Abstract
Mal secco is a tracheomycotic disease caused by the fungus Plenodomus tracheiphilus (Petri) Gruyter, Aveskamp, and Verkley that has caused severe damage and loss of yield in the citrus industry in the Mediterranean area, for 100 years. While the disease can affect different cultivated citrus species, lemon (C. × limon var. limon (L.) Burm. f.) and citron are the most susceptible. The identification of resistant or field-tolerant clones and hybrids is a major goal for lemon growers and breeders. To identify sources of resistance or tolerance to the disease, we performed a phenotypic survey on a lemon and lemon-like open-field germplasm planted at CREA (Research Centre for Olive, Fruit and Citrus Crops), Italy, in an area with high pathogen pressure. Phenotyping was performed visually, four times, for three consecutive years, on a total of 50 accessions, with two or three replicate trees per accession. Moreover, molecular screening based on real-time PCR was performed, for two consecutive years, on twigs, young leaves, and mature leaves of all plants, to detect the pathogen in the absence of clear symptoms. The accessions were categorized into seven groups based on the presence of visual symptoms, real-time PCR pathogen detection, and canopy volume. The results revealed sources of tolerance in lemon and citron hybrids. The molecular screening identified P. tracheiphilus in all lemon clones, with mean Ct values ranging from 17 to 39. The screening also identified P. tracheiphilus in clones without clear symptoms, indicating their ability to tolerate the disease. Moreover, a strong negative correlation was found between the Ct values in twigs and symptom severity (r = −0.72). This indicates that the DNA from twigs is the most appropriate for use in performing reliable phenotyping of mal secco susceptibility in adult plants. An autotetraploid lemon (Doppio Lentini) seems to be immune to the disease, under natural pressure, since P. tracheiphilus was not detected by real-time PCR and visual screening. Overall, the data obtained are a valuable resource for identifying both the most tolerant lemon varieties suitable for areas with high pathogen pressure and the best breeding parents for the introgression of resistance genes into lemon genotypes.
1. Introduction
Mal secco is a vascular disease caused by the quarantine fungus Plenodomus tracheiphilus Petri Gruyter, Aveskamp, and Verkley [1]. It was previously classified as Phoma tracheiphila and is now included in the A2 list of quarantine pests of the European and Mediterranean Plant Protection Organization [2,3].
This fungus was discovered in 1894 on two Aegean Greek islands, Chios and Poros, and later it spread to other Mediterranean and Black Sea countries. Recently, it was also found in Spain, although it is not present in Portugal, Morocco, Malta, and Croatia (https://gd.eppo.int/taxon/DEUTTR/distribution). Lemon is one of the most susceptible sensitive species to this pathogen. The fungus penetrates through wounds [2,4,5,6] caused by heavy rains, hail, and wind; these atmospheric conditions favor the spread of the disease.
The symptoms usually begin with leaf vein chlorosis and leaf drop. Afterward, the pathogen reaches twigs and branches, and it is possible to observe red discolored strands in the xylem of stems. This is followed by the dieback of twigs and branches and the eventual death of the tree.
The disease severity shows a seasonal fluctuation and varies in different growing areas, depending on the climatic conditions. Ruggieri [7] reported that, in the years from 1918 to 1953, mal secco disease (MSD) destroyed no less than 12,000 ha of lemon groves in Sicily, Italy. According to Salerno and Cutuli [8], the mean yield of the production of lemon orchards in Sicily was approximately 20 tons/ha in the presence of MSD, whereas, in lemon orchards not affected by MSD, the yield could reach 60–80 tons/ha.
The pathogenicity of the different isolates collected in different Mediterranean countries was characterized in many studies [4,9,10,11,12,13,14,15,16,17,18,19], and efficient protocols were optimized to detect fungal infection in different plant tissues [16,20].
Chemical treatments in commercial orchards can only be used to prevent infections [21]. Therefore, the selection of field-tolerant lemon varieties is the most effective strategy to control the disease [2,22].
Lemons have a narrow genetic base, since most of them are bud sports of a single ancestor, which is a hybrid between sour orange and citron [23,24]. Such a genetic background exposes the species to the threat of the pathogen and hampers the identification of resistant varieties. Although most lemons are susceptible to the disease, some sources of tolerance were observed in field conditions, specifically in Monachello [25,26,27], Interdonato [25,26,27], Santa Teresa [28,29], Quattrocchi [30], Zagara Bianca, and Continella [31,32,33]. Unfortunately, none of them combine high fruit quality and productivity with tolerance to the disease.
Many citrus and citrus relatives were classified as susceptible or resistant to MSD [2], but the classification was based on a comparison among few citrus species and lemon varieties by visual screening or artificial inoculum [34,35,36,37,38,39]. Most of the bibliographic information is based on observations of single or few cultivars grown in the same field, while phenotypic studies comparing several accessions in the same field block are lacking.
Obtaining a lemon cultivar with good qualitative and pomological traits, as well as resistance to MSD, is a major challenge for the Mediterranean citrus industry [22]. The use of genetic transformation might be useful to improve the resistance to MSD or other diseases [40,41], but the use of genetically modified organisms (GMOs) raises public concerns regarding their safety. Consequently, traditional breeding approaches are, so far, the only means of releasing improved cultivars. To achieve this aim, identifying and characterizing sources of tolerance in lemon germplasm is needed to provide growers with improved varieties that could be grown under high pathogen pressure, reducing the yield losses caused by MSD and achieving acceptable productivity and fruit quality [5]. Moreover, identifying sources of resistance within the lemon-like germplasm and, more generally, in other citrus species, is essential for the introgression of resistance genes into lemon commercial cultivars as a part of a long-term strategy.
In this study, we analyzed the behavior of a germplasm field collection, which mostly comprises lemons, in response to P. tracheiphilus natural infections by visual observation of symptoms and detection by real-time PCR. The objectives of the present study were (i) the identification of sources of MSD tolerance or resistance in the lemon and lemon-like germplasm by comparison of several clones and hybrids grown in the same field block under the same high pathogen pressure; (ii) the successful application of a fast and reliable method to detect P. tracheiphilus in natural infection conditions; and (iii) the identification of sources of resistance in other citrus species that could be used to introgress resistance genes into lemon interspecific hybrids.
2. Materials and Methods
2.1. Plant Material and Phenotyping
Phenotyping started in 2018, at the CREA (Research Centre for Olive, Fruit and Citrus Crops) germplasm collection of Acireale (Catania, Italy; 37°37′23″ N, 15°09′50″ E). The original collection was planted in 2002. Plants were grafted onto the sour orange (C. × aurantium L. var. aurantium), in which lemon clones were replicated three times, and the other genotypes were replicated two times. The plants were grown with standard cultural practices, allowing comparative evaluation of the MSD symptoms under similar natural pathogen pressure.
The studied germplasms included 1 citron clone, 27 lemon clones, 15 lemon and citron hybrids (most of them of unknown parentage), and 7 varieties belonging to other citrus species. The list of analyzed accessions and their reported parentage is included in Table 1. Information regarding yield and fruit quality of 18 of the 27 lemon clones was previously reported by Di Vaio et al. [42].

Table 1.
List of accessions phenotyped for mal secco disease (MSD) susceptibility at the CREA experimental farm of Acireale, Italy. Botanical names refer to the latest proposal of taxonomical classification by Ollitrault et al. [44]. Asterisks in the description of the citrus species refer to the species parentage, as revealed by Curk et al. [23] (*) and Wu et al. [45] (**).
Phenotyping was carried out through a visual screening, in four different periods, for three consecutive years, when the symptoms were more pronounced. Field evaluation was always performed by the same personnel. The wood of desiccated or defoliated twigs was examined for pink salmon discoloration, which is typical of MSD infection (Figure 1A), by removing the bark.

Figure 1.
Citrus plants of the CREA germplasm showing different MSD symptoms: (A) infected shoot shows a yellow or pink-salmon to reddish discoloration of the wood; (B) a plant of Khasi papeda that shows no symptoms of MSD, scored with 0; (C) a plant of Mascali seedless lemon that shows few symptoms of MSD, scored as 1; (D) a plant of Zagara Bianca M79 lemon that shows medium symptoms of MSD, scored as 2; (E) a plant Femminello Dosaco M503 lemon that shows strong symptoms of MSD, scored as 3; (F) a plant of Akragas lemon that died of MSD, scored as 4.
Phenotyping also included measurement of the canopy volume of each tree, because pruning was routinely performed to remove infected branches since the establishment of the collection field, influencing the canopy development of the most susceptible trees. Canopy volume was measured at the end of the last vegetative flush each year and was approximated as one-half prolate spheroid with the following formula [43]:
where h is the tree height, and d is the tree diameter.
V = 4/6πh(d/2)2
For each survey, symptom severity was scored according to an empirical scale based on the following assigned values:
- 0 = no symptoms—the plant did not show any twigs or branches with symptoms (Figure 1B);
- 1 = few symptoms—fewer than 5 twigs had visible symptoms (Figure 1C);
- 2 = medium symptoms—more than 5 twigs had visible symptoms (Figure 1D);
- 3 = strong symptoms—all branches had visible symptoms (Figure 1E);
- 4 = dead plant (Figure 1F).
2.2. DNA Extraction
Samples of plant tissues for real-time PCR analysis were collected in the four cardinal directions, for each plant, in July 2018 and July 2019. For each tree, three types of samples were collected: one consisted of bulks of 10 young leaves (less than 6 months), one consisted of bulks of 10 mature leaves (6–12 months), and one consisted of 5 twigs, for a total of 9 samples per accession (3 biological replicates per tissue type). When only two plants per accession were present, the third biological replicate consisted of bulks of tissues from the two plants. For accessions with one or two replicates that were missing due to plant death, the samples were taken from the survivor plant to obtain nine samples from each accession. A total of 828 samples were collected from the 84 surviving plants. All samples were first surface-sterilized in a solution of 2.5% sodium hypochlorite and then washed twice with sterile distilled water. Ten grams of each sample was homogenized, using liquid nitrogen, and less than 0.1 g was collected for DNA extraction [18]. The P. tracheiphilus Pt10 strain (kindly provided by Professor Vittoria Catara, Di3A, University of Catania) was cultured for DNA isolation, as a reference for the real-time PCR experiments. The fungus was cultured for 10 days, at 21 °C ± 2 °C, in Petri dishes containing potato dextrose agar medium. One hundred micrograms of mycelium were harvested with a sterile loop from the surface of the colony, placed into an Eppendorf tube, frozen at −80 °C, and homogenized with a grinder (TissueLyser—Qiagen, Hilden, Germany). DNA extraction of both plant and fungal tissues was performed by the CTAB method, as described in Caruso et al. [59], with slight modifications. Briefly, tubes containing 0.1 g of powdered plant tissues were mixed with 400 µL of extraction buffer (2% CTAB, 20 mM EDTA, 1.44 mM NaCl, 100 mM Tris HCl, pH 8) and 0.1% β-mercaptoethanol. Samples were vortexed and incubated at 65 °C, for 60 min, agitating for the first 5 min. After adding 300 µL of chloroform-isoamyl alcohol (24:1), the vials were vortexed for 15 s and finally centrifuged at 20,800× g for 10 min. The supernatant was recovered, 500 µL of 100% ethanol was added and incubated at −20 °C, for at least 30 min, or at 4 °C, overnight, followed by centrifugation at 20,800 g for 10 min. The pellet was rinsed with 1000 µL of 70% ethanol, resuspended in 50 µL of sterile distilled water, and stored at 4 °C until analysis. The quality and concentration of the isolated DNA were measured by using a Nanodrop 2000 spectrophotometer (Thermo Scientific™, Waltham, MA, USA). The 260/280 and 260/230 ratios were approximately 1.80 and 2.20, respectively, and the concentrations ranged from 50 to 300 ng µL−1. All the samples were diluted at 10 ng µL−1.
2.3. Real-Time PCR Analysis
Real-time PCR amplifications were performed according to the protocol described by Licciardello et al. [16], using GR70 forward primer (5′-GATCCGTACGCCTTGGGGAC-3′), GL1 reverse primer (5′-AGAAGCGTTTGGAGGAGAGAATG-3′), and the probe PP1 (5′-FAM-CACGCAATCTTGGCGACTGTCGTT-TAMRA-3′). Each sample was amplified, using the following mix: 2X real-time PCR master mix (TaqMan™ Fast Advanced Master Mix Applied Biosystems™), 200 nΜ forward primer, 200 nM reverse primer, 100 nM fluorogenic probe, and 40 ng/µL genomic DNA. Negative controls, using water in place of DNA, were routinely included. Amplifications were carried out in an ABI 7500 Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific, Foster City, CA, USA), using the following program: 50 °C for 2 min, 95 °C for 30 s, followed by 40 cycles at 95 °C for 10 s and 62 °C for 30 s.
Calibration of the standard curve for fungal DNA quantification by real-time PCR was assessed by using P. tracheiphilus DNA (100 µg mL−1) extracted from the Pt10 strain and serially diluted in sterile distilled water, as described in Licciardello et al. [16].
2.4. Statistical Analysis
To perform the comparison of means, a value of 40 was assigned to all runs where P. tracheiphilus was non-detectable. The correlation between the five variables measured (severity symptom scores, Ct value from young leaves, Ct values from old leaves, Ct values from twigs, and canopy volume) was performed, using Spearman’s method, at the 95% confidence level. Statistical analysis and the analysis of correlation among the variables were performed, using R software, version 3.6.3 [60], using the packages “corrplot” [61] and “corrgram” [62].
3. Results and Discussion
3.1. Field Phenotyping
In the present study, we evaluated the disease responses of 50 accessions belonging to the Citrus and Fortunella genera to natural MSD infections. The main purpose of this work was the identification of sources of tolerance and resistance in lemons and in other citrus species and hybrids. Information is essential to planning lemon-breeding programs based on hybridization and somaclonal variation, to generate new varieties with improved tolerance to the disease. The field trial was conducted in an area where the environmental conditions are particularly favorable to the disease. The field trial originally planted in 2002 included 50 accessions replicated two or three times, for a total of 123 plants. The accessions initially included in the field are listed in Table 1. Some plants died from MSD within the first years after planting [46], and were replanted in 2009 (Table 2). At the end of the survey (May 2020), just 84 plants belonging to 46 accessions survived. Specifically, 79 plants were the original plants (16 years old), and five were the replanted replicates (nine years old). All the trees of the following selections died before the beginning of the survey: Siracusano 2Kr lemon, Adamo VCR lemon, Fino VCR lemon, and Diamante citron. Attempts to replant them, to re-constitute the original collection, were made, but plants died again due to MSD, confirming their high susceptibility. The rest of the missing plants were replicates of other lemon clonal selections or susceptible citrus accessions, as indicated in Table 2.

Table 2.
Results of MSD germplasm phenotyping based on real-time PCR, visual observation of symptoms, and canopy volumes. Values of the real-time PCR refer to the mean values of the three replicates sampled in 2018 and the three replicates of 2019 approximated to the nearest integer. Values of symptoms represent the average of four scores recorded between May 2018 and May 2020. The table shows the number of original plants, the number of replicates replanted in 2009, and the number of surviving plants at the end of the survey. The list also includes the accessions that died of MSD before the beginning of the survey.
Visual screening was performed four times, starting from May 2018, for three consecutive years, to check the behavior of each plant in response to 18 years of natural infection and to follow the possible progression of the disease during the three years of observations.
The scores used for the estimation of symptom severity ranged between 0 (absence of symptoms) and 4 (plant death). Score 4 was also assigned to the replicates that died of MSD before May 2018 or during the visual screening. The mean scores of symptom severity recorded in the three years are shown in Table 2. Several accessions showed no symptoms and had a score of 0 (Table 3). This group includes only a true lemon, Quattrocchi, a clone very similar to Monachello, already known for its high tolerance to the disease. Other accessions with citron ancestry, such as Palestinian sweet lime, Ponderosa lemon, and Incomparabile, showed no symptoms, indicating that it is theoretically possible to generate mal secco–resistant lemon-like phenotypes through hybridization. In this group, the only accession not included in the genus Citrus was Changshou kumquat, which differs from the most common kumquat (Fortunella margarita) in its rounded shape. Interestingly, sour orange also had a score of 0. This species is reported as susceptible [2,5] and is often used to evaluate the pathogenicity of the P. tracheiphilus strains at the seedling stage [18,63,64,65,66,67]; however, we observed no symptoms during the three years of evaluation.

Table 3.
List of disease-severity groups based on visual observations, real-time PCR results of twig samples and canopy volumes, and accessions assigned to each group.
The genotypes with very few symptoms that had a score less than 1 were Spatafora (C × limon), Cardinale (C. × lumia), Vozza Vozza (C. × lumia), and Fantastico bergamot (C. × limon var. bergamia ined.). These genotypes are all lemon or citron hybrids and show a high tolerance to the disease. The accessions India CRC 2476 rangpur lime (C. × limonia Osbeck var. limonia), India CRC 2322 lemon (C. × limonia Osbeck var. limonia), Volkamer lemon (C. × limonia var. volkameriana Pasquale), Corrugated red lime rangpur lime (C. × limonia Osbeck var. limonia), Femminello bergamot (C. × limon var. bergamia ined.), and Limetta romana sweet lime (C. × limon var. limetta ined.) showed a range of symptoms, with a score from 2 to 3. These genotypes are all citron hybrids and showed susceptibility to MSD. Some lemon clones revealed field tolerance, such as Continella M84, Segesta, Interdonato, Zagara Bianca M79, Lo Porto, and Mascali seedless, with scores ranging between 0.6 and 2. Other lemon clones with scores between 2 and 3 were Femminello S, Kamarina, Dosaco M503, Selinunte, Scandurra, Sfusato Amalfitano, Ovale di Sorrento, Pink Fleshed, CNR L58, and Akragas. Higher scores among the two lemon clones were assigned to Erice and Cerza, with 3.3 and 3.4, respectively. Finally, the highest score was assigned to three lemons, Siracusano 2Kr, Adamo VCR, and Fino VCR, and citron Diamante, with a 0% survival rate before the beginning of the survey, which can be considered the most susceptible to the disease.
Among the autotetraploid lemon clones (Doppio Lentini, Doppio, 46321, 46245, and 46515), we generally noticed a high tolerance or the absence of symptoms, with some differences. Specifically, no symptoms were found in Doppio Lentini, Doppio, and 46321 (probably Monachello 4×) during the three years of visual monitoring, while few infected branches were observed in 46245 and 46515. In addition to symptom observation, we measured the canopy volume of all surviving plants, not as an indication of plant vigor, but as an additional parameter to describe the sensitivity of each accession to MSD. The canopy volume can be drastically reduced by pathogen attack and by pruning infected branches. Indeed, pruning is one of the few effective measures to contain the spread of the disease in lemon orchards. In our survey, we analyzed many different citrus species, and a lower canopy volume of some accessions was due to the different growth habits and not necessarily to MSD infections (Table 2). Specifically, some citrus species, such as Chandler pink pummelo or Tachibana, showed a very low canopy volume in the absence of MSD symptoms, probably because they are poorly adapted to the growing environment. Consequently, we found no general correlation (r = 0.01) between canopy volume and symptoms when analyzing the whole dataset (Figure 2). However, a higher correlation between canopy volume and symptoms (r = −0.40) was found when the comparison was limited to the lemon clonal selections (Figure 3). The accessions that had lower canopy volumes, such as CNR L58 lemon, Erice lemon, and Cerza lemon, were generally the ones that underwent severe pruning due to the presence of more symptoms. All the most susceptible clones had canopy volumes below 10 m3, whereas many tolerant clones showed values ranging from 11.00 and 75.63 m3, with some exceptions, such as Doppio, 46321, and Lo Porto.
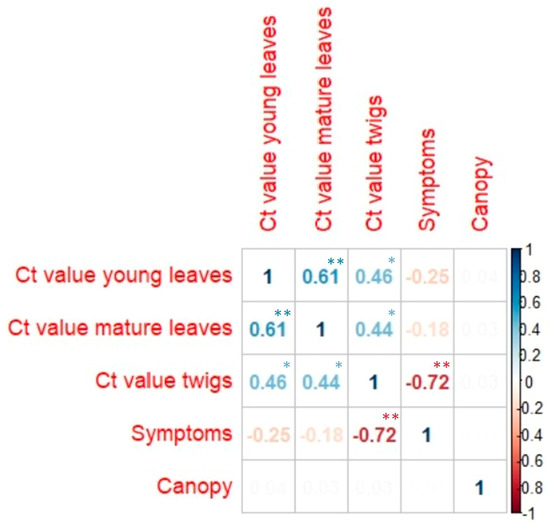
Figure 2.
Pairwise correlation matrix of five traits measured in the CREA germplasm. Blue numbers represent positive correlations, and red ones represent negative correlations. Faded numbers correspond to very low correlation values (<0.1). Correlation values were statistically significant with the following p-values: * <0.05 and ** <0.001. Values without asterisks were not statistically significant.
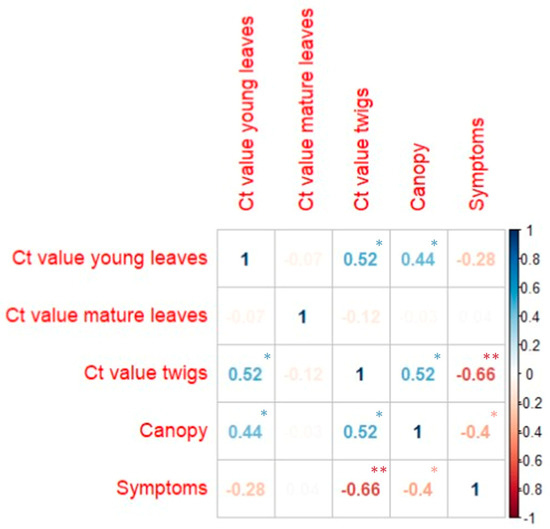
Figure 3.
Correlation index between the five variables of the lemon clones in the CREA germplasm. Blue numbers represent positive correlations, and red ones represent negative correlations. Faded numbers correspond to very low correlation values (<0.1). Correlation values were statistically significant with the following p-values: * <0.05 and ** <0.001. Values without asterisks were not statistically significant.
3.2. Real-Time PCR Detection of P. tracheiphilus
In addition to visual phenotyping, we performed molecular screening, to obtain a more exhaustive and reliable assessment of the P. tracheiphilus infection in all replicate trees of the germplasm collection, especially in the absence of clear symptoms. Sample collections for DNA isolation were conducted in the first week of July, because symptoms usually appear during spring and early summer [2]. Molecular detection was performed by using different types of tissues (twigs, young leaves, and mature leaves), amplifying a genomic region of the fungus by real-time PCR, as reported by Licciardello et al. [16]. In our experiment, Ct values ranged from 22 to 39 in young leaves, from 24 to 39 in mature leaves, and from 17 to 39 in DNA from twigs. Licciardello et al. [16] reported that the minimum amount of pathogen DNA that could be quantified accurately by using real-time PCR was 1 pg, corresponding to a Ct value of 37.93. Therefore, Ct values above 38 cannot reflect the occurrence of P. tracheiphilus infection (Table 2).
Leaf samples were included because the pathogen is able to penetrate through leaf wounds, so this survey could be potentially useful to identify early infections. However, the correlation between leaf Ct values and symptom scores was generally weak (Figure 2), and, in some cases, it was not useful to detect infections that were clearly visible in parts of the canopy, such as in the lemon clones CNR L58, Kamarina, Sfusato Amalfitano, Pink fleshed, and Cerza.
DNA samples from twigs were the most effective for P. tracheiphilus detection. The real-time PCR analysis of twig samples confirmed the presence of P. tracheiphilus in all genotypes where the symptoms were present. Furthermore, the molecular analysis detected P. tracheiphilus in xylem tissues of many accessions where no symptoms were present from any of the phenotyping data. This phenomenon occurred in Quattrocchi lemon and Chandler pink pummelo. These cases may include plants that were infected recently, so that symptoms were not yet visible or plants that showed some tolerance and that were able to block the movement of the pathogen and recover from the disease. In many replicates, the pathogen was detected only in the twigs and not in leaves, such as in Vozza Vozza, India CRC 2322, Corrugated red lime, Sour orange, ISA clementine, Tachibana, and Changshou Kumquat. Moreover, Ct values from twigs showed a high correlation (r = −0.72; p-values < 0.001) with the symptom scores (Figure 2). In particular, they ranged between 17 and 30 in the susceptible clones showing field symptoms, while they were higher (Ct value > 30) in tolerant clones and hybrids. Figure 4 shows the relationship between symptoms and Ct values in twigs and provides a view of the different degrees of susceptibility to MSD observed in the germplasm. The most susceptible accessions are in the lower right part of the plot, while the field tolerant or resistant accessions are grouped in the upper left part. Specifically, the right side of the plot includes all the lemon clones with the exceptions of Quattrocchi and Segesta, which are in the upper left side, grouped with some tetraploid lemons (Doppio Lentini, Doppio, and 46321), different citron and lemon hybrids (Vozza Vozza, Cardinale, Incomparabile, Spatafora, and Palestinian sweet lime), and other citrus species that are resistant to MSD (Khasi papeda, Doppio Sanguigno orange, Mangiagli lemon, Chandler pink pummelo, and ISA Clementine).
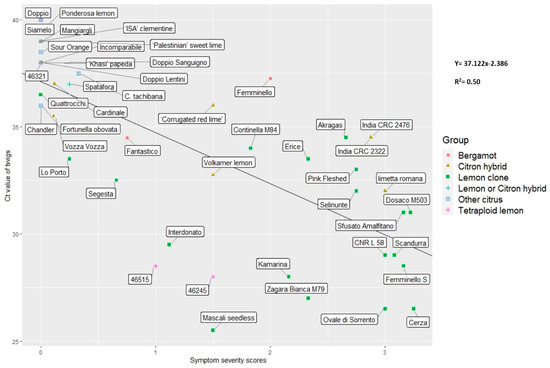
Figure 4.
Scatterplot with regression line showing the relationships between Ct value of twigs and symptoms of the germplasm collection of CREA considered in this study.
Significant correlations were also found analyzing the subset of the lemon clonal selections between Ct values of twigs and symptoms (r = −0.66; p-values < 0.001; Figure 3), and between Ct values of twigs and canopy volumes (r = 0.52; p-values < 0.05; Figure 3). A scatterplot revealing the relationship Ct values of twigs and canopies is shown in Figure 5.
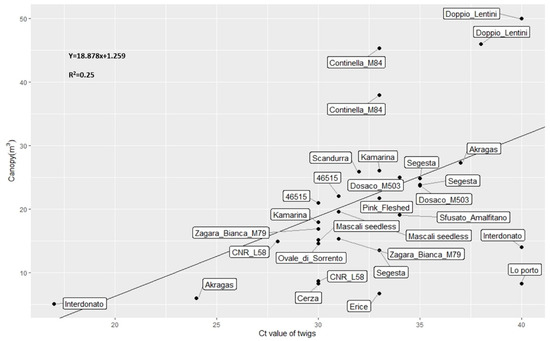
Figure 5.
Scatterplot with regression line showing the relationships between canopy of each surviving replicate of the lemon clonal selections and Ct values of twigs.
3.3. Assignment of the Accessions to Disease-Severity Groups
Based on the symptom-severity scores, canopy volumes, and real-time PCR results, we assigned the analyzed accessions to seven different disease-severity groups.
For determining disease groups, we considered the complete absence of the pathogen (immunity), the cases of very limited pathogen movement in the xylem with no visible symptoms (field resistance), the presence of very few symptoms with the ability of the plant to recover from infections (field tolerance), and successful colonization of the pathogen leading to clear disease symptom expression and, in some cases, to plant death (susceptibility). The groups and the list of accessions assigned to each group are listed in Table 3. Pictures of plants representative of each severity group are included in Supplementary Figures S1 to S6.
Group 1: In this group, we included the most susceptible accessions. Specifically, the lemon cultivars Siracusano 2Kr lemon, Adamo VCR lemon, Fino VCR lemon, and Diamante citron died before the beginning of the survey. Reforgiato Recupero and colleagues [46] reported that MSD was the cause of death of all replicates of the original field, and later attempts to replace the dead plants were not successful, since the new plants died again, due to MSD.
Group 2: Accessions in this category are susceptible to MSD and were also planted twice. Very severe symptoms were found on all plants, and some replicates died. This group includes the lemon clones Erice and Cerza.
Group 3: The accessions in this group showed a different range of symptoms, from medium to severe, and two of the three original replicates died of MSD. In some cases, a slight recovery of the plants during the three years of observation was recorded. This group includes Akragas lemon, Femminello S, Selinunte, Dosaco M503, Sfusato Amalfitano, Scandurra, Ovale di Sorrento, Mascali seedless, 46245, Pink Fleshed, Limetta romana India CRC 2476, India CRC 2322, and Femminello bergamot.
Group 4: This group includes accessions that can be considered tolerant to the disease, namely Continella M84 Lemon, Interdonato lemon, CNR L58 lemon, Zagara Bianca M79 lemon, Kamarina lemon, Lo Porto lemon, 46515 tetraploid lemon, and Corrugated red lime. The plants showed a different range of symptoms, from mild to severe, but real-time PCR showed medium levels of the pathogen, with Ct values between 30 and 31. Moreover, their canopy volume is generally higher than the accessions included in the previous groups, confirming their ability to tolerate the disease and guarantee canopy growth. This group also includes Volkamer lemon. This species was reported to exhibit a medium level of susceptibility by Russo [38], while other reports described it as highly susceptible [35,68]. We also observed different responses among the three replicates, with one healthy plant with very limited symptoms and the other two with severe dieback and reduced canopy volume.
Group 5: In this group, we can find just a lemon clone, Segesta. It showed high tolerance to the disease, and very few symptoms were detected during the visual screening. The plants had a high canopy volume, and real-time PCR confirmed the low level of infections in twigs, with a mean Ct value of 32. All replicates planted in 2002 are still alive.
Group 6: In this group, we were able to detect very few symptoms during the field phenotyping, but a low level of the fungus was detected periodically by real-time PCR (mean Ct values were between 35 and 37). Under field conditions, the pathogen could not be established in these hosts. The accessions are Chandler pink pummelo, Fantastico bergamot, Vozza Vozza, and Cardinale.
Group 7: This group includes all the accessions where the pathogen was detected in very low quantities, with a Ct value > 37, and the plants did not show any symptoms in the field. Therefore, these accessions showed resistance in the field conditions of natural pathogen pressure. The accessions in this group are Doppio Lentini tetraploid lemon, 46321 tetraploid lemon, Doppio tetraploid lemon, Palestinian sweet lime, sour orange, Khasi papeda, ISA Clementine, Doppio Sanguigno orange, Ponderosa lemon, Tachibana, Changshou kumquat, Quattrocchi lemon, Siamelo CRC 2586 tangelo, Spatafora, Incomparabile, and Mangiagli lemon. The possible resistance of these accessions needs confirmation on a larger number of replicates, since some of these genotypes, such as a mandarin hybrid or sweet orange, showed sporadically mild infections; however, the pathogen caused the typical symptoms of “mal nero”, a form of the disease where the fungus enters the plant through the roots [69,70,71]. Sour orange is reported to be very sensitive to the disease [2,5,30,34,35,72,73], but in our study, no symptoms or pathogens were detected by phenotyping or real-time PCR, respectively. This might be due to different degrees of susceptibility to different clonal selections. Some sour orange clones are reported as being resistant to MSD, as already confirmed by Reforgiato Recupero [74] and Nigro [75]. It is also well-known that plant age is a determinant of susceptibility, since adult plants are more tolerant than young seedlings [5].
4. Conclusions
This survey was useful to discriminate many citrus accessions belonging to true and derived species based on their field tolerance to MSD. Many accessions were found to be immune or resistant to the disease under natural pathogen pressure, but a broad degree of tolerance was also observed. Several degrees of field tolerance cannot be explained by a single gene involved in the resistance [38]. The presence of many genes involved in host–pathogen interaction was also supported by Reforgiato Recupero et al. [76].
We found that DNA isolation from twigs, coupled with real-time PCR detection, is a reliable method for field phenotyping. This method could be routinely used to validate phenotyping of mapping populations or germplasm collections, to better understand the genetic basis of MSD resistance.
A putative source of resistance was found in Doppio Lentini (autotetraploid lemon) and 46321 (probably a tetraploid Monachello), since no symptoms were found during the three years of visual monitoring, and no pathogen was detected by real-time PCR analysis. This resistance seems not to be exclusively related to tetraploidy [77], since other autotetraploids included in the phenotyping, namely 46515 and 46245, showed clear symptoms confirmed by real-time PCR detection.
This survey was also useful to identify sources of resistance among other citrus species that could be used to introgress resistance genes into the lemon genome. Therefore, based on the phenotyping results, two monoembryonic mal secco-resistant species, namely Khasi papeda and Clementine, were chosen as female parents and crossed with Femminello Siracusano 2Kr, a very susceptible lemon clone, to create two populations that might be helpful in the future for studying the segregation of MSD susceptibility and for identifying candidate genes and QTLs associated with the disease.
Supplementary Materials
The following materials are available online, at https://www.mdpi.com/2073-4395/10/11/1806/s1. Pictures of plants representative of each disease severity group (Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Figure S6).
Author Contributions
Investigation, formal analysis, and writing—original draft preparation, R.R.; conceptualization, funding acquisition, and writing—review and editing, M.C.; investigation, C.A.; writing—review and editing, A.R.L.P.; supervision and writing—review and editing, E.N.; conceptualization, methodology, supervision, funding acquisition, and writing—review and editing, S.D.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by FREECLIMB—Fruit Crops Resilience to Climate Change in the Mediterranean Basin, a project funded in framework of the Partnership for Research and Innovation in the Mediterranean Area (PRIMA) and INNO.VI.A—Innovative products and processes in the horticultural nursery and agri-food sectors- a project founded in Sicilian operative program—European Social Found 2014–2020.
Acknowledgments
We are grateful to Vittoria Catara, University of Catania, who provided P. tracheiphilus Pt10 strain, to Luciano Consagra for the help during the sampling, and to Benjamin Merritt for text editing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- De Gruyter, J.; Woudenberg, J.H.C.; Aveskamp, M.M.; Verkley, G.J.M.; Groenewald, J.Z.; Crous, P.W. Redisposition of phoma-like anamorphs in pleosporales. Stud. Mycol. 2013, 75, 1–36. [Google Scholar] [CrossRef]
- Nigro, F.; Ippolito, A.; Salerno, M.G. Mal secco disease of citrus: A journey through a century of research. J. Plant Pathol. 2011, 93, 523–560. [Google Scholar] [CrossRef]
- Pérez-Tornero, O.; Córdoba, F.; Moreno, M.; Yuste, L.; Porras, I. Classic methods and biotechnical tools in lemon breeding: Preliminary results. Acta Hortic. 2012, 259–263. [Google Scholar] [CrossRef]
- Butera, N.; Cupperi, L.; Zuckher, W.V.; Catara, A.; Grasso, S. Observation on the localization of Phoma tracheihila in canopy infections. In Integrated Pest Control in Citrus-Groves, Proceedings of the Experts’ Meeting, Acireale, Italy, 26–29 March 1985; AA Balkema: Rotterdam, The Netherlands, 1986. [Google Scholar]
- Migheli, Q.; Cacciola, S.; Balmas, V.; Pane, A.; Ezra, D.; Magnano di San Lio, G. Mal secco disease caused by Phoma tracheiphila: A potential threat to lemon industry worldwide. Plant Dis. 2009, 93, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Zucker, W.V.; Catara, A. Osservazioni al microscopio elettronico a scansione sulla penetrazione fogliare di Phoma tracheiphila. Inf. Fitopatol. 1985, 35, 33–35. [Google Scholar]
- Ruggieri, G. Periodicità nelle infezioni di “mal secco” e fondamentali orientamenti di lotta. G. Agric. 1953, 34, 1–8. [Google Scholar]
- Salerno, M.; Cutuli, G. The management of fungal and bacterial disease of citrus in Italy. In Proceedings of the International Society of Citriculture/International Citrus Congress, Tokyo, Japan, 9–12 November 1981; pp. 360–362. [Google Scholar]
- Salerno, M.; Perrotta, G. Ricerche sul “Mal secco” degli Agrumi (Deuterophoma tracheiphila Petri). Virulenza e caratteri colturali del fungo in Sicilia. Riv. Patol. Veg. 1966, 2, 303–312. [Google Scholar]
- Kroitor-Keren, T.; Liarzi, O.; Gat, T.; Skovorodnikova, J.; Belausov, E.; Ezra, D. Identification and characterization of Phoma tracheiphila mutants impaired in pathogenicity following Agrobacterium-mediated mutagenesis. Phytoparasitica 2013, 41, 491–502. [Google Scholar] [CrossRef]
- Ziadi, S.; Chebil, S.; Melki, I.; Ippolito, A.; Mliki, A. Virulence spectra and geographical distribution of Mal Secco disease of citrus caused by Phoma tracheiphila in the Mediterranean countries: Tunisia and Italy. Eur. J. Plant Pathol. 2014, 138, 123–131. [Google Scholar] [CrossRef]
- Graniti, A. Host-parasite relations in citrus diseases as exemplified by Phytophthora gummosis and Deuterophoma mal secco. In Proceedings of the 1st International Citrus Symposium, Riverside, CA, USA, 16–26 March 1968; Volume 3, pp. 1187–1200. [Google Scholar]
- Surico, G.; Jacobellis, N.S. Produzione di fitotossine di Phoma tracheiphila (Petri) Kanc. et Ghik. I. Influenza delle condizioni colturali e ricerca di idonei saggi biologici. Mediterr. Phytopathol. 1980, 19, 173–174. [Google Scholar]
- Cacciola, S.O.; Perrotta, G.; Graniti, A.; Magnano di San Lio, G. Esame preliminare di ceppi di Phoma tracheiphila (Petri) Kanc. et Ghick. mediante elettroforesi. In Atti del Convegno “Il Recente Contributo della Ricerca allo Sviluppo dell’Agrumicoltura Italiana”; Università di Sassari: Sassari, Italy, 1986; pp. 687–691. [Google Scholar]
- Balmas, V.; Migheli, Q.; Scherm, B.; Ghignone, S.; Salem, A.O.M.; Cacciola, S.O. Characterisation of Phoma tracheiphila by RAPD-PCR, microsatellite-primed PCR and ITS rDNA sequencing and development of specific primers for in planta PCR detection. Eur. J. Plant Pathol. 2005, 111, 235–247. [Google Scholar] [CrossRef]
- Licciardello, G.; Grasso, F.M.; Bella, P.; Cirvilleri, G.; Grimaldi, V.; Catara, V. Identification and Detection of Phoma tracheiphila, Causal Agent of Citrus Mal Secco Disease, by Real-Time Polymerase Chain Reaction. Plant Dis. 2006, 90, 1523–1530. [Google Scholar] [CrossRef]
- Ezra, D.; Kroitor, T.; Sadowsky, A. Molecular characterization of Phoma tracheiphila, causal agent of Mal secco disease of citrus, in Israel. Eur. J. Plant Pathol. 2007, 118, 183–191. [Google Scholar] [CrossRef]
- Kalai, L.; Mnari-Hattab, M.; Hajlaoui, M.R. Molecular diagnostics to assess the progression of Phoma tracheiphila in Citrus aurantium seedlings and analysis of genetic diversity of isolates recovered from different citrus species in Tunisia. J. Plant Pathol. 2010, 92, 629–636. [Google Scholar] [CrossRef]
- Kalai-Grami, L.; Bahri, A.; Rabeh Hajlaoui, M.; Boubaker, A. Comparison of Growth, Sporulation, Germination and Pathogenicity of Two Tunisian Isolates of Phoma tracheiphila, the Causal Agent of Mal Secco. Tunis. J. Plant Prot. 2012, 7, 1–17. [Google Scholar]
- Demontis, M.A.; Cacciola, S.O.; Orrù, M.; Balmas, V.; Chessa, V.; Maserti, B.E.; Mascia, L.; Raudino, F.; Magnano di San Lio, G.; Migheli, Q. Development of real-time PCR systems based on SYBR® Green I and TaqMan® technologies for specific quantitative detection of Phoma tracheiphila in infected Citrus. Eur. J. Plant Pathol. 2008, 120, 339–351. [Google Scholar] [CrossRef]
- Salerno, M.; Perrotta, G. Lo stato della lotta chimica contro il Mal secco degli Agrumi. Tec. Agric. Catania 1978, 30, 307–316. [Google Scholar]
- Khanchouch, K.; Pane, A.; Chriki, A.; Cacciola, S.O. Major and Emerging Fungal Diseases of Citrus in the Major and Emerging Fungal Diseases of Citrus in the Mediterranean Region. In Citrus Phatology; Gill, H., Garg, H., Eds.; IntechOpen: London, UK, 2017. [Google Scholar]
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef]
- Barry, G.H.; Caruso, M.; Gmitter, F.G., Jr. Commercial scion varieties. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing: Duxford, UK, 2020; pp. 83–104. [Google Scholar]
- Ruggieri, G. Sopra i presunti rapporti genetici col Limone e col Cedro di una particolare varietà di Limone assai resistente alla ‘Deuterophoma tracheiphila’ Petri. Boll. Staz. Pat. Veg. 1935, 15, 490–499. [Google Scholar]
- Ruggieri, G. Varietà di limoni resistenti al “mal secco”. G. Agric. Domen 1936, 20, 4. [Google Scholar]
- Ruggieri, G. Indagini sulla varieta di Limone ‘Monachello’. Boll. Staz. Pat. Veg. 1937, 17, 293–304. [Google Scholar]
- Ruggieri, G. Richerche sull’affinita d’innesto del limone” Monachello” con altri citrus. Boll. Staz. Pat. Veg. Roma 1937, 17, 79–86. [Google Scholar]
- Ruggieri, G. Mal secco degli agrumi e attuali mezzi di lotta. Riv. Agrumic. 1956, 1, 201–206. [Google Scholar]
- Ruggieri, G. I portinnesti degli agrumi in relazione alla resistenza alle malattie, all’adattamento, alle condizioni ambientali, all’affinità d’innesto ed alle reciproche influenze con le forme orticole. Nuovi Ann. Agric. 1940, 1, 27–28. [Google Scholar]
- Damigella, P.; Continella, G. Il Miglioramento Genetico Del Limone. Osservazioni Comparative su Alcune Selezioni Clonali. Available online: https://www.cabdirect.org/cabdirect/abstract/19720301743 (accessed on 15 November 2020).
- Damigella, P.; Continella, G. Il Miglioramento Genetico Del Limone. Osservazioni Comparative su Alcune Selezioni Clonali. Available online: https://agris.fao.org/agris-search/search.do?recordID=US201302255375 (accessed on 15 November 2020).
- Salerno, M.; Cutuli, G. Guida Illustrata di Patologia Degli Agrumi; Edagricole: Bologna, Italy, 1992. [Google Scholar]
- Ruggieri, G. Fattori che condizionano o contribuiscono allo sviluppo del “mal secco” degli agrumi e metodi di lotta contro il medesimo. Ann. Sper. Agrar. 1948, 2, 255–305. [Google Scholar]
- Catara, A.; Cutuli, G. Osservazioni sulla suscettibilità di alcune Rutacee alle infezioni epigee di Phoma tracheiphila. Ann. Ist. Sper. Agrumic. Acireale 1972, 5, 29–43. [Google Scholar]
- Crescimano, F.G.; Somma, V.; Calabrese, F. Preliminary research on resistance of rootstocks to mal secco. Proc. Int. Soc. Citric. 1973, 2, 119–120. [Google Scholar]
- Solel, Z.; Oren, Y. Outbreak of mal secco disease in Israel on normally tolerant citrus cultivars. Plant Dis. Rep. 1975, 59, 945–946. [Google Scholar]
- Russo, F. Il miglioramento genetico per la resistenza al “mal secco” del limone in Italia. Ann. Ist. Sper. Agrumic. Acireale 1977, 9, 231–243. [Google Scholar]
- Cutuli, G.; Laviola, C.; Perrotta, G.; Salerno, M.; Spina, P. Il Mal Secco Degli Agrumi. In Proceedings of the Seminario Internazionale di Studio Organizzato Nell’ambito del Programma di Ricerche Agrimed, Capo d’Orlando, Italy, 30 May–1 June 1984; p. 133. [Google Scholar]
- Gentile, A.; Deng, Z.; La Malfa, S.; Distefano, G.; Domina, F.; Vitale, A.; Polizzi, G.; Lorito, M.; Tribulato, E. Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 2007, 126, 146–151. [Google Scholar] [CrossRef]
- Muccilli, V.; Vitale, A.; Sheng, L.; Gentile, A.; Cardullo, N.; Tringali, C.; Oliveri, C.; La Rosa, R.; Di Guardo, M.; La Malfa, S.; et al. Substantial Equivalence of a Transgenic Lemon Fruit Showing Postharvest Fungal Pathogens Resistance. J. Agric. Food Chem. 2020, 68, 3806–3816. [Google Scholar] [CrossRef] [PubMed]
- Di Vaio, C.; Nocerino, S.; Graziani, G.; Gaspari, A.; Marallo, N.; Ritieni, A. Scelta varietale del limone in Campania: Produzioni, qualità e bio-attività. Frutticoltura 2010, 9, 58–63. [Google Scholar]
- Turrell, F.M. Tables of Surfaces and Volumes of Spheres and of Prolate and Oblate Spheroids, and Spheroidal Coefficients; University of California Press: Oakland, CA, USA, 1946. [Google Scholar]
- Ollitrault, P.; Curk, F.; Krueger, R. Citrus taxonomy. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing: Duxford, UK, 2020; pp. 57–81. [Google Scholar]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Reforgiato Recupero, G.; Russo, G.; Recupero, S.; Di Vaio, C. The culture of lemon in italy. In Proceedings of the VI Simposio Internacional Citrícola Colima, Colima, Mexico, 4–6 November 2010; pp. 207–216. [Google Scholar]
- Calabrese, F.; De Michele, A.; Barone, F.; Peri, G.; Somma, V. Cinque nuove cultivar di limone apirene e tolleranti il “mal secco”. Frutticoltura 2001, 63, 33–36. [Google Scholar]
- Abbata, L.; Mercati, F.; Fatta Del Bosco, S. An Overview on Citrus Mal Secco Disease: Approaches and Strategies to Select Tolerant Genotypes in C. limon. Crop Breed. Genet. Genom. 2019, 1–29. [Google Scholar] [CrossRef]
- University of California Riverside Citrus Variety Collection. Available online: https://citrusvariety.ucr.edu/citrus/alphabetical.html (accessed on 30 September 2020).
- Gentile, A.; Tribulato, E.; Deng, Z.N.; Vardi, A. In vitro selection of nucellar lemon callus and regeneration of plants tolerant to Phoma tracheiphila toxin. Adv. Hortic. Sci. 1992, 6, 151–154. [Google Scholar] [CrossRef]
- Starrantino, A.; Russo, F. Coltura ‘in vitro’ di nucelle isolate da frutticini di limone irraggiati con CO60. Tentativi per ottenere semenzali nucellari resistenti al ‘mal secco’. Ann. Sper. Agrar. 1977, 9–10, 209–211. [Google Scholar]
- Carrante, V.; Bottari, V. Miglioramento genetico del Limone e ricerca di varietà resistenti al “mal secco”. Ann. Sper. Agrar. 1952, 6, 323–346. [Google Scholar]
- Deng, Z.N.; Gentile, A.; Nicolosi, E.; Continella, G.; Tribulato, E. Caratterizzazione del germoplasma di agrumi: Dalle descrizioni degli antichi citrologi all’analisi del DNA. Italus Hortus 1995, 5–6, 68–79. [Google Scholar]
- Arlotta, C.; Ciacciulli, A.; Strano, M.C.; Cafaro, V.; Salonia, F.; Caruso, P.; Licciardello, C.; Russo, G.; Smith, M.W.; Cuenca, J.; et al. Disease Resistant Citrus Breeding Using Newly Developed High Resolution Melting and CAPS Protocols for Alternaria Brown Spot Marker Assisted Selection. Agronomy 2020, 10, 1368. [Google Scholar] [CrossRef]
- Singh, B. Establishment of First Gene Sanctuary in India for Citrus in Garo Hills; Concept Publishing Company: New Delhi, India, 1981; p. 182. [Google Scholar]
- Dutta, S. Origin and history of Citrus fruits of Assam. Indian J. Hort. 1958, 15, 146–153. [Google Scholar]
- Swingle, W.T. The botany of Citrus and its wild relatives in the orange subfamily. Citrus Ind. 1943, 1, 128–474. [Google Scholar]
- Caruso, M.; Ferlito, F.; Licciardello, C.; Allegra, M.; Strano, M.C.; Di Silvestro, S.; Russo, M.P.; Pietro Paolo, D.; Caruso, P.; Las Casas, G.; et al. Pomological diversity of the Italian blood orange germplasm. Sci. Hortic. 2016, 213, 331–339. [Google Scholar] [CrossRef]
- Caruso, M.; Distefano, G.; Pietro Paolo, D.; La Malfa, S.; Russo, G.; Gentile, A.; Reforgiato Recupero, G. High resolution melting analysis for early identification of citrus hybrids: A reliable tool to overcome the limitations of morphological markers and assist rootstock breeding. Sci. Hortic. 2014, 180, 199–206. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: https://www.R-project.org/ (accessed on 30 September 2020).
- Wei, T.; Simko, V. R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). Available online: https://github.com/taiyun/corrplot. (accessed on 30 September 2020).
- Wright, K. Corrgram: Plot a Correlogram. R Package Version 1.13. Available online: https://CRAN.R-project.org/package=corrgram (accessed on 30 September 2020).
- Magnano di San Lio, G.; Cacciola, S.O.; Pane, A.; Grasso, S. Relationship between xylem colonization and symptom expression in mal secco infected sour orange seedlings. Proc. Int. Soc. Citric. 1992, 2, 873–876. [Google Scholar]
- Traversa, E.; Ippolito, A.; De Cicco, V. Epidemiological investigation on Citrus mal secco (Phoma tracheiphila). Presence of the pathogen in the leaves of infected twigs. Phytopathol. Mediterr. 1992, 31, 103–106. [Google Scholar]
- Raimondo, F.; Raudino, F.; Cacciola, S.O.; Salleo, S.; Lo Gullo, M.A. Impairment of leaf hydraulics in young plants of Citrus aurantium (sour orange) infected by Phoma tracheiphila. Funct. Plant Biol. 2007, 34, 720–729. [Google Scholar] [CrossRef]
- Raimondo, F.; Nardini, A.; Salleo, S.; Cacciola, S.O.; Lo Gullo, M.A. A tracheomycosis as a tool for studying the impact of stem xylem dysfunction on leaf water status and gas exchange in Citrus aurantium L. Trees Struct. Funct. 2010, 24, 327–333. [Google Scholar] [CrossRef]
- Kalai-Grami, L.; Ben Slimane, I.; Mnari-Hattab, M.; Rezgui, S.; Aouani, M.A.; Hajlaoui, M.R.; Limam, F. Protective effect of Bacillus amyloliquefaciens against infections of Citrus aurantium seedlings by Phoma tracheiphila. World J. Microbiol. Biotechnol. 2014, 30, 529–538. [Google Scholar] [CrossRef]
- Salerno, M.; Catara, A.; Pacetto, M. Ricerche sul comportamento di Citrus volkameriana alle infezioni di Phoma e osservazioni sulla suscettibilità ad altre malattie. Tec. Agric. Catania 1967, 19, 228–237. [Google Scholar]
- Grasso, S. Infezioni di phoma su clementine riscontrate in Sicilia. Tec. Agric. Catania 1973, 25, 157–162. [Google Scholar]
- Hajlaoui, M.R.; Kalai, L.; Mnari-Hattab, M.; Guermech, A.; Ben Abdelaal, N. Occurrence of ‘mal nero’ disease on mandarin and orange trees in Tunisia. Plant Pathol. 2008, 57, 784. [Google Scholar] [CrossRef]
- Karapapa, V.; Doudoumis, V.; Tsiamis, G. First report of Phoma tracheiphila causing severe mal secco disease on a mandarin hybrid (cv. Ortanique) grafted onto Citrumelo rootstock in western Greece. New Dis. Rep. 2015, 31, 20. [Google Scholar] [CrossRef]
- Salerno, M. Ricerche sul “Mal secco” degli Agrumi (Deuterophoma tracheiphila Petri). I.- Influenza della temperatura sulla crescita del fungo e sulla produzione dei picnoconidi. Riv. Patol. Veg. 1964, 4, 289–299. [Google Scholar]
- Nigro, F.; Ippolito, A.; Lima, G.; Salerno, M. Field Trials on the Behaviour of Potential Lemon Rootstocks towards Mal Secco Disease. Basal and Root Infection ofLemon Grafted and Ungrafted Rootstocks. Proc. Int. Soc. Citric. 1996, 1, 440–444. [Google Scholar]
- Reforgiato Recupero, G. Osservazioni sulla suscettibilita al mal secco di semenzali di alcuni Citrus e generi affini. In Proceedings of the Atti del “II Seminario di Studio sul Miglioramento Genetico del Limone”, Giovinazzo, Italy, 5–6 April 1979; pp. 81–86. [Google Scholar]
- Nigro, F.; Ippolito, A.; Salerno, M.G. Searching for Citrus Rootstocks Resistant To Mal Secco Disease: A Review. Acta Hortic. 2015, 987–991. [Google Scholar] [CrossRef]
- Recupero, G.R.; Gentile, A.; Russo, M.P.; Domina, F. Genetic analysis of resistance to Phoma tracheiphila in three Citrus and Poncirus progenies. Plant Breed. 1997, 116, 198–200. [Google Scholar] [CrossRef]
- Grosser, J.W.; Barthe, G.A.; Castle, B.; Gmitter, F.G.; Lee, O. The development of improved tetraploid citrus rootstocks to facilitate advanced production systems and sustainable citriculture in Florida. Acta Hortic. 2015, 1065, 319–328. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).