3D Printed Microfluidic Cell for SAXS Time-Resolved Measurements of the Structure of Protein Crystallization Solutions
Abstract
1. Introduction
2. Materials and Methods
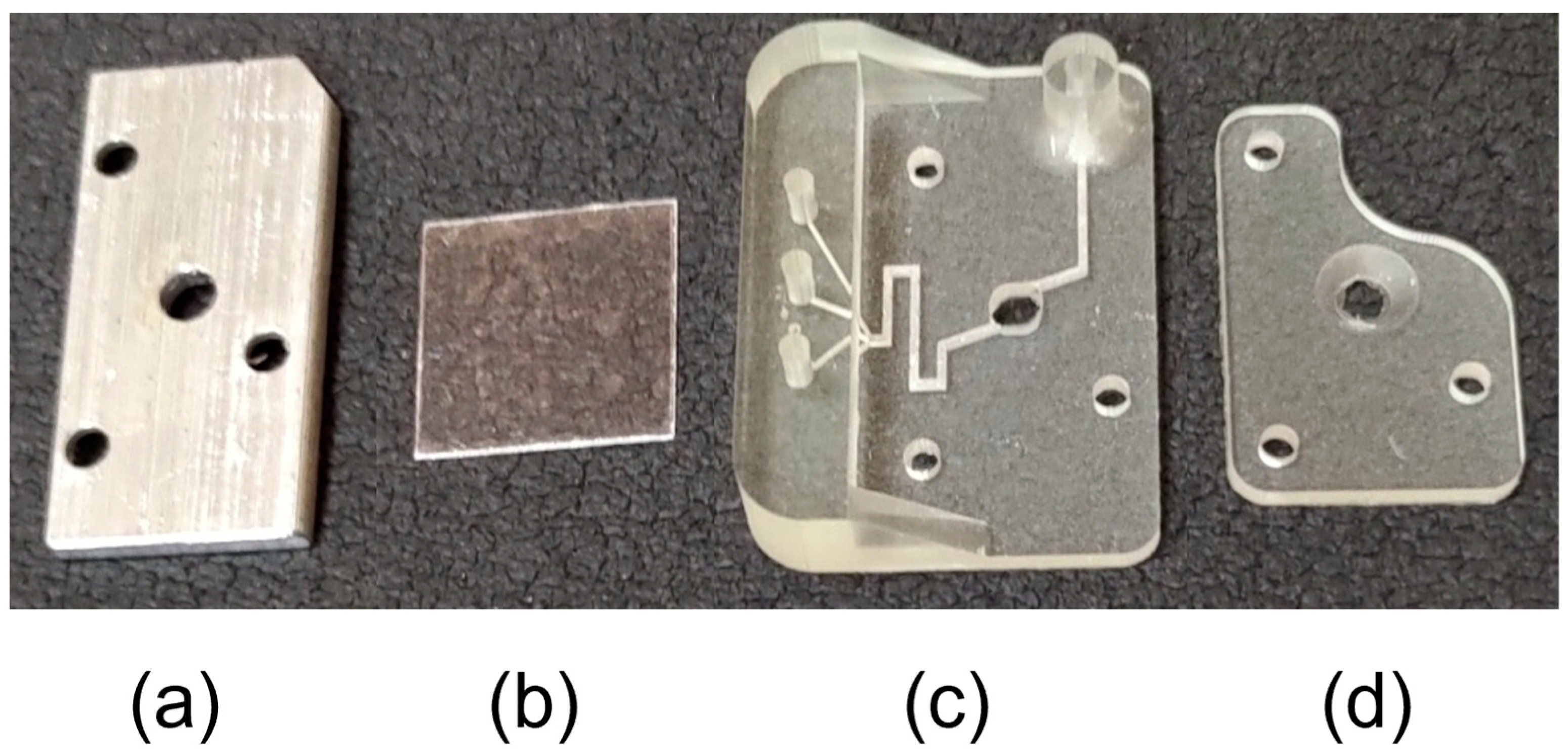
2.1. Production of Microfluidic Cell
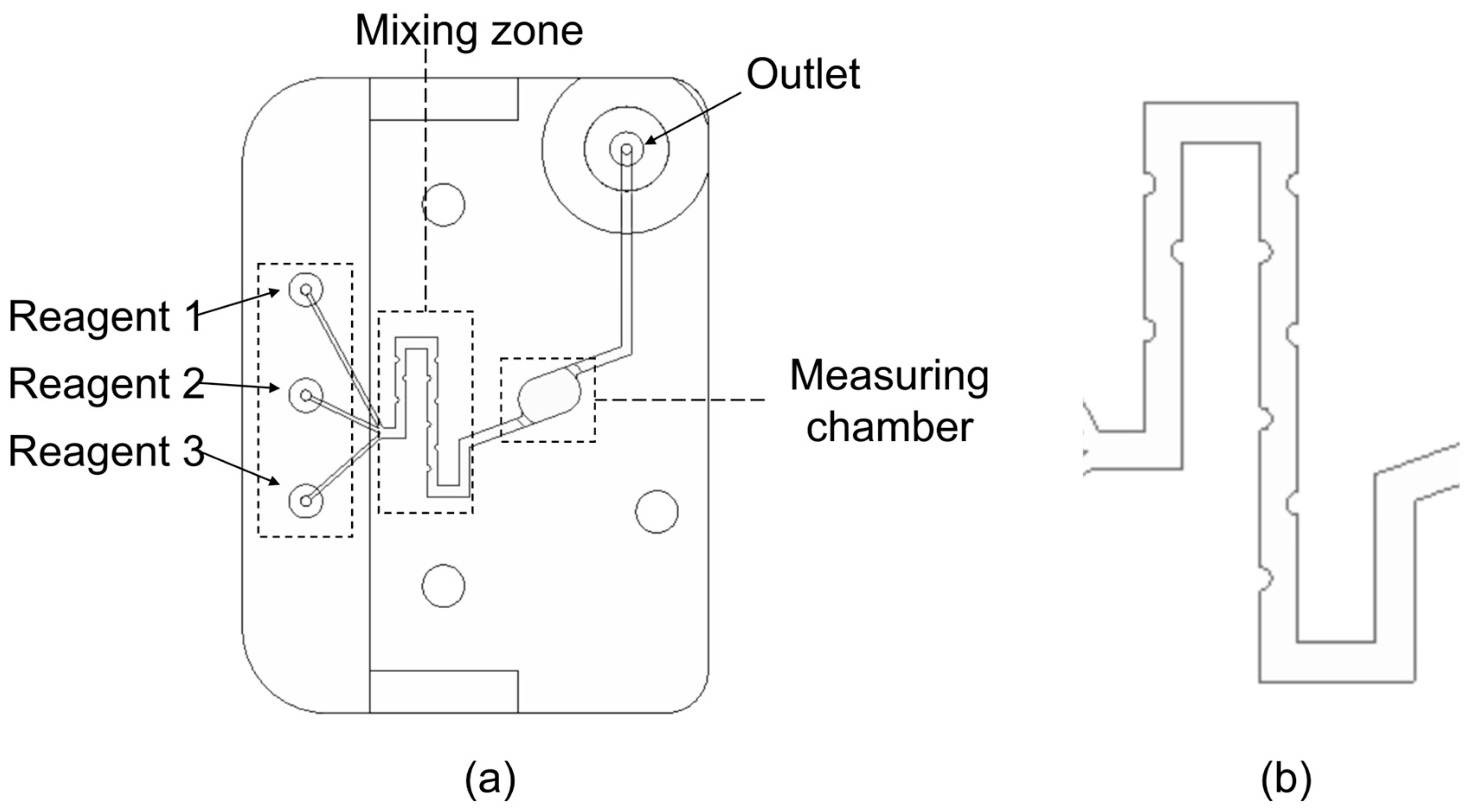
2.2. Description of the Microfluidic Cell
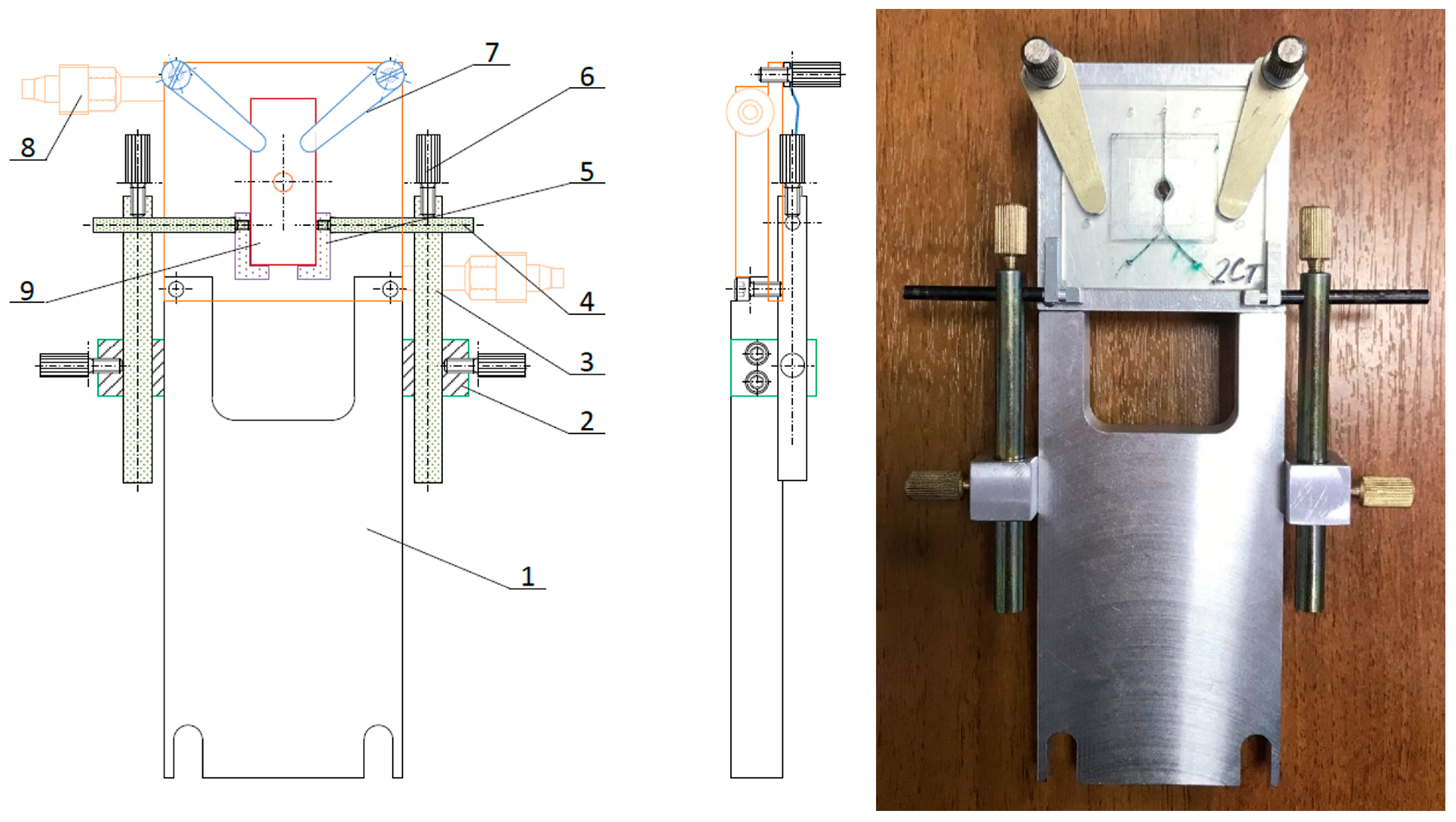
2.3. Holder
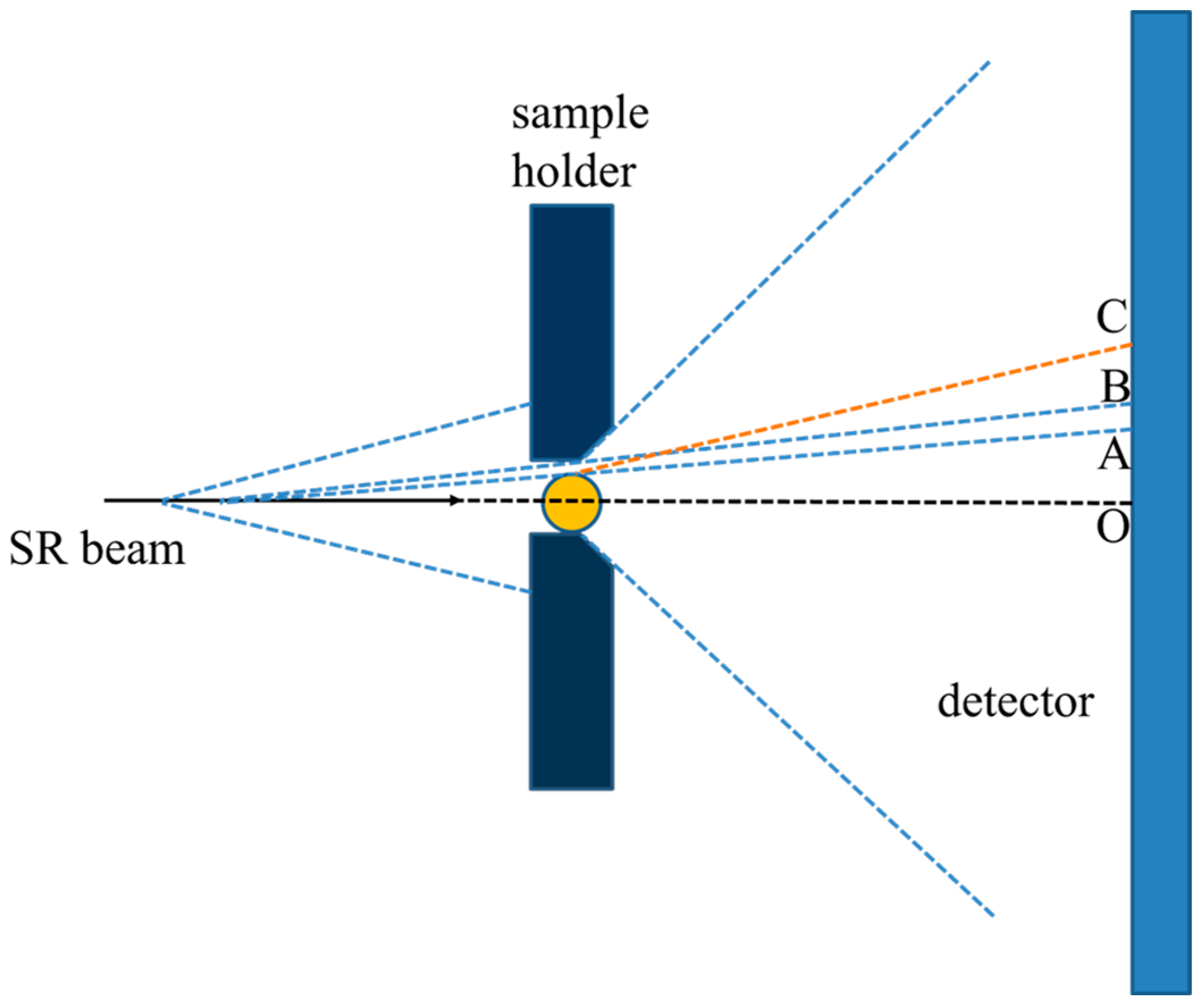
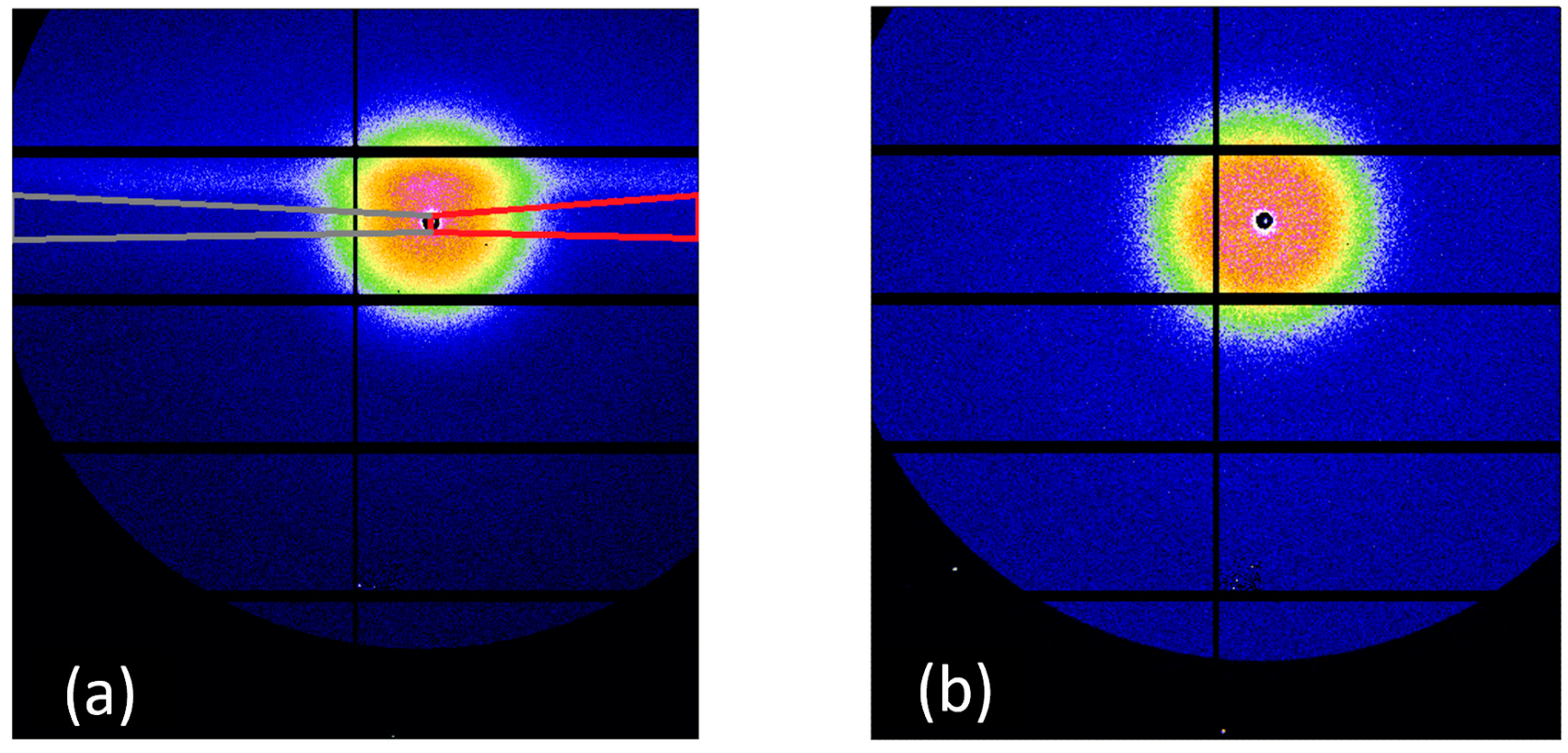
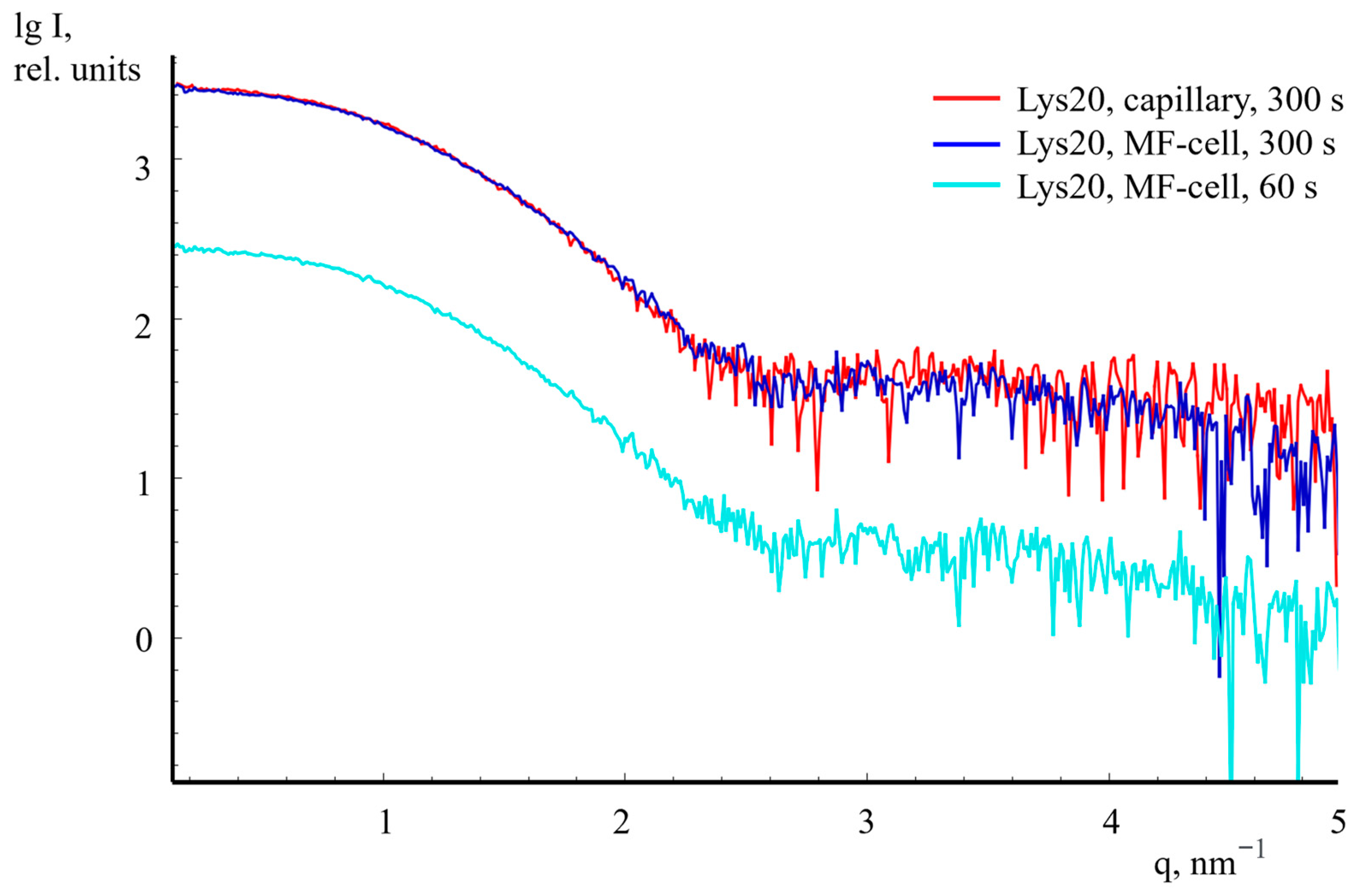
2.4. SAXS Measurement
2.5. Materials and Sample Preparation
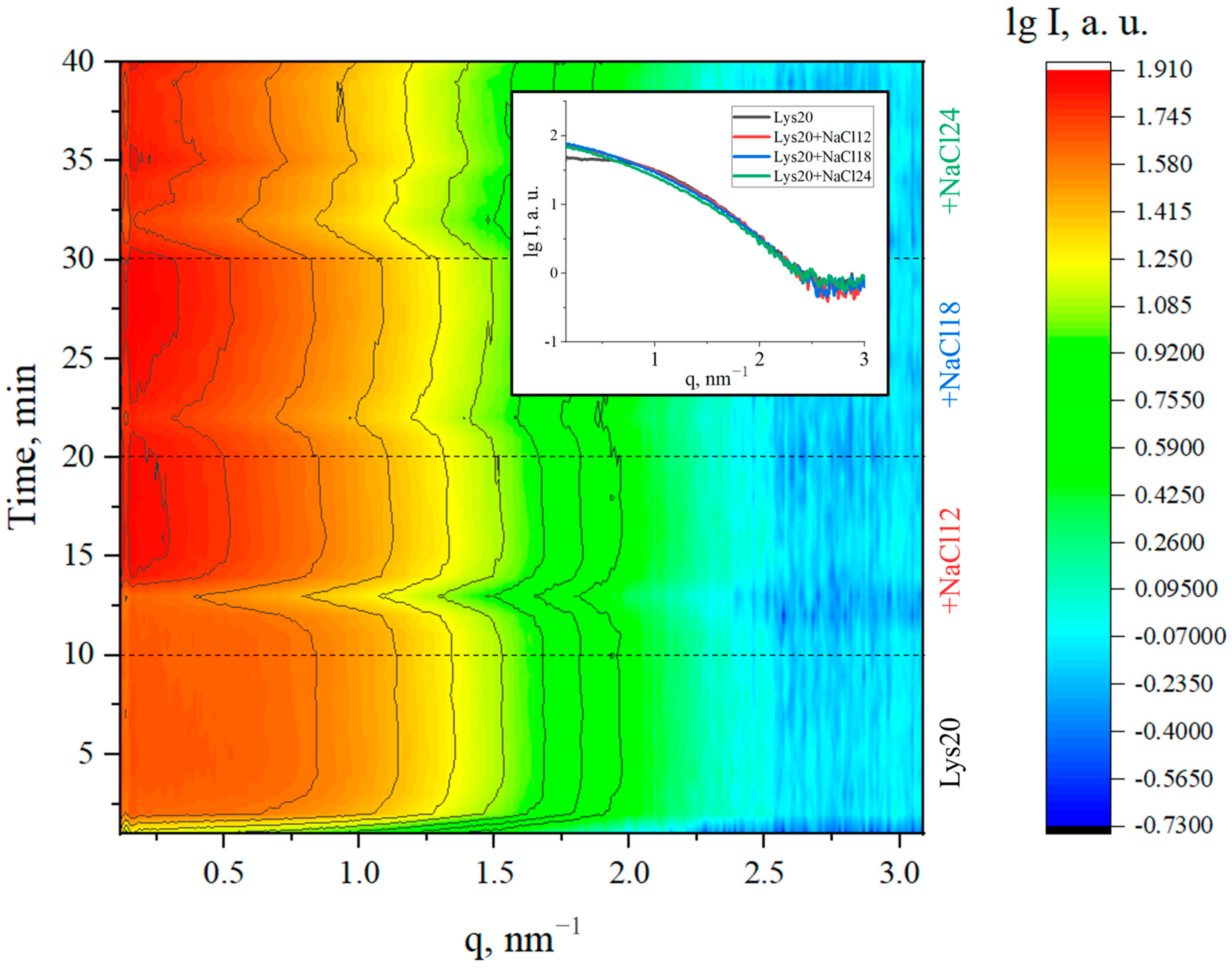
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kozyr, E.G.; Njoroge, P.N.; Chapek, S.V.; Shapovalov, V.V.; Skorynina, A.A.; Pnevskaya, A.Y.; Bulgakov, A.N.; Soldatov, A.V.; Pellegrino, F.; Groppo, E.; et al. Operando Laboratory X-ray Absorption Spectroscopy and UV–Vis Study of Pt/TiO2 Photocatalysts during Photodeposition and Hydrogen Evolution Reactions. Catalysts 2023, 13, 414. [Google Scholar] [CrossRef]
- Schwemmer, F.; Blanchet, C.E.; Spilotros, A.; Kosse, D.; Zehnle, S.; Mertens, H.D.T.; Graewert, M.A.; Rössle, M.; Paust, N.; Svergun, D.I.; et al. LabDisk for SAXS: A Centrifugal Microfluidic Sample Preparation Platform for Small-Angle X-ray Scattering. Lab Chip 2016, 16, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Schewa, S.; Schroer, M.A.; Zickmantel, T.; Song, Y.-H.; Blanchet, C.E.; Gruzinov, A.Y.; Katona, G.; Svergun, D.I.; Roessle, M. A THz Transparent 3D Printed Microfluidic Cell for Small Angle X-ray Scattering. Rev. Sci. Instrum. 2020, 91, 084101. [Google Scholar] [CrossRef] [PubMed]
- García-Lojo, D.; Modin, E.; Gómez-Graña, S.; Impéror-Clerc, M.; Chuvilin, A.; Pastoriza-Santos, I.; Pérez-Juste, J.; Constantin, D.; Hamon, C. Structure and Formation Kinetics of Millimeter-Size Single Domain Supercrystals. Adv. Funct. Mater. 2021, 31, 2101869. [Google Scholar] [CrossRef]
- Anaraki, N.I.; Sadeghpour, A.; Iranshahi, K.; Toncelli, C.; Cendrowska, U.; Stellacci, F.; Dommann, A.; Wick, P.; Neels, A. New Approach for Time-Resolved and Dynamic Investigations on Nanoparticles Agglomeration. Nano Res. 2020, 13, 2847–2856. [Google Scholar] [CrossRef]
- Lange, T.; Charton, S.; Bizien, T.; Testard, F.; Malloggi, F. OSTE+ for in Situ SAXS Analysis with Droplet Microfluidic Devices. Lab Chip 2020, 20, 2990–3000. [Google Scholar] [CrossRef] [PubMed]
- Guda, A.A.; Kirichkov, M.V.; Shapovalov, V.V.; Muravlev, A.I.; Pashkov, D.M.; Guda, S.A.; Bagliy, A.P.; Soldatov, S.A.; Chapek, S.V.; Soldatov, A.V. Machine Learning Analysis of Reaction Parameters in UV-Mediated Synthesis of Gold Nanoparticles. J. Phys. Chem. C 2023, 127, 1097–1108. [Google Scholar] [CrossRef]
- Khvostichenko, D.S.; Kondrashkina, E.; Perry, S.L.; Pawate, A.S.; Brister, K.; Kenis, P.J.A. An X-ray Transparent Microfluidic Platform for Screening of the Phase Behavior of Lipidic Mesophases. Analyst 2013, 138, 5384. [Google Scholar] [CrossRef]
- Ghazal, A.; Lafleur, J.P.; Mortensen, K.; Kutter, J.P.; Arleth, L.; Jensen, G.V. Recent advances in X-ray compatible microfluidics for applications in soft materials and life sciences. Lab Chip 2016, 16, 4263–4295. [Google Scholar] [CrossRef]
- Silva, B.F. SAXS on a chip: From dynamics of phase transitions to alignment phenomena at interfaces studied with microfluidic devices. Phys. Chem. Chem. Phys. 2017, 19, 23690–23703. [Google Scholar] [CrossRef]
- Khaliqi, K.; Ghazal, A.; Azmi, I.D.M.; Amenitsch, H.; Mortensen, K.; Salentinig, S.; Yaghmur, A. Direct monitoring of lipid transfer on exposure of citrem nanoparticles to an ethanol solution containing soybean phospholipids by combining synchrotron SAXS with microfluidics. Analyst 2017, 142, 3118–3126. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.P.; Brooks, N.J.; Seddon, J.M.; Luckham, P.F.; Terrill, N.J.; Kowalski, A.J.; Cabral, J.T. Microfluidic processing of concentrated surfactant mixtures: Online SAXS, microscopy and rheology. Soft Matter 2016, 12, 1750–1758. [Google Scholar] [CrossRef] [PubMed]
- Karthika, S.; Radhakrishnan, T.K.; Kalaichelvi, P. A Review of Classical and Nonclassical Nucleation Theories. Cryst. Growth Des. 2016, 16, 6663–6681. [Google Scholar] [CrossRef]
- Ivanov, V.K.; Fedorov, P.P.; Baranchikov, A.Y.; Osiko, V. V Oriented Attachment of Particles: 100 Years of Investigations of Non-Classical Crystal Growth. Russ. Chem. Rev. 2014, 83, 1204–1222. [Google Scholar] [CrossRef]
- De Yoreo, J.J.; Gilbert, P.U.P.A.; Sommerdijk, N.A.J.M.; Penn, R.L.; Whitelam, S.; Joester, D.; Zhang, H.; Rimer, J.D.; Navrotsky, A.; Banfield, J.F.; et al. Crystallization by Particle Attachment in Synthetic, Biogenic, and Geologic Environments. Science 2015, 349, aaa6760. [Google Scholar] [CrossRef] [PubMed]
- Krauss, I.R.; Merlino, A.; Vergara, A.; Sica, F.; Russo Krauss, I.; Merlino, A.; Vergara, A.; Sica, F. An Overview of Biological Macromolecule Crystallization. Int. J. Mol. Sci. 2013, 14, 11643–11691. [Google Scholar] [CrossRef]
- McPherson, A. Introduction to Protein Crystallization. Methods 2004, 34, 254–265. [Google Scholar] [CrossRef]
- Berman, H.M. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Kovalchuk, M.V.; Blagov, A.E.; Dyakova, Y.A.; Gruzinov, A.Y.; Marchenkova, M.A.; Peters, G.S.; Pisarevsky, Y.V.; Timofeev, V.I.; Volkov, V.V. Investigation of the Initial Crystallization Stage in Lysozyme Solutions by Small-Angle X-ray Scattering. Cryst. Growth Des. 2016, 16, 1792–1797. [Google Scholar] [CrossRef]
- Marchenkova, M.A.; Volkov, V.V.; Blagov, A.E.; Dyakova, Y.A.; Ilina, K.B.; Tereschenko, E.Y.; Timofeev, V.I.; Pisarevsky, Y.V.; Kovalchuk, M.V. In Situ Study of the State of Lysozyme Molecules at the Very Early Stage of the Crystallization Process by Small-Angle X-ray Scattering. Crystallogr. Rep. 2016, 61, 5–10. [Google Scholar] [CrossRef]
- Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Konarev, P.V.; Kryukova, A.E.; Kuklin, A.I.; Marchenkova, M.A.; Nabatov, B.V.; Blagov, A.E.; Pisarevsky, Y.V.; et al. Octamer Formation in Lysozyme Solutions at the Initial Crystallization Stage Detected by Small-Angle Neutron Scattering. Acta Crystallogr. Sect. D 2017, 73, 591–599. [Google Scholar] [CrossRef] [PubMed]
- Boikova, A.S.; D’yakova, Y.A.; Il’ina, K.B.; Konarev, P.V.; Kryukova, A.E.; Marchenkova, M.A.; Blagov, A.E.; Pisarevskii, Y.V.; Koval’chuk, M.V. Small-Angle X-ray Scattering Study of the Influence of Solvent Replacement (from H2O to D2O) on the Initial Crystallization Stage of Tetragonal Lysozyme. Crystallogr. Rep. 2017, 62, 837–842. [Google Scholar] [CrossRef]
- Dyakova, Y.A.; Boikova, A.S.; Ilina, K.B.; Konarev, P.V.; Marchenkova, M.A.; Pisarevsky, Y.V.; Timofeev, V.I.; Kovalchuk, M.V. Study of the Influence of a Precipitant Cation on the Formation of Oligomers in Crystallization Solutions of Lysozyme Protein. Crystallogr. Rep. 2019, 64, 11–15. [Google Scholar] [CrossRef]
- Marchenkova, M.A.; Konarev, P.V.; Boikova, A.S.; Ilina, K.B.; Pisarevsky, Y.V.; Kovalchuk, M.V. Influence of Chlorides of Mono- and Divalent Metals on the Oligomeric Composition of Lysozyme Crystallization Solutions and Further Crystal Growth. Crystallogr. Rep. 2021, 66, 751–757. [Google Scholar] [CrossRef]
- Kovalchuk, M.V.; Alekseeva, O.A.; Blagov, A.E.; Ilyushin, G.D.; Il’ina, K.B.; Konarev, P.V.; Lomonov, V.A.; Pisarevsky, Y.V.; Peters, G.S. Investigation of the Structure of Crystal-Forming Solutions of Potassium Dihydrogen Phosphate K(H2PO4) (KDP Type) on the Basis of Modeling Precursor Clusters and According to Small-Angle X-ray Scattering Data. Crystallogr. Rep. 2019, 64, 6–10. [Google Scholar] [CrossRef]
- Kovalchuk, M.V.; Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Konarev, P.V.; Kryukova, A.E.; Marchenkova, M.A.; Pisarevsky, Y.V.; Timofeev, V.I. Pre-Crystallization Phase Formation of Thermolysin Hexamers in Solution Close to Crystallization Conditions. J. Biomol. Struct. Dyn. 2019, 37, 3058–3064. [Google Scholar] [CrossRef]
- Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Konarev, P.V.; Kryukova, A.E.; Marchenkova, M.A.; Pisarevsky, Y.V.; Kovalchuk, M.V. Investigation of the Pre-Crystallization Stage of Proteinase K in Solution (Influence of Temperature and Precipitant Type) by Small-Angle X-ray Scattering. Crystallogr. Rep. 2018, 63, 865–870. [Google Scholar] [CrossRef]
- Marchenkova, M.A.; Konarev, P.V.; Rakitina, T.V.; Timofeev, V.I.; Boikova, A.S.; Dyakova, Y.A.; Ilina, K.B.; Korzhenevskiy, D.A.; Yu Nikolaeva, A.; Pisarevsky, Y.V.; et al. Dodecamers Derived from the Crystal Structure Were Found in the Pre-Crystallization Solution of the Transaminase from the Thermophilic Bacterium Thermobaculum Terrenum by Small-Angle X-ray Scattering. J. Biomol. Struct. Dyn. 2020, 38, 2939–2944. [Google Scholar] [CrossRef]
- van der Linden, P.J.E.M.; Popov, A.M.; Pontoni, D. Accurate and Rapid 3D Printing of Microfluidic Devices Using Wavelength Selection on a DLP Printer. Lab Chip 2020, 20, 4128–4140. [Google Scholar] [CrossRef] [PubMed]
- Peters, G.S.; Zakharchenko, O.A.; Konarev, P.V.; Karmazikov, Y.V.; Smirnov, M.A.; Zabelin, A.V.; Mukhamedzhanov, E.H.; Veligzhanin, A.A.; Blagov, A.E.; Kovalchuk, M.V. The Small-Angle X-ray Scattering Beamline BioMUR at the Kurchatov Synchrotron Radiation Source. Nucl. Instrum. Methods Phys. Res. A 2019, 945, 162616. [Google Scholar] [CrossRef]
- Hammersley, A.P. FIT2D: A Multi-Purpose Data Reduction, Analysis and Visualization Program. J. Appl. Crystallogr. 2016, 49, 646–652. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-Based System for Small-Angle Scattering Data Analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]


| Precipitant | 0 mg/mL | 12 mg/mL (60 mg/mL) | 18 mg/mL (60 mg/mL) | 24 mg/mL (60 mg/mL) | |
|---|---|---|---|---|---|
| Protein | |||||
| 0 mg/mL | Vprot = 0 | Vprot = 0 | Vprot = 0 | Vprot = 0 | |
| Vpret = 0 | Vpret = 0.2 | Vpret = 0.3 | Vpret = 0.4 | ||
| Vbuf = 1 | Vbuf = 0.8 | Vbuf = 0.7 | Vbuf = 0.6 | ||
| 10 mg/mL (20 mg/mL) | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | |
| Vpret = 0 | Vpret = 0.2 | Vpret = 0.3 | Vpret = 0.4 | ||
| Vbuf = 0.5 | Vbuf = 0.3 | Vbuf = 0.2 | Vbuf = 0.3 | ||
| 20 mg/mL (40 mg/mL) | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | |
| Vpret = 0 | Vpret = 0.2 | Vpret = 0.3 | Vpret = 0.4 | ||
| Vbuf = 0.5 | Vbuf = 0.3 | Vbuf = 0.2 | Vbuf = 0.3 | ||
| 40 mg/mL (80 mg/mL) | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | Vprot = 0.5 | |
| Vpret = 0 | Vpret = 0.2 | Vpret = 0.3 | Vpret = 0.4 | ||
| Vbuf = 0.5 | Vbuf = 0.3 | Vbuf = 0.2 | Vbuf = 0.3 | ||
| Lysozyme Concentration, mg/mL | Concentration of NaCl Precipitant, mg/mL | Rg, nm | Proportion of Monomers,% | Proportion of Dimers, % | Percentage of Octamers,% |
|---|---|---|---|---|---|
| 10 | 0 | 1.41 ± 0.03 | 100 | 0 | 0 |
| 10 | 12 | 1.69 ± 0.03 | 98.7 ± 0.5 | 0 | 1.3 ± 0.1 |
| 10 | 18 | 1.72 ± 0.03 | 98.4 ± 0.5 | 0 | 1.6 ± 0.1 |
| 10 | 24 | 1.85 ± 0.03 | 97.6 ± 0.5 | 0 | 2.4 ± 0.1 |
| 20 | 0 | 1.42 ± 0.03 | 100 | 0 | 0 |
| 20 | 12 | 1.80 ± 0.03 | 96.4 ± 0.5 | 1.8 ± 0.1 | 1.8 ± 0.1 |
| 20 | 18 | 2.04 ± 0.03 | 94.5 ± 0.5 | 1.7 ± 0.1 | 3.8 ± 0.1 |
| 20 | 24 | 2.16 ± 0.03 | 93.1 ± 0.5 | 1.8 ± 0.1 | 5.1 ± 0.1 |
| 40 | 0 | 1.43 ± 0.03 | 100 | 0 | 0 |
| 40 | 12 | 1.94 ± 0.03 | 96.6 ± 0.5 | 0.8 ± 0.1 | 2.6 ± 0.1 |
| 40 | 18 | 2.43 ± 0.03 | 89.9 ± 0.5 | 1.2 ± 0.1 | 8.9 ± 0.1 |
| 40 | 24 | 2.53 ± 0.03 | 87.1 ± 0.5 | 2.0 ± 0.1 | 10.9 ± 0.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marchenkova, M.A.; Chapek, S.V.; Konarev, P.V.; Ilina, K.B.; Peters, G.S.; Pisarevsky, Y.V.; Shishkov, V.A.; Soldatov, A.V.; Kovalchuk, M.V. 3D Printed Microfluidic Cell for SAXS Time-Resolved Measurements of the Structure of Protein Crystallization Solutions. Crystals 2023, 13, 938. https://doi.org/10.3390/cryst13060938
Marchenkova MA, Chapek SV, Konarev PV, Ilina KB, Peters GS, Pisarevsky YV, Shishkov VA, Soldatov AV, Kovalchuk MV. 3D Printed Microfluidic Cell for SAXS Time-Resolved Measurements of the Structure of Protein Crystallization Solutions. Crystals. 2023; 13(6):938. https://doi.org/10.3390/cryst13060938
Chicago/Turabian StyleMarchenkova, Margarita A., Sergei V. Chapek, Petr V. Konarev, Ksenia B. Ilina, Georgy S. Peters, Yury V. Pisarevsky, Vladimir A. Shishkov, Alexander V. Soldatov, and Mikhail V. Kovalchuk. 2023. "3D Printed Microfluidic Cell for SAXS Time-Resolved Measurements of the Structure of Protein Crystallization Solutions" Crystals 13, no. 6: 938. https://doi.org/10.3390/cryst13060938
APA StyleMarchenkova, M. A., Chapek, S. V., Konarev, P. V., Ilina, K. B., Peters, G. S., Pisarevsky, Y. V., Shishkov, V. A., Soldatov, A. V., & Kovalchuk, M. V. (2023). 3D Printed Microfluidic Cell for SAXS Time-Resolved Measurements of the Structure of Protein Crystallization Solutions. Crystals, 13(6), 938. https://doi.org/10.3390/cryst13060938









