Ethoxylate Polymer-Based 96-Well Screen for Protein Crystallization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Applied Agents for Crystallization
2.2. Applied Proteins
2.3. Crystallization Method and X-ray Diffraction Measurements
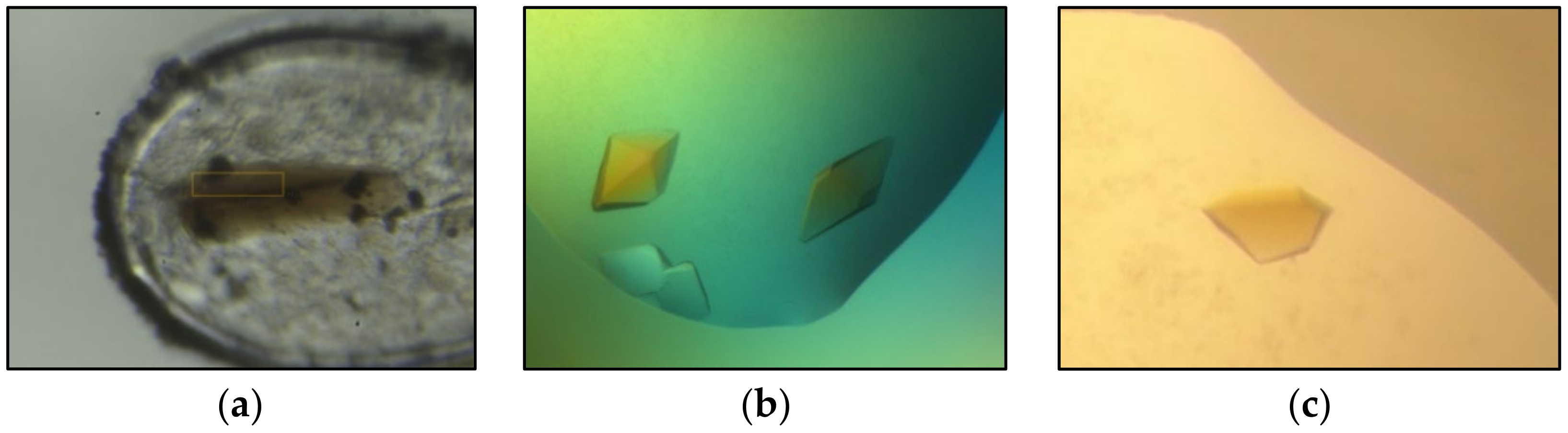
3. Results and Discussion
3.1. The Composition of the Crystallization Screen
3.2. Application to Proteins
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Gorrec, F. A beginner’s guide to macromolecular crystallization. Biochemist 2021, 43, 36–43. [Google Scholar] [CrossRef]
- Jancarik, J.; Kim, S.H. Sparse-Matrix Sampling—A Screening Method for Crystallization of Proteins. J. Appl. Crystallogr. 1991, 24, 409–411. [Google Scholar] [CrossRef]
- Gorrec, F. The MORPHEUS protein crystallization screen. J. Appl. Crystallogr. 2009, 42, 1035–1042. [Google Scholar] [CrossRef] [PubMed]
- Newman, J.; Egan, D.; Walter, T.S.; Meged, R.; Berry, I.; Ben Jelloul, M.; Sussman, J.L.; Stuart, D.I.; Perrakis, A. Towards rationalization of crystallization screening for small- to medium-sized academic laboratories: The PACT/JCSG+ strategy. Acta Crystallogr. D Biol. Crystallogr. 2005, 61, 1426–1431. [Google Scholar] [CrossRef]
- Grimm, C.; Chari, A.; Reuter, K.; Fischer, U. A crystallization screen based on alternative polymeric precipitants. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 685–697. [Google Scholar] [CrossRef] [PubMed]
- Gorrec, F. The MORPHEUS II protein crystallization screen. Acta Crystallogr. F Struct. Biol. Commun. 2015, 71, 831–837. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A., Jr. Crystallization of proteins from polyethylene glycol. J. Biol. Chem. 1976, 251, 6300–6303. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Cudney, B.; McPherson, A. Polymeric precipitants for the crystallization of macromolecules. Biochem. Biophys. Res. Commun. 1995, 207, 819–828. [Google Scholar] [CrossRef] [PubMed]
- Skalova, T.; Duskova, J.; Hasek, J.; Kolenko, P.; Stepankova, A.; Dohnalek, J. Alternative polymer precipitants for protein crystallization. J. Appl. Crystallogr. 2010, 43, 737–742. [Google Scholar] [CrossRef]
- Gulick, A.M.; Horswill, A.R.; Thoden, J.B.; Escalante-Semerena, J.C.; Rayment, I. Pentaerythritol propoxylate: A new crystallization agent and cryoprotectant induces crystal growth of 2-methylcitrate dehydratase. Acta Crystallogr. D Biol. Crystallogr. 2002, 58, 306–309. [Google Scholar] [CrossRef] [PubMed]
- McPherson, A.; Cudney, B. Searching for silver bullets: An alternative strategy for crystallizing macromolecules. J. Struct. Biol. 2006, 156, 387–406. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, J.K.; Bhat, R. Why Is Trehalose an Exceptional Protein Stabilizer?: An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J. Biol. Chem. 2003, 278, 26458–26465. [Google Scholar] [CrossRef]
- Sauter, C.; Ng, J.D.; Lorber, B.; Keith, G.; Brion, P.; Hosseini, M.W.; Lehn, J.M.; Giege, R. Additives for the crystallization of proteins and nucleic acids. J. Cryst. Growth 1999, 196, 365–376. [Google Scholar] [CrossRef]
- Willistein, M.; Haas, J.; Fuchs, J.; Estelmann, S.; Ferlaino, S.; Müller, M.; Ludeke, S.; Boll, M. Enantioselective Enzymatic Naphthoyl Ring Reduction. Chem.-Eur. J. 2018, 24, 12505–12508. [Google Scholar] [CrossRef]
- Schneider, C.; Konjik, V.; Kissling, L.; Mack, M. The novel phosphatase RosC catalyzes the last unknown step of roseoflavin biosynthesis in Streptomyces davaonensis. Mol. Microbiol. 2020, 114, 609–625. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, C.; Goerke, M.; Bezold, D.; Jessen, H.; Boll, M. A fully reversible 25-hydroxy steroid kinase involved in oxygen-independent cholesterol side-chain oxidation. J. Biol. Chem. 2021, 297, 101105. [Google Scholar] [CrossRef] [PubMed]
- Kung, J.W.; Meier, A.K.; Willistein, M.; Weidenweber, S.; Demmer, U.; Ermler, U.; Boll, M. Structural Basis of Cyclic 1,3-Diene Forming Acyl-Coenzyme A Dehydrogenases. Chembiochem 2021, 22, 3173–3177. [Google Scholar] [CrossRef]
- Tiedt, O.; Fuchs, J.; Eisenreich, W.; Boll, M. A catalytically versatile benzoyl-CoA reductase, key enzyme in the degradation of methyl- and halobenzoates in denitrifying bacteria. J. Biol. Chem. 2018, 293, 10264–10274. [Google Scholar] [CrossRef]
- Mergelsberg, M.; Willistein, M.; Meyer, H.; Stark, H.J.; Bechtel, D.F.; Pierik, A.J.; Boll, M. Phthaloyl-coenzyme A decarboxylase from Thauera chlorobenzoica: The prenylated flavin-, K+- and Fe2+-dependent key enzyme of anaerobic phthalate degradation. Environ. Microbiol. 2017, 19, 3734–3744. [Google Scholar] [CrossRef] [PubMed]
- Jeoung, J.H.; Nicklisch, S.; Dobbek, H. Structural basis for coupled ATP-driven electron transfer in the double-cubane cluster protein. Proc. Natl. Acad. Sci. USA 2022, 119, e2203576119. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, O.N.; Wagner, T. Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum. BBA-Bioenerg. 2021, 1862, 148330. [Google Scholar] [CrossRef] [PubMed]
- Lemaire, O.N.; Müller, M.C.; Kahnt, J.; Wagner, T. Structural Rearrangements of a Dodecameric Ketol-Acid Reductoisomerase Isolated from a Marine Thermophilic Methanogen. Biomolecules 2021, 11, 1679. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Qiu, T.L.; Yin, X.B.; Wu, X.L.; Hu, G.Q.; Deng, Y.; Zhang, H. Methermicoccus shengliensis gen. nov., sp nov., a thermophilic, methylotrophic methanogen isolated from oil-production water, and proposal of Methermicoccaceae fam. nov. Int. J. Syst. Evol. Micr. 2007, 57, 2964–2969. [Google Scholar] [CrossRef] [PubMed]
- Izard, T.; Sygusch, J. Induced fit movements and metal cofactor selectivity of class II aldolases—Structure of thermus aquaticus fructose-1,6-bisphosphate aldolase. J. Biol. Chem. 2004, 279, 11825–11833. [Google Scholar] [CrossRef]
- Grabarse, W.; Vaupel, M.; Vorholt, J.A.; Shima, S.; Thauer, R.K.; Wittershagen, A.; Bourenkov, G.; Bartunik, H.D.; Ermler, U. The crystal structure of methenyltetrahydromethanopterin cyclohydrolase from the hyperthermophilic archaeon Methanopyrus kandleri. Structure 1999, 7, 1257–1268. [Google Scholar] [CrossRef] [PubMed]
- Song, R.Q.; Colfen, H. Additive controlled crystallization. Crystengcomm 2011, 13, 1249–1276. [Google Scholar] [CrossRef]
- McPherson, A.; Nguyen, C.; Cudney, R.; Larson, S.B. The Role of Small Molecule Additives and Chemical Modification in Protein Crystallization. Cryst. Growth Des. 2011, 11, 1469–1474. [Google Scholar] [CrossRef]
- Trakhanov, S.; Quiocho, F.A. Influence of Divalent-Cations in Protein Crystallization. Protein Sci. 1995, 4, 1914–1919. [Google Scholar] [CrossRef]
- Sousa, R. Use of Glycerol, Polyols and Other Protein-Structure Stabilizing Agents in Protein Crystallization. Acta Crystallogr. D 1995, 51, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Vagenende, V.; Yap, M.G.; Trout, B.L. Mechanisms of protein stabilization and prevention of protein aggregation by glycerol. Biochemistry 2009, 48, 11084–11096. [Google Scholar] [CrossRef]
- Vera, L.; Czarny, B.; Georgiadis, D.; Dive, V.; Stura, E.A. Practical Use of Glycerol in Protein Crystallization. Cryst. Growth Des. 2011, 11, 2755–2762. [Google Scholar] [CrossRef]
- Singh, S.; Singh, J. Effects of polyols on the conformational stability and biological activity of a model protein lysozyme. AAPS PharmSciTech 2003, 4, E42. [Google Scholar] [CrossRef] [PubMed]
- Douzou, P. Aqueous-organic solutions of enzymes at sub-zero temperatures. Biochimie 1971, 53, 1135–1145. [Google Scholar] [CrossRef]
- Sankha, B. Cryoprotectants and Their Usage in Cryopreservation Process. In Cryopreservation Biotechnology in Biomedical and Biological Sciences; Yusuf, B., Ed.; IntechOpen: Rijeka, Croatia, 2018; Chapter 2; pp. 7–19. [Google Scholar] [CrossRef]
- Golovanov, A.P.; Hautbergue, G.M.; Wilson, S.A.; Lian, L.Y. A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 2004, 126, 8933–8939. [Google Scholar] [CrossRef] [PubMed]
- Shaikh, A.R.; Shah, D. Arginine-Amino Acid Interactions and Implications to Protein Solubility and Aggregation. J. Eng. Res. TJER 2015, 12, 1–14. [Google Scholar] [CrossRef]
- Arakawa, T.; Ejima, D.; Tsumoto, K.; Obeyama, N.; Tanaka, Y.; Kita, Y.; Timasheff, S.N. Suppression of protein interactions by arginine: A proposed mechanism of the arginine effects. Biophys. Chem. 2007, 127, 1–8. [Google Scholar] [CrossRef]
- Yancey, P.H. Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J. Exp. Biol. 2005, 208, 2819–2830. [Google Scholar] [CrossRef]
- Yancey, P.H.; Gerringer, M.E.; Drazen, J.C.; Rowden, A.A.; Jamieson, A. Marine fish may be biochemically constrained from inhabiting the deepest ocean depths. Proc. Natl. Acad. Sci. USA 2014, 111, 4461–4465. [Google Scholar] [CrossRef] [PubMed]
- Russo, A.T.; Rösgen, J.; Bolen, D.W. Osmolyte Effects on Kinetics of FKBP12 C22A Folding Coupled with Prolyl Isomerization. J. Mol. Biol. 2003, 330, 851–866. [Google Scholar] [CrossRef] [PubMed]
- Galinski, E.A.; Pfeiffer, H.P.; Trüper, H.G. 1,4,5,6-Tetrahydro-2-methyl-4-pyrimidinecarboxylic acid. A novel cyclic amino acid from halophilic phototrophic bacteria of the genus Ectothiorhodospira. Eur. J. Biochem. 1985, 149, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Lippert, K.; Galinski, E.A. Enzyme stabilization be ectoine-type compatible solutes: Protection against heating, freezing and drying. Appl. Microbiol. Biotechnol. 1992, 37, 61–65. [Google Scholar] [CrossRef]
- Salmannejad, F.; Nafissi-Varcheh, N. Ectoine and hydroxyectoine inhibit thermal-induced aggregation and increase thermostability of recombinant human interferon Alfa2b. Eur. J. Pharm. Sci. 2017, 97, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Harjes, S.; Scheidig, A.; Bayer, P. Expression, purification and crystallization of human 3′-phosphoadenosine-5′-phosphosulfate synthetase 1. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 350–352. [Google Scholar] [CrossRef]
- Expert-Bezancon, N.; Rabilloud, T.; Vuillard, L.; Goldberg, M.E. Physical-chemical features of non-detergent sulfobetaines active as protein-folding helpers. Biophys. Chem. 2003, 100, 469–479. [Google Scholar] [CrossRef]
- Vuillard, L.; Baalbaki, B.; Lehmann, M.; Norager, S.; Legrand, P.; Roth, M. Protein crystallography with non-detergent sulfobetaines. J. Cryst. Growth 1996, 168, 150–154. [Google Scholar] [CrossRef]

| No. | Precipitant * | Buffer | Additives | No. | Precipitant * | Buffer | Additives |
|---|---|---|---|---|---|---|---|
| 1 | 15 % TMPE 1014 | 0.1 M MES pH 6.5 | − | 49 | 15 % GE 1000 | 0.1 M Mes pH 6.5 | − |
| 2 | 25 % TMPE 1014 | 0.1 M MES pH 6.5 | − | 50 | 25 % GE 1000 | 0.1 M Mes pH 6.5 | − |
| 3 | 35 % TMPE 1014 | 0.1 M MES pH 6.5 | − | 51 | 35 % GE 1000 | 0.1 M Mes pH 6.5 | − |
| 4 | 45 % TMPE 1014 | 0.1 M MES pH 6.5 | − | 52 | 45 % GE 1000 | 0.1 M Mes pH 6.5 | − |
| 5 | 15 % TMPE 1014 | 0.1 M HEPES pH 7.5 | − | 53 | 15 % GE 1000 | 0.1 M Hepes pH 7.5 | − |
| 6 | 25 % TMPE 1014 | 0.1 M HEPES pH 7.5 | − | 54 | 25 % GE 1000 | 0.1 M HEPES pH 7.5 | − |
| 7 | 35 % TMPE 1014 | 0.1 M HEPES pH 7.5 | − | 55 | 35 % GE 1000 | 0.1 M HEPES pH 7.5 | − |
| 8 | 45 % TMPE 1014 | 0.1 M HEPES pH 7.5 | − | 56 | 45 % GE 1000 | 0.1 M HEPES pH 7.5 | − |
| 9 | 15 % TMPE 1014 | 0.1 M TRIS pH 8.5 | − | 57 | 15 % GE 1000 | 0.1 M HEPES pH 8.5 | − |
| 10 | 25 % TMPE 1014 | 0.1 M TRIS pH 8.5 | − | 58 | 25 % GE 1000 | 0.1 M TRIS pH 8.5 | − |
| 11 | 35 % TMPE 1014 | 0.1 M TRIS pH 8.5 | − | 59 | 35 % GE 1000 | 0.1 M TRIS pH 8.5 | − |
| 12 | 45 % TMPE 1014 | 0.1 M TRIS pH 8.5 | − | 60 | 45 % GE 1000 | 0.1 M TRIS pH 8.5 | − |
| 13 | 15 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.05 M MgCl2 | 61 | 15 % GE 1000 | 0.1 M MES pH 6.5 | 0.05 M MgCl2 |
| 14 | 25 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.1 M CaCl2 | 62 | 25 % GE 1000 | 0.1 M MES pH 6.5 | 0.1 M CaCl2 |
| 15 | 35 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.2 M (NH4)2SO4 | 63 | 35 % GE 1000 | 0.1 M MES pH 6.5 | 0.2 M (NH4)2SO4 |
| 16 | 45 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.4 M KCl | 64 | 45 % GE 1000 | 0.1 M MES pH 6.5 | 0.4 M KCl |
| 17 | 15 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2 | 65 | 15 % GE 1000 | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2 |
| 18 | 25 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 | 66 | 25 % GE 1000 | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 19 | 35 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.2 M (NH4)2SO4 | 67 | 35 % GE 1000 | 0.1 M HEPES pH 7.5 | 0.2 M (NH4)2SO4 |
| 20 | 45 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.4 M KCl | 68 | 45 % GE 1000 | 0.1 M HEPES pH 7.5 | 0.4 M KCl |
| 21 | 15 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.05 M MgCl2 | 69 | 15 % GE 1000 | 0.1 M TRIS pH 8.5 | 0.05 M MgCl2 |
| 22 | 25 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.1 M CaCl2 | 70 | 25 % GE 1000 | 0.1 M TRIS pH 8.5 | 0.1 M CaCl2 |
| 23 | 35 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.2 M (NH4)2SO4 | 71 | 35 % GE 1000 | 0.1 M TRIS pH 8.5 | 0.2 M (NH4)2SO4 |
| 24 | 45 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.4 M KCl | 72 | 45 % GE 1000 | 0.1 M TRIS pH 8.5 | 0.4 M KCl |
| 25 | 15 % TMPE 1014 | 0.1 M MES pH 6.5 | 10 % (v/v) Propane-1,2-diol | 73 | 15 % GEPT 2600 | 0.1 M MES pH 6.5 | − |
| 26 | 25 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.2 M KSCN | 74 | 25 % GEPT 2600 | 0.1 M MES pH 6.5 | − |
| 27 | 35 % TMPE 1014 | 0.1 M MES pH 6.5 | 5 % Glycerol | 75 | 35 % GEPT 2600 | 0.1 M MES pH 6.5 | − |
| 28 | 45 % TMPE 1014 | 0.1 M MES pH 6.5 | 50 mM L-Arg, 50 mM L-Glu | 76 | 45 % GEPT 2600 | 0.1 M MES pH 6.5 | − |
| 29 | 15 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 10 % (v/v) Propane-1,2-diol | 77 | 15 % GEPT 2600 | 0.1 M HEPES pH 7.5 | − |
| 30 | 25 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.2 M KSCN | 78 | 25 % GEPT 2600 | 0.1 M HEPES pH 7.5 | − |
| 31 | 35 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 5 % Glycerol | 79 | 35 % GEPT 2600 | 0.1 M HEPES pH 7.5 | − |
| 32 | 45 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 50 mM L-Arg, 50 mM L-Glu | 80 | 45 % GEPT 2600 | 0.1 M HEPES pH 7.5 | − |
| 33 | 15 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 10 % (v/v) Propane-1,2-diol | 81 | 15 % GEPT 2600 | 0.1 M TRIS pH 8.5 | − |
| 34 | 25 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.2 M KSCN | 82 | 25 % GEPT 2600 | 0.1 M TRIS pH 8.5 | − |
| 35 | 35 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 5 % Glycerol | 83 | 35 % GEPT 2600 | 0.1 M TRIS pH 8.5 | − |
| 36 | 45 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 50 mM L-Arg, 50 mM L-Glu | 84 | 45 % GEPT 2600 | 0.1 M TRIS pH 8.5 | − |
| 37 | 15 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.2 M TMAO | 85 | 15 % GEPT 2600 | 0.1 M MES pH 6.5 | 0.05 M MgCl2 |
| 38 | 25 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.4 M D-(+)-Trehalose | 86 | 25 % GEPT 2600 | 0.1 M MES pH 6.5 | 0.1 M CaCl2 |
| 39 | 35 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.25 M Ectoine | 87 | 35 % GEPT 2600 | 0.1 M MES pH 6.5 | 0.2 M (NH4)2SO4 |
| 40 | 45 % TMPE 1014 | 0.1 M MES pH 6.5 | 0.15 M NDSB-256 | 88 | 45 % GEPT 2600 | 0.1 M MES pH 6.5 | 0.4 M KCl |
| 41 | 15 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.2 M TMAO | 89 | 15 % GEPT 2600 | 0.1 M HEPES pH 7.5 | 0.05 M MgCl2 |
| 42 | 25 % TMPE 1014 | 0.1 M HEEPS pH 7.5 | 0.4 M D-(+)-Trehalose | 90 | 25 % GEPT 2600 | 0.1 M HEPES pH 7.5 | 0.1 M CaCl2 |
| 43 | 35 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.25 M Ectoine | 91 | 35 % GEPT 2600 | 0.1 M HEPES pH 7.5 | 0.2 M (NH4)2SO4 |
| 44 | 45 % TMPE 1014 | 0.1 M HEPES pH 7.5 | 0.15 M NDSB-256 | 92 | 45 % GEPT 2600 | 0.1 M HEPES pH 7.5 | 0.4 M KCl |
| 45 | 15 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.2 M TMAO | 93 | 15 % GEPT 2600 | 0.1 M TRIS pH 8.5 | 0.05 M MgCl2 |
| 46 | 25 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.4 M D-(+)-Trehalose | 94 | 25 % GEPT 2600 | 0.1 M TRIS pH 8.5 | 0.1 M CaCl2 |
| 47 | 35 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.25 M Ectoine | 95 | 35 % GEPT 2600 | 0.1 M TRIS pH 8.5 | 0.2 M (NH4)2SO4 |
| 48 | 45 % TMPE 1014 | 0.1 M TRIS pH 8.5 | 0.15 M NDSB-256 | 96 | 45 % GEPT 2600 | 0.1 M TRIS pH 8.5 | 0.4 M KCl |
| Protein | BCR-QONP | Ch1-2en-DH | CML | CODH/ACS | DCCP | MsKARI | MsFruA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Added ligands | 5 mM Benzoyl-CoA 5 mM ADP 5 mM MgCl2 | 0.1 mM FAD | 5 mM AMPPNP | – | – | – | – | |||||||
| Most successful ethoxylate precipitant | GE 1000 | GE 1000 | GEPT 2600 | TMPE 1014 | TMPE 1014 | |||||||||
| Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | |
| Ethoxylate based screen | • | xx/2.5 Å (GE 1000) | • | • | • | xx/2.9 Å (GE 1000) | • | xxx/1.6 Å (GEPT 2600) | • | xx | • | xxx/2.0Å (TMPE 1014) | ||
| JCSG++ | • | x | • | xx/2.9 Å (NH4)2SO4 | ||||||||||
| JCSG Core Suite I | • | x/2.1 Å (PEG 3350) | ||||||||||||
| JBS Classic 1-4 | • | x | • | x | • | x/3.0 Å (PEG 4000) | ||||||||
| JBS Classic 5-8 | • | – | • | xx | • | x | ||||||||
| Midas | • | x | ||||||||||||
| Morpheus I | • | x | • | – | • | – | ||||||||
| Morpheus II | • | x | • | x | • | – | ||||||||
| PACT++ | • | – | • | x | • | – | • | xxx | • | xxx | ||||
| JBS Pentaerythritol | • | x | • | x | • | xx | • | xx | ||||||
| ProPlex | • | – | • | x | ||||||||||
| SaltRx | • | – | ||||||||||||
| SG1 | • | xxx | • | xxx/1.9 Å (PEG 3350) | ||||||||||
| Wizard Classic 1 | • | – | • | x | ||||||||||
| Wizard Classic 2 | • | x | • | x | ||||||||||
| Wizard Classic 3 | • | x | • | xx | ||||||||||
| Wizard Classic 4 | • | x | • | xx | ||||||||||
| Protein | MsMch | MsMer | PCD | RosC-E107Q | 25-PSL | S25-PT | ||||||||
| Added ligands | – | – | 5 mM CN-benzoyl-CoA | 5 mM AFP | – | 5 mM ADP | ||||||||
| Most successful ethoxylate precipitant | TMPE 1014 | GEPT 2600 | GE 1000 | GE 1000 | TMPE 1014 | TMPE 1014 | ||||||||
| Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | Screens used | Hits/best resolution/ precipitant | |||
| Ethoxylate based screen | • | xx/2.7 Å (TMPE 1014) | • | xxx | • | x | • | xx/3.5 Å (GE 1000) | • | x | • | xx/2.1 Å (TMPE 1014) | ||
| JCSG++ | • | xx/2.2 Å (PEG 3350) | ||||||||||||
| JCSG Core Suite I | ||||||||||||||
| JBS Classic 1-4 | • | x | • | x | • | x | • | x | ||||||
| JBS Classic 5-8 | • | x | • | x/3.0 Å (PEG 8000) | • | – | ||||||||
| Midas | ||||||||||||||
| Morpheus I | • | xx/2.0 Å (PEG 8000) | • | xx | ||||||||||
| Morpheus II | • | xx | • | x | ||||||||||
| PACT++ | • | x | • | x/2.9 Å (PEG 3350) | • | xxx | • | – | ||||||
| JBS Pentaerythritol | • | x | • | x | • | x | • | – | ||||||
| ProPlex | ||||||||||||||
| SaltRx | • | x | • | – | ||||||||||
| SG1 | • | xx/2.0 Å (PEG 3350) | • | xx | ||||||||||
| Wizard Classic 1 | ||||||||||||||
| Wizard Classic 2 | ||||||||||||||
| Wizard Classic 3 | ||||||||||||||
| Wizard Classic 4 | ||||||||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demmer, U.; Lemaire, O.N.; Belhamri, M.; Ermler, U. Ethoxylate Polymer-Based 96-Well Screen for Protein Crystallization. Crystals 2023, 13, 1519. https://doi.org/10.3390/cryst13101519
Demmer U, Lemaire ON, Belhamri M, Ermler U. Ethoxylate Polymer-Based 96-Well Screen for Protein Crystallization. Crystals. 2023; 13(10):1519. https://doi.org/10.3390/cryst13101519
Chicago/Turabian StyleDemmer, Ulrike, Olivier N. Lemaire, Mélissa Belhamri, and Ulrich Ermler. 2023. "Ethoxylate Polymer-Based 96-Well Screen for Protein Crystallization" Crystals 13, no. 10: 1519. https://doi.org/10.3390/cryst13101519
APA StyleDemmer, U., Lemaire, O. N., Belhamri, M., & Ermler, U. (2023). Ethoxylate Polymer-Based 96-Well Screen for Protein Crystallization. Crystals, 13(10), 1519. https://doi.org/10.3390/cryst13101519






