Motility Suppression and Trapping Bacteria by ZnO Nanostructures
Abstract
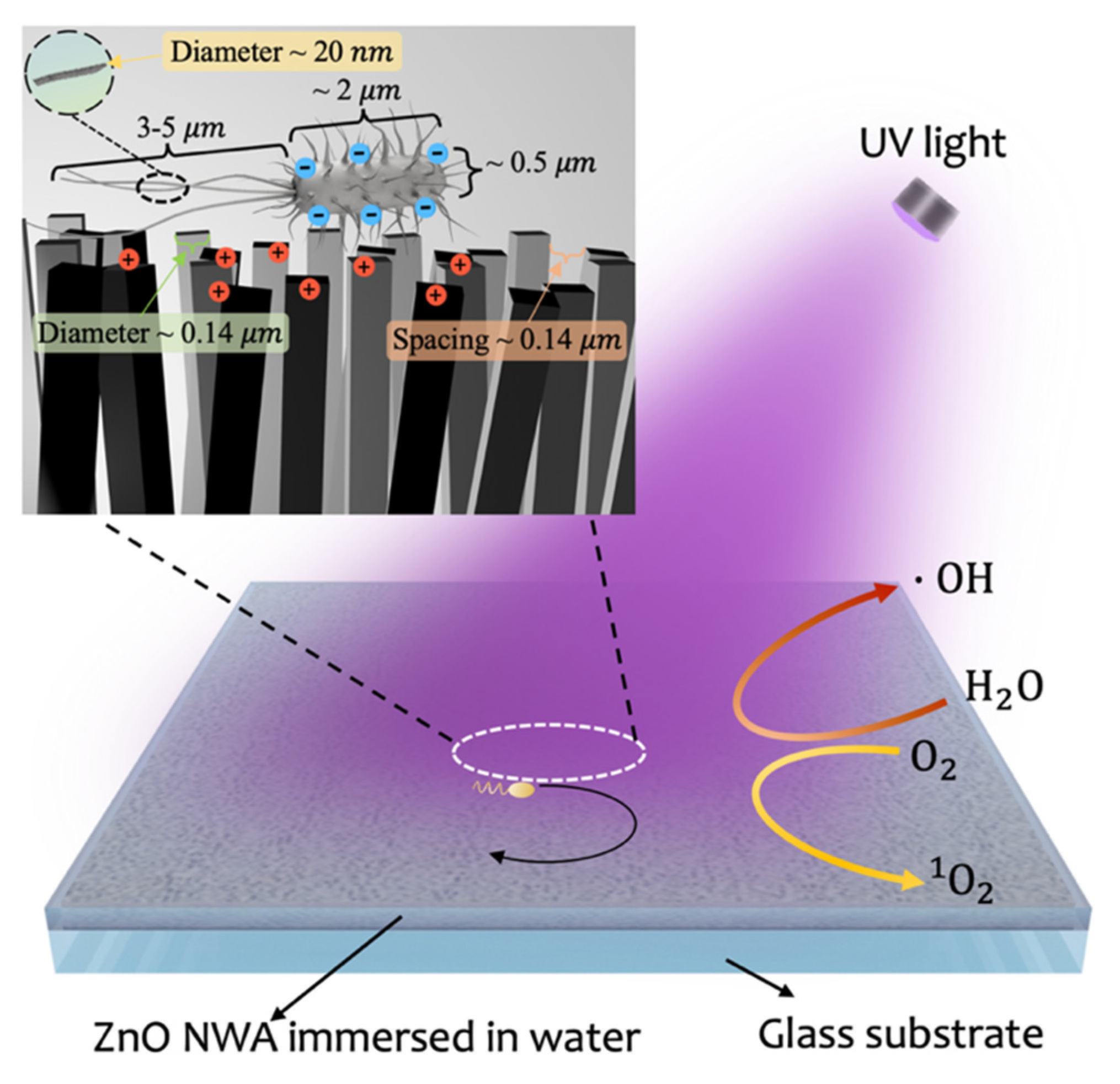
:1. Introduction
2. Materials and Methods
2.1. Bacteria Culturing
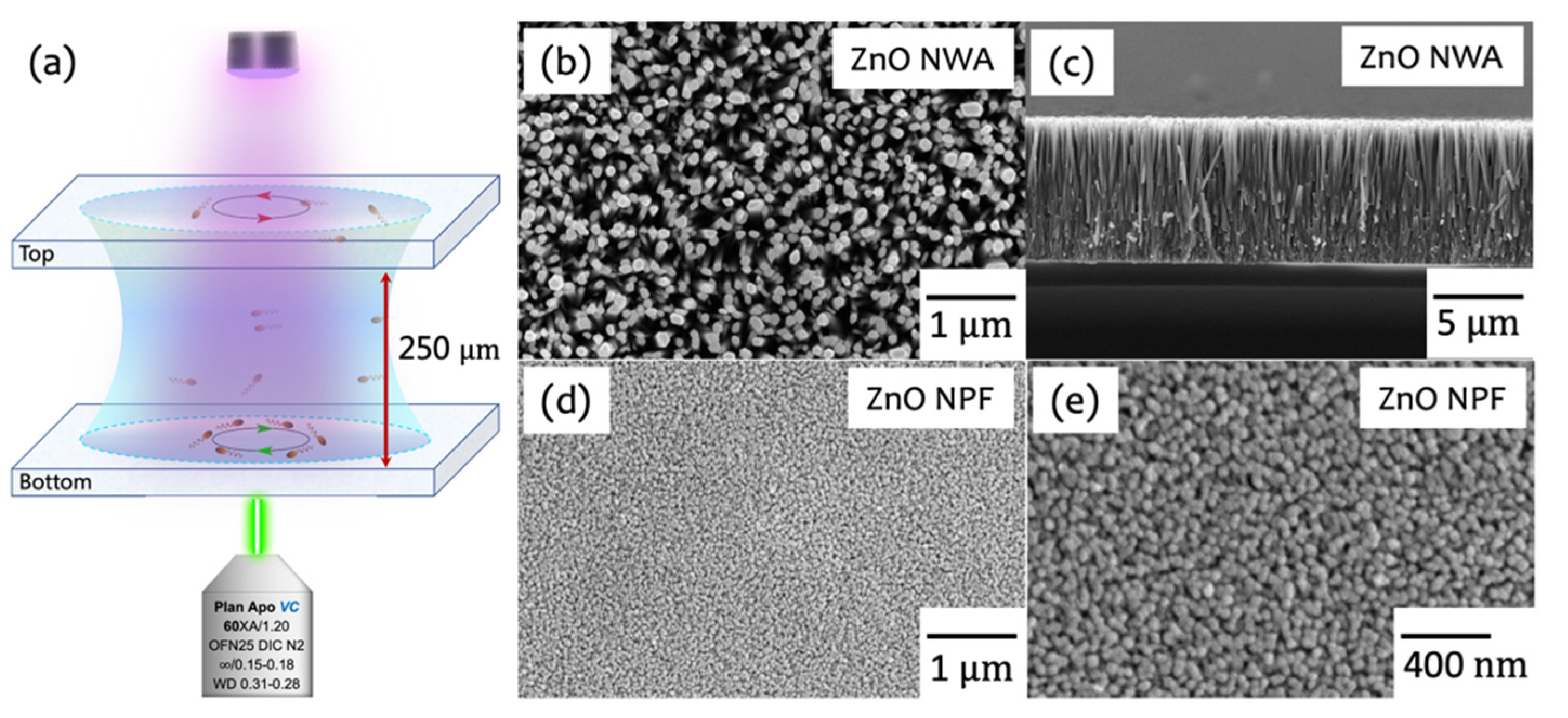
2.2. ZnO Nanostructures Preparation
2.3. Observation Setup
2.4. Bacteria Tracking
3. Results and Discussion
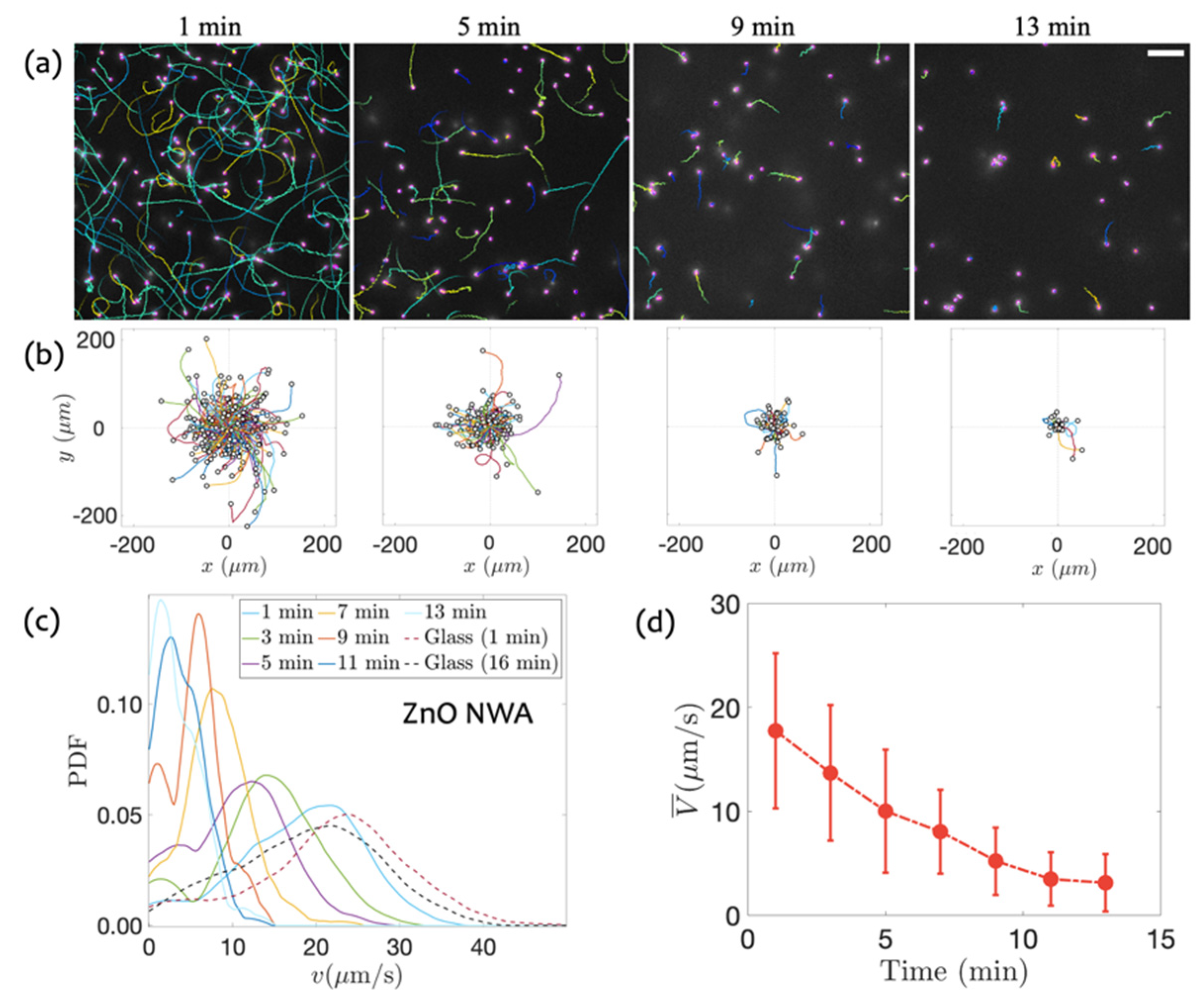
3.1. Swimming on ZnO NWA
3.2. Swimming on ZnO NWA in Response to UV Irradiation
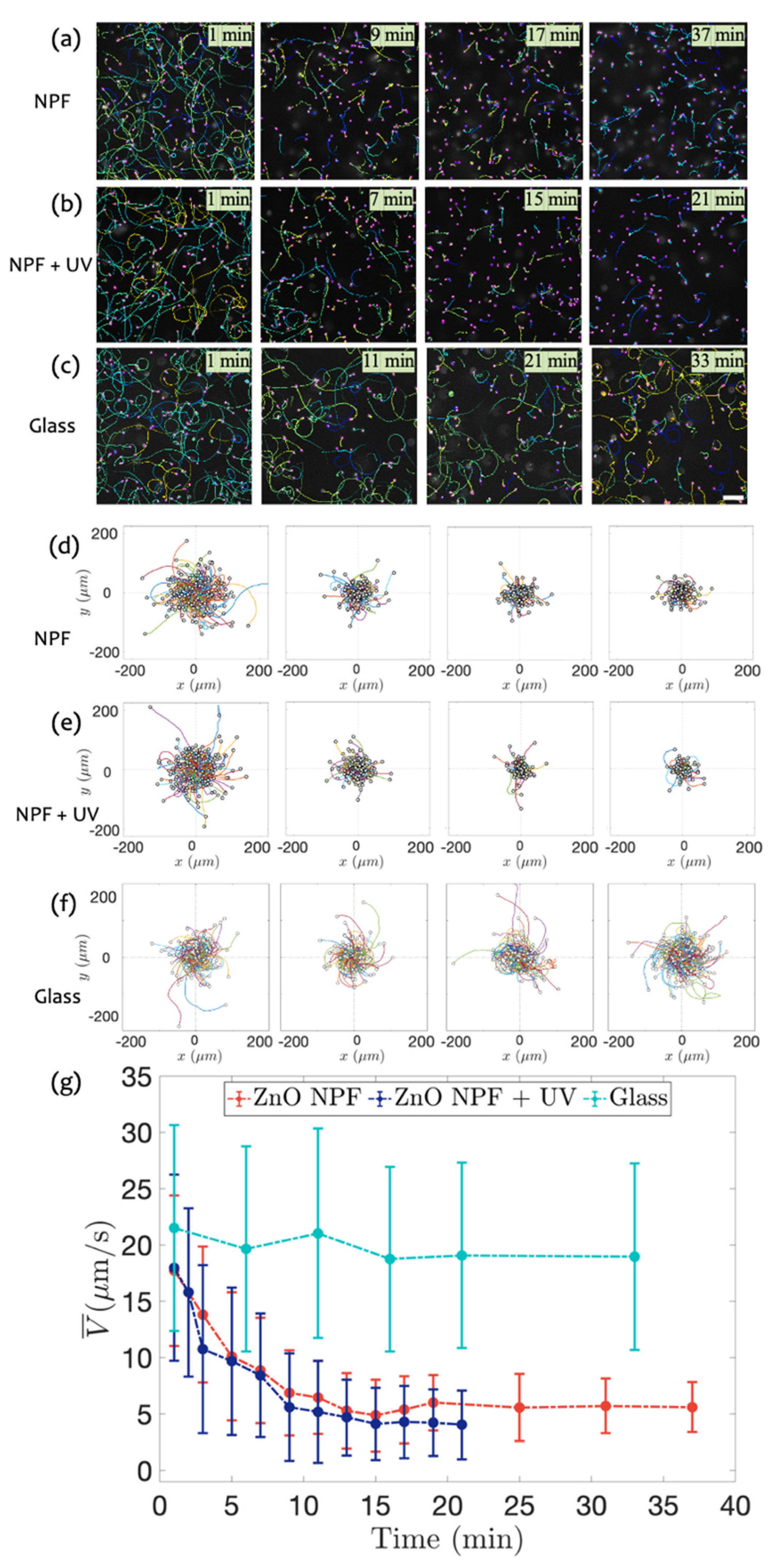
3.3. Swimming on ZnO NPF with and without UV Radiation
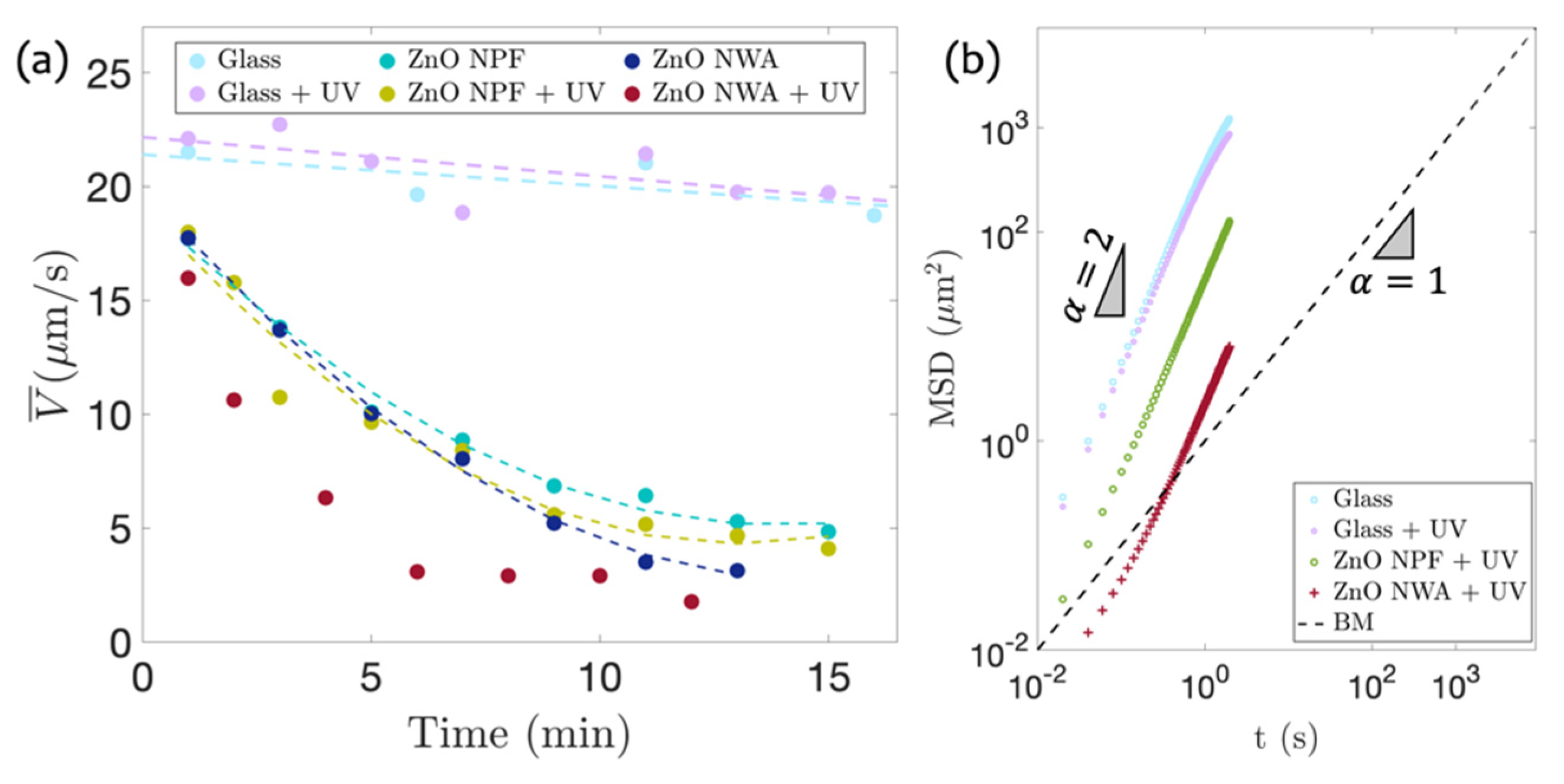
3.4. Comparison of Bacterial Motility
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Knights, H.E.; Jorrin, B.; Haskett, T.L.; Poole, P.S. Deciphering bacterial mechanisms of root colonization. Environ. Microbiol. Rep. 2021, 13, 428–444. [Google Scholar] [CrossRef] [PubMed]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Siegel, S.J.; Weiser, J.N. Mechanisms of bacterial colonization of the respiratory tract. Annu. Rev. Microbiol. 2015, 69, 425–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ribet, D.; Cossart, P. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 2015, 17, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Bosecker, K. Bioleaching: Metal solubilization by microorganisms. FEMS Microbiol. Rev. 1997, 20, 591–604. [Google Scholar] [CrossRef]
- Kolencik, M.; Vojtkova, H.; Urik, M.; Caplovicova, M.; Pistora, J.; Cada, M.; Babicova, A.; Feng, H.; Qian, Y.; Ramakanth, I. Heterotrophic bacterial leaching of zinc and arsenic from artificial adamite. Water Air Soil Pollut. 2017, 228, 1–11. [Google Scholar] [CrossRef]
- Vaccari, L.; Molaei, M.; Niepa, T.H.; Lee, D.; Leheny, R.L.; Stebe, K.J. Films of bacteria at interfaces. Adv. Colloid Interface Sci. 2017, 247, 561–572. [Google Scholar] [CrossRef]
- Alapan, Y.; Yasa, O.; Yigit, B.; Yasa, I.C.; Erkoc, P.; Sitti, M. Microrobotics and microorganisms: Biohybrid autonomous cellular robots. Annu. Rev. Control Robot. Auton. Syst. 2019, 2, 205–230. [Google Scholar] [CrossRef]
- Raina, J.-B.; Fernandez, V.; Lambert, B.; Stocker, R.; Seymour, J.R. The role of microbial motility and chemotaxis in symbiosis. Nat. Rev. Microbiol. 2019, 17, 284–294. [Google Scholar] [CrossRef]
- Lauga, E. Bacterial hydrodynamics. Annu. Rev. Fluid Mech. 2016, 48, 105–130. [Google Scholar] [CrossRef] [Green Version]
- Berg, H.C.; Turner, L. Chemotaxis of bacteria in glass capillary arrays. Escherichia coli, motility, microchannel plate, and light scattering. Biophys. J. 1990, 58, 919–930. [Google Scholar] [CrossRef] [Green Version]
- Frymier, P.D.; Ford, R.M.; Berg, H.C.; Cummings, P.T. Three-dimensional tracking of motile bacteria near a solid planar surface. Proc. Natl. Acad. Sci. USA 1995, 92, 6195–6199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frymier, P.D.; Ford, R.M. Analysis of bacterial swimming speed approaching a solid–liquid interface. AIChE J. 1997, 43, 1341–1347. [Google Scholar] [CrossRef]
- Lauga, E.; DiLuzio, W.R.; Whitesides, G.M.; Stone, H.A. Swimming in circles: Motion of bacteria near solid boundaries. Biophys. J. 2006, 90, 400–412. [Google Scholar] [CrossRef] [Green Version]
- Lopez, D.; Lauga, E. Dynamics of swimming bacteria at complex interfaces. Phys. Fluids 2014, 26, 400–412. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Bensson, J.; Nisimova, L.; Munger, D.; Mahautmr, P.; Tang, J.X.; Maxey, M.R.; Brun, Y.V. Accumulation of swimming bacteria near a solid surface. Phys. Rev. E 2011, 84, 041932. [Google Scholar] [CrossRef] [Green Version]
- Vigeant, M.A.-S.; Ford, R.M.; Wagner, M.; Tamm, L.K. Reversible and irreversible adhesion of motile Escherichia coli cells analyzed by total internal reflection aqueous fluorescence microscopy. Appl. Environ. Microbiol. 2002, 68, 2794–2801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Tam, L.-K.; Tang, J.X. Amplified effect of Brownian motion in bacterial near-surface swimming. Proc. Natl. Acad. Sci. USA 2008, 105, 18355–18359. [Google Scholar] [CrossRef] [Green Version]
- Kantsler, V.; Dunkel, J.; Polin, M.; Goldstein, R.E. Ciliary contact interactions dominate surface scattering of swimming eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 1187–1192. [Google Scholar] [CrossRef] [Green Version]
- Vigeant, M.; Ford, R.M. Interactions between motile Escherichia coli and glass in media with various ionic strengths, as observed with a three-dimensional-tracking microscope. Appl. Environ. Microbiol. 1997, 63, 3474–3479. [Google Scholar] [CrossRef] [Green Version]
- Bos, R.; Van der Mei, H.C.; Busscher, H.J. Physico-chemistry of initial microbial adhesive interactions–its mechanisms and methods for study. FEMS Microbiol. Rev. 1999, 23, 179–230. [Google Scholar] [CrossRef]
- Hermansson, M. The DLVO theory in microbial adhesion. Colloids Surf. B Biointerfaces 1999, 14, 105–119. [Google Scholar] [CrossRef]
- Zhao, W.; Walker, S.L.; Huang, Q.; Cai, P. Adhesion of bacterial pathogens to soil colloidal particles: Influences of cell type, natural organic matter, and solution chemistry. Water Res. 2014, 53, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, S.; Saglimbeni, F.; Frangipane, G.; Dell’Arciprete, D.; Di Leonardo, R. 3D dynamics of bacteria wall entrapment at a water–air interface. Soft Matter 2019, 15, 3397–3406. [Google Scholar] [CrossRef]
- Kim, D.; Kim, Y.; Lim, S. Effects of swimming environment on bacterial motility. Phys. Fluids 2022, 34, 031907. [Google Scholar] [CrossRef]
- Hu, J.; Wysocki, A.; Winkler, R.G.; Gompper, G. Physical sensing of surface properties by microswimmers–directing bacterial motion via wall slip. Sci. Rep. 2015, 5, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Gottenbos, B.; Grijpma, D.W.; van der Mei, H.C.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef]
- Zheng, S.; Bawazir, M.; Dhall, A.; Kim, H.-E.; He, L.; Heo, J.; Hwang, G. Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 2021, 9, 82. [Google Scholar] [CrossRef]
- Schwibbert, K.; Menzel, F.; Epperlein, N.; Bonse, J.; Krüger, J. Bacterial adhesion on femtosecond laser-modified polyethylene. Materials 2019, 12, 3107. [Google Scholar] [CrossRef] [Green Version]
- Chien, H.-W.; Chen, X.-Y.; Tsai, W.-P.; Lee, M. Inhibition of biofilm formation by rough shark skin-patterned surfaces. Colloids Surf. B Biointerfaces 2020, 186, 110738. [Google Scholar] [CrossRef]
- Tang, M.; Chen, C.; Zhu, J.; Allcock, H.R.; Siedlecki, C.A.; Xu, L.-C. Inhibition of bacterial adhesion and biofilm formation by a textured fluorinated alkoxyphosphazene surface. Bioact. Mater. 2021, 6, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Wang, C.; Zhang, T.; Yi, X.; Liang, J.; Wang, H. Reduced bacterial adhesion on zirconium-based bulk metallic glasses by femtosecond laser nanostructuring. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2020, 234, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Lutey, A.H.; Gemini, L.; Romoli, L.; Lazzini, G.; Fuso, F.; Faucon, M.; Kling, R. Towards laser-textured antibacterial surfaces. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ding, Y.; Ge, X.; Leng, Y. Control of bacterial adhesion and growth on honeycomb-like patterned surfaces. Colloids Surf. B Biointerfaces 2015, 135, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Helbig, R.; Günther, D.; Friedrichs, J.; Rößler, F.; Lasagni, A.; Werner, C. The impact of structure dimensions on initial bacterial adhesion. Biomater. Sci. 2016, 4, 1074–1078. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.-R.; Weeks, E.R.; Ducker, W.A. Surface topography hinders bacterial surface motility. ACS Appl. Mater. Interfaces 2018, 10, 9225–9234. [Google Scholar] [CrossRef]
- Kolodziejczak-Radzimska, A.; Jesionowski, T. Zinc oxide—from synthesis to application: A review. Materials 2014, 7, 2833–2881. [Google Scholar] [CrossRef] [Green Version]
- Hecht, A.; Endy, D.; Salit, M.; Munson, M.S. When wavelengths collide: Bias in cell abundance measurements due to expressed fluorescent proteins. ACS Synth. Biol. 2016, 5, 1024–1027. [Google Scholar] [CrossRef] [Green Version]
- Yin, Z.; Shan, Y.; Yu, M.; Yang, L.; Song, J.; Hu, P.; Teng, F. Enhanced performance of UV photodetector based on ZnO nanorod arrays via TiO2 as electrons trap layer. Mater. Sci. Semicond. Processing 2022, 148, 106813. [Google Scholar] [CrossRef]
- Son, K.; Brumley, D.R.; Stocker, R. Live from under the lens: Exploring microbial motility with dynamic imaging and microfluidics. Nat. Rev. Microbiol. 2015, 13, 761–775. [Google Scholar] [CrossRef]
- Wu, X.-L.; Libchaber, A. Particle diffusion in a quasi-two-dimensional bacterial bath. Phys. Rev. Lett. 2000, 84, 3017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiland, G.; Kunstmann, P. Polar surfaces of zinc oxide crystals. Surf. Sci. 1969, 13, 72–84. [Google Scholar] [CrossRef]
- Ching, K.-L.; Li, G.; Ho, Y.-L.; Kwok, H.-S. The role of polarity and surface energy in the growth mechanism of ZnO from nanorods to nanotubes. CrystEngComm 2016, 18, 779–786. [Google Scholar] [CrossRef]
- Luo, H.; Ma, J.; Wang, P.; Bai, J.; Jing, G. Two-step wetting transition on ZnO nanorod arrays. Appl. Surf. Sci. 2015, 347, 868–874. [Google Scholar] [CrossRef]
- Pacchioni, G. Oxygen vacancy: The invisible agent on oxide surfaces. ChemPhysChem 2003, 4, 1041–1047. [Google Scholar] [CrossRef]
- Scorza, E.; Birkenheuer, U.; Pisani, C. The oxygen vacancy at the surface and in bulk MgO: An embedded-cluster study. J. Chem. Phys. 1997, 107, 9645–9658. [Google Scholar] [CrossRef] [Green Version]
- Güler, S.; Oruç, Ç. Comparison of the behavior of negative electrically charged E. coli and E. faecalis bacteria under electric field effect. Colloids Surf. B Biointerfaces 2021, 208, 112097. [Google Scholar] [CrossRef]
- Yi, C.; Yu, Z.; Ren, Q.; Liu, X.; Wang, Y.; Sun, X.; Yin, S.; Pan, J.; Huang, X. Nanoscale ZnO-based photosensitizers for photodynamic therapy. Photodiagnosis Photodyn. Ther. 2020, 30, 101694. [Google Scholar] [CrossRef]
- Jyothi, M.; Nayak, V.; Padaki, M.; Balakrishna, R.G.; Ismail, A. The effect of UV irradiation on PSf/TiO2 mixed matrix membrane for chromium rejection. Desalination 2014, 354, 189–199. [Google Scholar] [CrossRef]
- Bono, N.; Ponti, F.; Punta, C.; Candiani, G. Effect of UV irradiation and TiO2-photocatalysis on airborne bacteria and viruses: An overview. Materials 2021, 14, 1075. [Google Scholar] [CrossRef]
- Fakhar-e-Alam, M.; Kishwar, S.; Willander, M. Photodynamic effects of zinc oxide nanowires in skin cancer and fibroblast. Lasers Med. Sci. 2014, 29, 1189–1194. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, N.; Luo, H.; Liu, Y.; Yu, H.; Jing, G. Motility Suppression and Trapping Bacteria by ZnO Nanostructures. Crystals 2022, 12, 1027. https://doi.org/10.3390/cryst12081027
Yan N, Luo H, Liu Y, Yu H, Jing G. Motility Suppression and Trapping Bacteria by ZnO Nanostructures. Crystals. 2022; 12(8):1027. https://doi.org/10.3390/cryst12081027
Chicago/Turabian StyleYan, Ningzhe, Hao Luo, Yanan Liu, Haiping Yu, and Guangyin Jing. 2022. "Motility Suppression and Trapping Bacteria by ZnO Nanostructures" Crystals 12, no. 8: 1027. https://doi.org/10.3390/cryst12081027
APA StyleYan, N., Luo, H., Liu, Y., Yu, H., & Jing, G. (2022). Motility Suppression and Trapping Bacteria by ZnO Nanostructures. Crystals, 12(8), 1027. https://doi.org/10.3390/cryst12081027






