/Heidelberg, Germany, 1992; Volume 17, ISBN 978-3-642-84085-2. [Google Scholar]
- Froelicher, Y.; Luro, F.; Ollitrault, P. Analysis of meiotic behavior of the tetraploid Clausena excavata species by molecular marker segregation studies. In Proceedings of the 9th ISC International Society of Citriculture Congress, Orlando, FL, USA, 3–7 December 2000. [Google Scholar]
- Sanford, J.C. Ploidy manipulations. In Methods in Fruit Breeding; Moore, J.N., Janick, J., Eds.; Purdue University Press: West Lafayette, IN, USA, 1983; pp. 100–123. [Google Scholar]
- Chen, C.L.; Guo, W.W.; Yi, H.L.; Deng, X.X. Cytogenetic analysis of two interspecific Citrus allotetraploid somatic hybrids and their diploid fusion parents. Plant Breed. 2004, 123, 332–337. [Google Scholar] [CrossRef]
- Cameron, J.W.; Garber, M.J. Identical-Twin Hybrids of Citrus x Poncirus from Strictly Sexual Seed Parents. Am. J. Bot. 1968, 55, 199–205. [Google Scholar] [CrossRef]
- Curole, J.P.; Hedgecock, D. Estimation of Preferential Pairing Rates in Second-Generation Autotetraploid Pacific Oysters (Crassostrea gigas). Genetics 2005, 171, 855–859. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, G.L. Variation and Evolution in Plants; Columbia University Press: New York, NY, USA, 1950; p. 643. [Google Scholar]
- Svačina, R.; Sourdille, P.; Kopecký, D.; Bartoš, J. Chromosome Pairing in Polyploid Grasses. Front. Plant Sci. 2020, 11, 1056. [Google Scholar] [CrossRef] [PubMed]
- Waines, J.G. A Model for the Origin of Diploidizing Mechanisms in Polyploid Species. Am. Nat. 1976, 110, 415–430. [Google Scholar] [CrossRef]
- Jenczewski, E.; Alix, K. From Diploids to Allopolyploids: The Emergence of Efficient Pairing Control Genes in Plants. Crit. Rev. Plant Sci. 2004, 23, 21–45. [Google Scholar] [CrossRef]
- Riley, R.; Law, C.N. Genetic Variation in Chromosome Pairing. In Advances in Genetics; Caspari, E.W., Thoday, J.M., Eds.; Academic Press: Cambridge, England, 1965; Volume 13, pp. 57–114. ISBN 0065-2660. [Google Scholar]
- McGuire, P.E.; Dvořák, J. Genetic regulation of heterogenetic chromosome pairing in polyploid species of the genus triticum. Can. J. Genet. Cytol. 1982, 24, 57–82. [Google Scholar] [CrossRef]
- Sears, E.R. Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 1976, 10, 31–51. [Google Scholar] [CrossRef]
- Parker, J.S.; Palmer, R.W.; Whitehorn, M.A.F.; Edgar, L.A. Chiasma frequency effects of structural chromosome change. Chromosoma 1982, 85, 673–686. [Google Scholar] [CrossRef]
- Chambers, S.R.; Hunter, N.; Louis, E.J.; Borts, R.H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol. Cell. Biol. 1996, 16, 6110–6120. [Google Scholar] [CrossRef][Green Version]
- Liharska, T.; van Wordragen, M.; van Kammen, A.; Zabel, P.; Koornneef, M. Tomato chromosome 6: Effect of alien chromosomal segments on recombinant frequencies. Genome 1996, 39, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Chetelat, R.T.; Meglic, V.; Cisneros, P. A Genetic Map of Tomato Based on BC1 Lycopersicon esculentum × Solanum lycopersicoides Reveals Overall Synteny but Suppressed Recombination Between These Homeologous Genomes. Genetics 2000, 154, 857. [Google Scholar] [PubMed]
- Opperman, R.; Emmanuel, E.; Levy, A.A. The Effect of Sequence Divergence on Recombination Between Direct Repeats in Arabidopsis. Genetics 2004, 168, 2207–2215. [Google Scholar] [CrossRef]
- Li, L.; Jean, M.; Belzile, F. The impact of sequence divergence and DNA mismatch repair on homeologous recombination in Arabidopsis. Plant J. 2006, 45, 908–916. [Google Scholar] [CrossRef] [PubMed]
- Lyon, M.T. A Genomic Genetic Map of the Common Sweet Orange and Poncirus Trifoliata. Ph.D. Thesis, University of California, Riverside, CA, USA, 2008. [Google Scholar]
- Imai, A.; Yoshioka, T.; Hayashi, T. Quantitative trait locus (QTL) analysis of fruit-quality traits for mandarin breeding in Japan. Tree Genet. Genomes 2017, 13, 79. [Google Scholar] [CrossRef]
- Guo, F.; Yu, H.; Tang, Z.; Jiang, X.; Wang, L.; Wang, X.; Xu, Q.; Deng, X. Construction of a SNP-based high-density genetic map for pummelo using RAD sequencing. Tree Genet. Genomes 2015, 11, 2. [Google Scholar] [CrossRef]
- Ollitrault, P.; Navarro, L. Citrus. In Fruit Breeding; Badenes, M.L., Byrne, D.H., Eds.; Springer: Boston, MA, USA, 2012; pp. 623–662. ISBN 978-1-4419-0762-2. [Google Scholar]

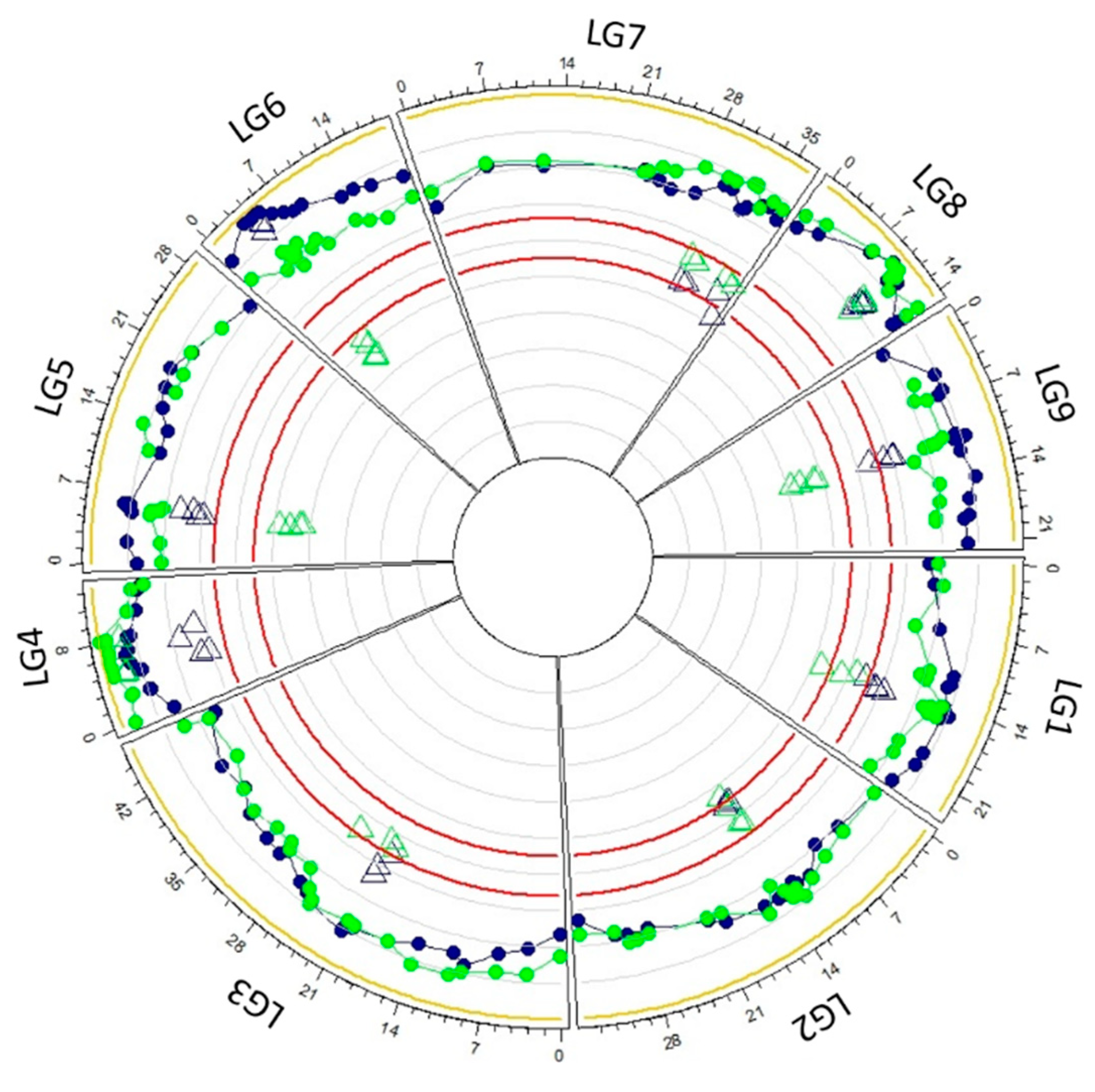
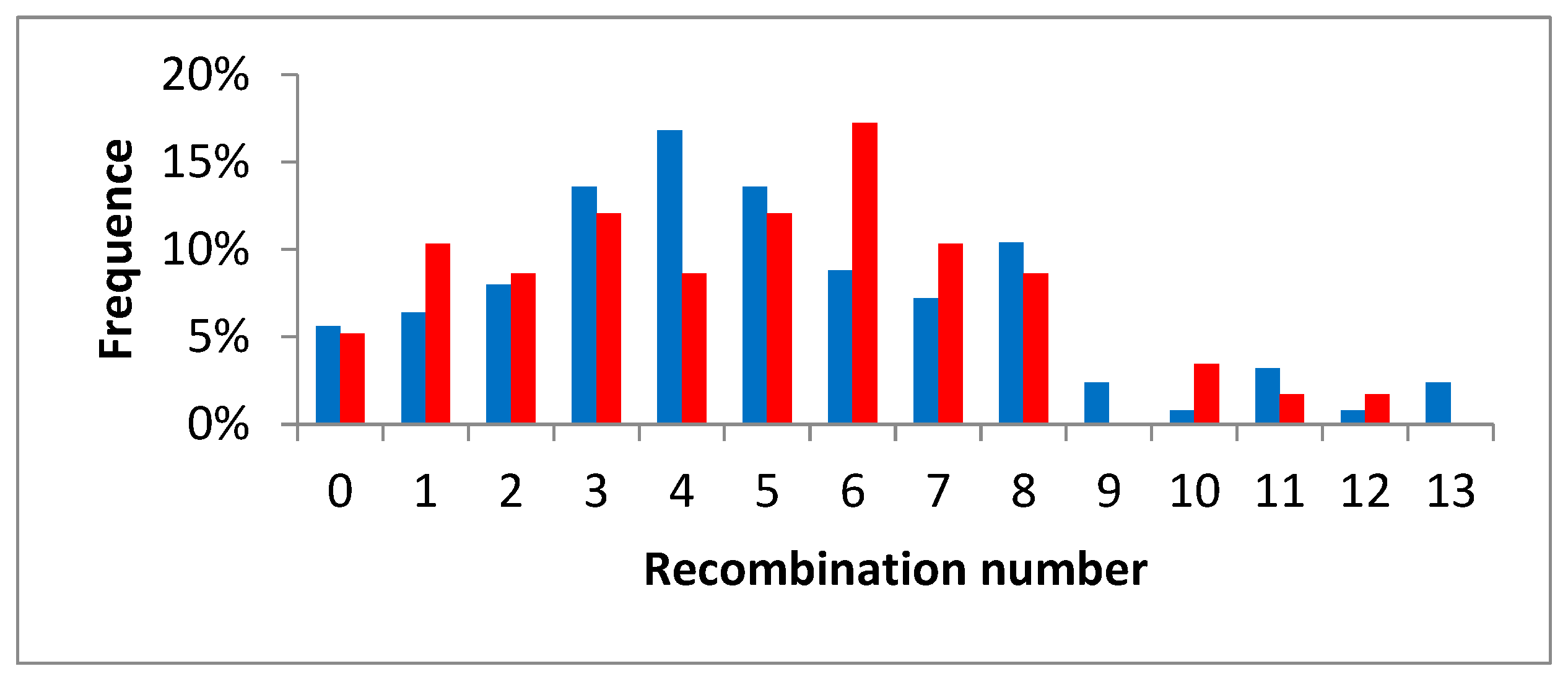

| Chr1 | Chr2 | Chr3 | Chr4 | Chr5 | Chr6 | Chr7 | Chr8 | Chr9 | Total | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C. medica | He | 0.035 | 0.035 | 0.038 | 0.036 | 0.042 | 0.033 | 0.031 | 0.035 | 0.036 | 0.036 |
| Ho | 0.038 | 0.035 | 0.039 | 0.034 | 0.049 | 0.035 | 0.035 | 0.039 | 0.046 | 0.039 | |
| C. reticulata | He | 0.055 | 0.072 | 0.060 | 0.067 | 0.061 | 0.047 | 0.054 | 0.077 | 0.061 | 0.062 |
| Ho | 0.071 | 0.083 | 0.076 | 0.078 | 0.074 | 0.057 | 0.065 | 0.083 | 0.070 | 0.074 | |
| C. maxima | He | 0.072 | 0.068 | 0.069 | 0.060 | 0.073 | 0.067 | 0.069 | 0.078 | 0.078 | 0.070 |
| Ho | 0.090 | 0.084 | 0.088 | 0.075 | 0.096 | 0.085 | 0.083 | 0.097 | 0.088 | 0.087 | |
| C. micrantha | He | 0.049 | 0.046 | 0.048 | 0.043 | 0.057 | 0.046 | 0.051 | 0.045 | 0.050 | 0.049 |
| Ho | 0.099 | 0.092 | 0.096 | 0.087 | 0.115 | 0.093 | 0.101 | 0.090 | 0.100 | 0.097 | |
| Citrus secondary species | He | 0.175 | 0.184 | 0.185 | 0.183 | 0.185 | 0.189 | 0.184 | 0.180 | 0.186 | 0.183 |
| Ho | 0.243 | 0.226 | 0.248 | 0.246 | 0.241 | 0.268 | 0.258 | 0.243 | 0.250 | 0.246 | |
| P. trifoliata | He | 0.048 | 0.050 | 0.047 | 0.047 | 0.057 | 0.044 | 0.050 | 0.046 | 0.048 | 0.049 |
| Ho | 0.081 | 0.086 | 0.082 | 0.077 | 0.099 | 0.072 | 0.082 | 0.080 | 0.085 | 0.083 | |
| Inter-generic hybrids | He | 0.196 | 0.195 | 0.200 | 0.195 | 0.195 | 0.203 | 0.196 | 0.196 | 0.202 | 0.197 |
| Ho | 0.319 | 0.304 | 0.307 | 0.312 | 0.298 | 0.314 | 0.300 | 0.302 | 0.314 | 0.308 | |
| Gst Poncirus/Citrus | 0.268 | 0.259 | 0.266 | 0.262 | 0.250 | 0.273 | 0.247 | 0.260 | 0.261 | 0.261 | |
| Number P. trifoliata dSNPs | 902 | 1006 | 1441 | 860 | 868 | 703 | 642 | 658 | 802 | 7882 | |
| LG | N.M | L.G. Size (cM) | G. Size (Mb) | RR (cM/Mb) |
|---|---|---|---|---|
| 1 | 17 | 23.12 | 28.462 | 0.812 |
| 2 | 20 | 34.91 | 35.658 | 0.979 |
| 3 | 23 | 46.17 | 49.034 | 0.942 |
| 4 | 14 | 13.3 | 22.898 | 0.581 |
| 5 | 17 | 28.67 | 42.483 | 0.675 |
| 6 | 18 | 18.86 | 23.487 | 0.803 |
| 7 | 16 | 36.38 | 18.473 | 1.969 |
| 8 | 16 | 14.18 | 23.361 | 0.607 |
| 9 | 18 | 21.22 | 29.960 | 0.708 |
| Total | 159 | 236.81 | 273.816 | 0.865 |
| Chromosome | Citrumelo | Citrandarin | ||
|---|---|---|---|---|
| PHR (rank) | PP (rank) | PHR (rank) | PP (rank) | |
| Chr1 | 0.865 ± 0.021 (de) | 0.683 ± 0.024 (d) | 0.817 ± 0.018 (cd) | 0.565 ± 0.048 (d) |
| Chr2 | 0.824 ± 0.016 (f) | 0.561 ± 0.007 (e) | 0.831 ± 0.016 (bcd) | 0.587 ± 0.042 (cd) |
| Chr3 | 0.842 ± 0.020 (ef) | 0.685 ± 0.039 (d) | 0.848 ± 0.020 (bc) | 0.633 ± 0.010 (c) |
| Chr4 | 0.902 ± 0.014 (bc) | 0.738 ± 0.029 (c) | 0.957 ± 0.023 (a) | 0.946 ± 0.002 (a) |
| Chr5 | 0.882 ± 0.015 (cd) | 0.722 ± 0.027 (c) | 0.842 ± 0.018 (bcd) | 0.447 ± 0.029 (e) |
| Chr6 | 0.942 ± 0.022 (a) | 0.942 ± 0.011 (a) | 0.812 ± 0.013 (d) | 0.488 ± 0.029 (e) |
| Chr7 | 0.821 ± 0.016 (f) | 0.558 ± 0.027 (e) | 0.855 ± 0.018 (b) | 0.631 ± 0.006 (c) |
| Chr8 | 0.926 ± 0.019 (ab) | 0.821 ± 0.011 (b) | 0.935 ± 0.010 (a) | 0.825 ± 0.023 (b) |
| Chr9 | 0.885 ± 0.013 (cd) | 0.678 ± 0.031 (d) | 0.810 ± 0.013 (d) | 0.451 ± 0.034 (e) |
| Whole genome | 0.874 ± 0.008 | 0.71 ± 0.039 | 0.854 ± 0.009 | 0.620 ± 0.055 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvez, L.; Dereeper, A.; Mournet, P.; Froelicher, Y.; Bruyère, S.; Morillon, R.; Ollitrault, P. Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding. Agronomy 2020, 10, 1961. https://doi.org/10.3390/agronomy10121961
Calvez L, Dereeper A, Mournet P, Froelicher Y, Bruyère S, Morillon R, Ollitrault P. Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding. Agronomy. 2020; 10(12):1961. https://doi.org/10.3390/agronomy10121961
Chicago/Turabian StyleCalvez, Leny, Alexis Dereeper, Pierre Mournet, Yann Froelicher, Saturnin Bruyère, Raphaël Morillon, and Patrick Ollitrault. 2020. "Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding" Agronomy 10, no. 12: 1961. https://doi.org/10.3390/agronomy10121961
APA StyleCalvez, L., Dereeper, A., Mournet, P., Froelicher, Y., Bruyère, S., Morillon, R., & Ollitrault, P. (2020). Intermediate Inheritance with Disomic Tendency in Tetraploid Intergeneric Citrus × Poncirus Hybrids Enhances the Efficiency of Citrus Rootstock Breeding. Agronomy, 10(12), 1961. https://doi.org/10.3390/agronomy10121961





