Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization
Abstract
1. Introduction
2. Materials and Methods
2.1. In Vitro Studies
2.1.1. Turbidimetric Assay
2.1.2. Thioflavin T Fluorescence Assay (ThT)
2.1.3. ANS Competitive Binding Assay
Recombinant Expression of TTR in E. coli
ANS Competitive Binding Assay
2.2. Crystallogenesis and Structure Resolution
2.3. 2,2-Diphenyl-1-Picrylhydrazyl (DPPH) Radical Scavenging
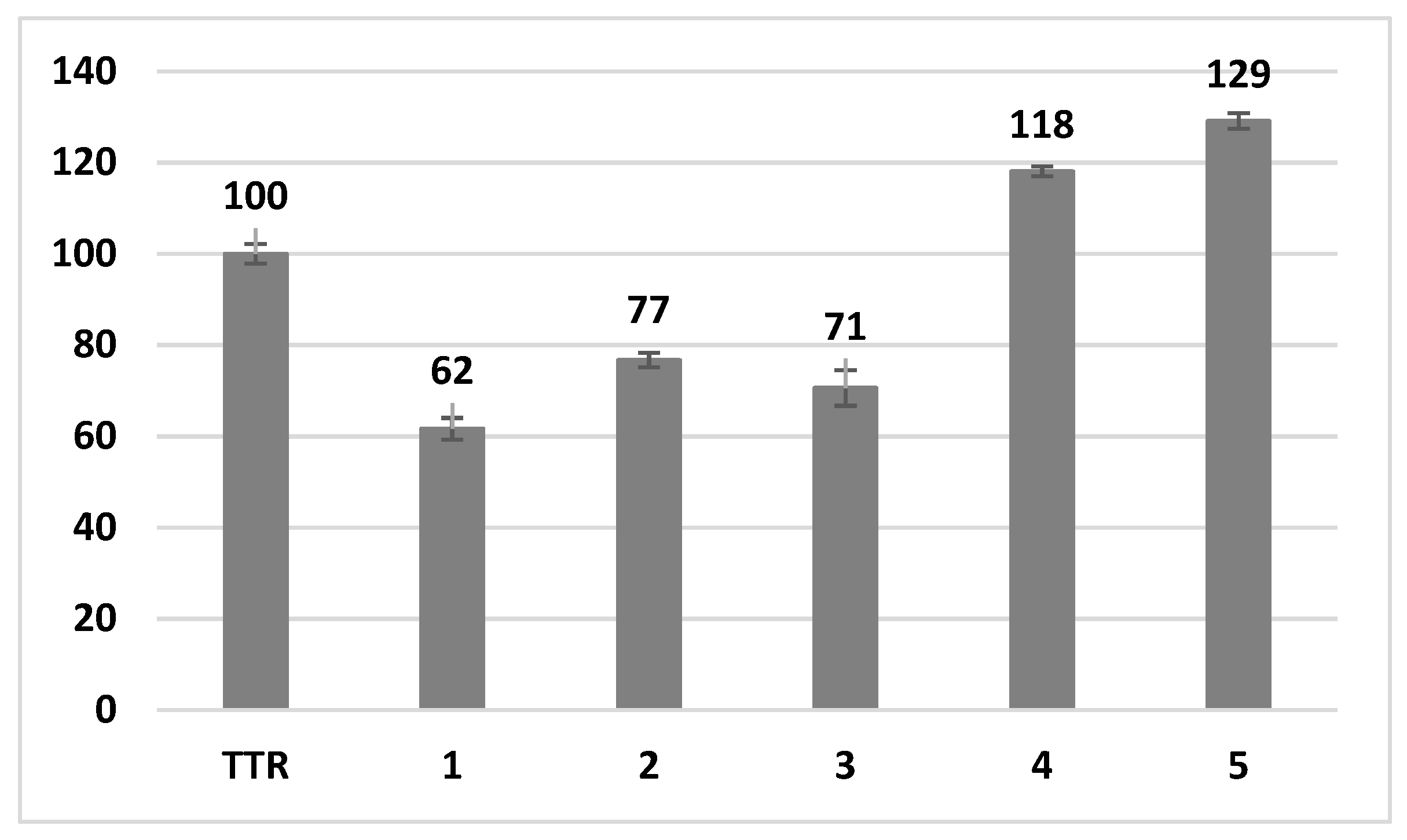
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Souyoul, S.A.; Saussy, K.P.; Lupo, M.P. Nutraceuticals: A Review. Dermatol. Ther. 2018, 8, 5–16. [Google Scholar] [CrossRef]
- Lu, J.; Wang, Z.; Li, S.; Xin, Q.; Yuan, M.; Li, H.; Song, X.; Gao, H.; Pervaiz, N.; Sun, X.; et al. Quercetin Inhibits the Migration and Invasion of HCCLM3 Cells by Suppressing the Expression of P-Akt1, Matrix Metalloproteinase (MMP) MMP-2, and MMP-9. Med. Sci. Monit. 2018, 24, 2583–2589. [Google Scholar] [CrossRef]
- Kawabata, K.; Mukai, R.; Ishisaka, A. Quercetin and Related Polyphenols: New Insights and Implications for Their Bioactivity and Bioavailability. Food Funct. 2015, 6, 1399–1417. [Google Scholar] [CrossRef]
- Salehi, B.; Machin, L.; Monzote, L.; Sharifi-Rad, J.; Ezzat, S.M.; Salem, M.A.; Merghany, R.M.; El Mahdy, N.M.; Kılıç, C.S.; Sytar, O.; et al. Therapeutic Potential of Quercetin: New Insights and Perspectives for Human Health. ACS Omega 2020, 5, 11849–11872. [Google Scholar] [CrossRef]
- Ciccone, L.; Vandooren, J.; Nencetti, S.; Orlandini, E. Natural Marine and Terrestrial Compounds as Modulators of Matrix Metalloproteinases-2 (MMP-2) and MMP-9 in Alzheimer’s Disease. Pharmaceuticals 2021, 14, 86. [Google Scholar] [CrossRef]
- Alizadeh, S.R.; Ebrahimzadeh, M.A. O-Glycoside Quercetin Derivatives: Biological Activities, Mechanisms of Action, and Structure–Activity Relationship for Drug Design, a Review. Phytother. Res. 2022, 36, 778–807. [Google Scholar] [CrossRef]
- Wang, D.D.; Li, Y.; Bhupathiraju, S.N.; Rosner, B.A.; Sun, Q.; Giovannucci, E.L.; Rimm, E.B.; Manson, J.E.; Willett, W.C.; Stampfer, M.J.; et al. Fruit and Vegetable Intake and Mortality. Circulation 2021, 143, 1642–1654. [Google Scholar] [CrossRef]
- van der Merwe, M. Gut Microbiome Changes Induced by a Diet Rich in Fruits and Vegetables. Int. J. Food Sci. Nutr. 2021, 72, 665–669. [Google Scholar] [CrossRef]
- López-González, L.; Becerra-Tomás, N.; Babio, N.; Martínez-González, M.Á.; Díaz-López, A.; Corella, D.; Goday, A.; Romaguera, D.; Vioque, J.; Alonso-Gómez, Á.M.; et al. Variety in Fruits and Vegetables, Diet Quality and Lifestyle in an Older Adult Mediterranean Population. Clin. Nutr. 2021, 40, 1510–1518. [Google Scholar] [CrossRef]
- Bieschke, J. Natural Compounds May Open New Routes to Treatment of Amyloid Diseases. Neurotherapeutics 2013, 10, 429–439. [Google Scholar] [CrossRef]
- Ferreira, N.; Saraiva, M.J.; Almeida, M.R. Natural Polyphenols Inhibit Different Steps of the Process of Transthyretin (TTR) Amyloid Fibril Formation. FEBS Lett. 2011, 585, 2424–2430. [Google Scholar] [CrossRef]
- Ortore, G.; Orlandini, E.; Braca, A.; Ciccone, L.; Rossello, A.; Martinelli, A.; Nencetti, S. Targeting Different Transthyretin Binding Sites with Unusual Natural Compounds. ChemMedChem 2016, 11, 1865–1874. [Google Scholar] [CrossRef]
- Pagano, K.; Tomaselli, S.; Molinari, H.; Ragona, L. Natural Compounds as Inhibitors of Aβ Peptide Aggregation: Chemical Requirements and Molecular Mechanisms. Front. Neurosci. 2020, 14, 1384. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Younis, K.; Philippe, J.; Aschner, M.; Khan, H. Strategic Approaches to Target the Enzymes using Natural Compounds for the Management of Alzheimer’s Disease: A Review. Available online: https://www.ingentaconnect.com/content/ben/cnsnddt/pre-prints/content-34382514 (accessed on 15 March 2022).
- Buxbaum, J.N.; Reixach, N. Transthyretin: The Servant of Many Masters. Cell. Mol. Life Sci. 2009, 66, 3095–3101. [Google Scholar] [CrossRef]
- Liz, M.A.; Mar, F.M.; Franquinho, F.; Sousa, M.M. Aboard Transthyretin: From Transport to Cleavage. IUBMB Life 2010, 62, 429–435. [Google Scholar] [CrossRef]
- Vieira, M.; Saraiva, M.J. Transthyretin: A Multifaceted Protein. BioMolecular Concepts 2014, 5, 45–54. [Google Scholar] [CrossRef]
- Garai, K.; Posey, A.E.; Li, X.; Buxbaum, J.N.; Pappu, R.V. Inhibition of Amyloid Beta Fibril Formation by Monomeric Human Transthyretin. Protein Sci. 2018, 27, 1252–1261. [Google Scholar] [CrossRef]
- Ciccone, L.; Fruchart-Gaillard, C.; Mourier, G.; Savko, M.; Nencetti, S.; Orlandini, E.; Servent, D.; Stura, E.A.; Shepard, W. Copper Mediated Amyloid- β Binding to Transthyretin. Sci. Rep. 2018, 8, 13744. [Google Scholar] [CrossRef]
- Ciccone, L.; Shi, C.; di Lorenzo, D.; Van Baelen, A.-C.; Tonali, N. The Positive Side of the Alzheimer’s Disease Amyloid Cross-Interactions: The Case of the Aβ 1-42 Peptide with Tau, TTR, CysC, and ApoA1. Molecules 2020, 25, 2439. [Google Scholar] [CrossRef]
- Ashrafian, H.; Zadeh, E.H.; Khan, R.H. Review on Alzheimer’s Disease: Inhibition of Amyloid Beta and Tau Tangle Formation. Int. J. Biol. Macromol. 2021, 167, 382–394. [Google Scholar] [CrossRef]
- Alemi, M.; Gaiteiro, C.; Ribeiro, C.A.; Santos, L.M.; Gomes, J.R.; Oliveira, S.M.; Couraud, P.-O.; Weksler, B.; Romero, I.; Saraiva, M.J.; et al. Transthyretin Participates in Beta-Amyloid Transport from the Brain to the Liver- Involvement of the Low-Density Lipoprotein Receptor-Related Protein 1? Sci. Rep. 2016, 6, 20164. [Google Scholar] [CrossRef]
- Fong, V.H.; Vieira, A. Cytotoxic Effects of Transthyretin Aggregates in an Epidermoid Cell Line. PAT 2017, 84, 218–222. [Google Scholar] [CrossRef]
- Teixeira, P.F.; Cerca, F.; Santos, S.D.; Saraiva, M.J. Endoplasmic Reticulum Stress Associated with Extracellular Aggregates: Evidence from Transthyretin Deposition in Familial Amyloid Polyneuropathy. J. Biol. Chem. 2006, 281, 21998–22003. [Google Scholar] [CrossRef]
- Wieczorek, E.; Ożyhar, A. Transthyretin: From Structural Stability to Osteoarticular and Cardiovascular Diseases. Cells 2021, 10, 1768. [Google Scholar] [CrossRef]
- Pereira, C.D.; Minamino, N.; Takao, T. Free Thiol of Transthyretin in Human Plasma Most Accessible to Modification/Oxidation. Anal. Chem. 2015, 87, 10785–10791. [Google Scholar] [CrossRef]
- Hanyu, N.; Shimizu, T.; Yamauchi, K.; Okumura, N.; Hidaka, H. Characterization of Cysteine and Homocysteine Bound to Human Serum Transthyretin. Clin. Chim. Acta 2009, 403, 70–75. [Google Scholar] [CrossRef]
- Sharma, M.; Khan, S.; Rahman, S.; Singh, L.R. The Extracellular Protein, Transthyretin Is an Oxidative Stress Biomarker. Front. Physiol. 2019, 10, 5. [Google Scholar] [CrossRef]
- Obici, L.; Mussinelli, R. Current and Emerging Therapies for Hereditary Transthyretin Amyloidosis: Strides Towards a Brighter Future. Neurotherapeutics 2021, 18, 2286–2302. [Google Scholar] [CrossRef]
- Park, G.Y.; Jamerlan, A.; Shim, K.H.; An, S.S.A. Diagnostic and Treatment Approaches Involving Transthyretin in Amyloidogenic Diseases. Int. J. Mol. Sci. 2019, 20, 2982. [Google Scholar] [CrossRef]
- Elliott, P.; Drachman, B.M.; Gottlieb, S.S.; Hoffman, J.E.; Hummel, S.L.; Lenihan, D.J.; Ebede, B.; Gundapaneni, B.; Li, B.; Sultan, M.B.; et al. Long-Term Survival With Tafamidis in Patients With Transthyretin Amyloid Cardiomyopathy. Circ. Heart Fail. 2022, 15, e008193. [Google Scholar] [CrossRef]
- Nelson, L.T.; Paxman, R.J.; Xu, J.; Webb, B.; Powers, E.T.; Kelly, J.W. Blinded Potency Comparison of Transthyretin Kinetic Stabilisers by Subunit Exchange in Human Plasma. Amyloid 2021, 28, 24–29. [Google Scholar] [CrossRef]
- Lill, M.A.; Danielson, M.L. Computer-Aided Drug Design Platform Using PyMOL. J. Comput. Aided Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef]
- Ciccone, L.; Tonali, N.; Nencetti, S.; Orlandini, E. Natural Compounds as Inhibitors of Transthyretin Amyloidosis and Neuroprotective Agents: Analysis of Structural Data for Future Drug Design. J. Enzym. Inhib. Med. Chem. 2020, 35, 1145–1162. [Google Scholar] [CrossRef]
- Guo, X.; Liu, Z.; Zheng, Y.; Li, Y.; Li, L.; Liu, H.; Chen, Z.; Wu, L. Review on the Structures and Activities of Transthyretin Amyloidogenesis Inhibitors. DDDT 2020, 14, 1057–1081. [Google Scholar] [CrossRef]
- Trivella, D.B.B.; dos Reis, C.V.; Lima, L.M.T.R.; Foguel, D.; Polikarpov, I. Flavonoid Interactions with Human Transthyretin: Combined Structural and Thermodynamic Analysis. J. Struct. Biol. 2012, 180, 143–153. [Google Scholar] [CrossRef]
- Cianci, M.; Folli, C.; Zonta, F.; Florio, P.; Berni, R.; Zanotti, G. Structural Evidence for Asymmetric Ligand Binding to Transthyretin. Acta Cryst. D 2015, 71, 1582–1592. [Google Scholar] [CrossRef]
- Nilsson, L.; Larsson, A.; Begum, A.; Iakovleva, I.; Carlsson, M.; Brännström, K.; Sauer-Eriksson, A.E.; Olofsson, A. Modifications of the 7-Hydroxyl Group of the Transthyretin Ligand Luteolin Provide Mechanistic Insights into Its Binding Properties and High Plasma Specificity. PLoS ONE 2016, 11, e0153112. [Google Scholar] [CrossRef]
- Genistein, a Natural Product from Soy, Is a Potent Inhibitor of Transthyretin Amyloidosis. Available online: https://www.pnas.org/doi/10.1073/pnas.0501609102 (accessed on 7 March 2022).
- Azeem, M.; Hanif, M.; Mahmood, K.; Ameer, N.; Chughtai, F.R.S.; Abid, U. An Insight into Anticancer, Antioxidant, Antimicrobial, Antidiabetic and Anti-Inflammatory Effects of Quercetin: A Review. Polym. Bull. 2022. [Google Scholar] [CrossRef]
- Tian, C.; Liu, X.; Chang, Y.; Wang, R.; Lv, T.; Cui, C.; Liu, M. Investigation of the Anti-Inflammatory and Antioxidant Activities of Luteolin, Kaempferol, Apigenin and Quercetin. South Afr. J. Bot. 2021, 137, 257–264. [Google Scholar] [CrossRef]
- Ulusoy, H.G.; Sanlier, N. A Minireview of Quercetin: From Its Metabolism to Possible Mechanisms of Its Biological Activities. Crit. Rev. Food Sci. Nutr. 2020, 60, 3290–3303. [Google Scholar] [CrossRef]
- Pawlowska, A.M.; Camangi, F.; Braca, A. Quali-Quantitative Analysis of Flavonoids of Cornus mas L. (Cornaceae) Fruits. Food Chem. 2010, 119, 1257–1261. [Google Scholar] [CrossRef]
- Bruzual De Abreu, M.; Temraz, A.; Malafronte, N.; Gonzalez-Mujica, F.; Duque, S.; Braca, A. Phenolic Derivatives from Ruprechtia polystachya and their Inhibitory Activities on the Glucose-6-Phosphatase System. Chem. Biodivers. 2011, 8, 2126–2134. [Google Scholar] [CrossRef]
- Iannuzzi, A.M.; Giacomelli, C.; Leo, M.D.; Russo, L.; Camangi, F.; Tommasi, N.D.; Braca, A.; Martini, C.; Trincavelli, M.L. Cornus sanguinea Fruits: A Source of Antioxidant and Antisenescence Compounds Acting on Aged Human Dermal and Gingival Fibroblasts. Planta Med. 2021, 87, 879–891. [Google Scholar] [CrossRef]
- Razavi, S.M.; Zahri, S.; Zarrini, G.; Nazemiyeh, H.; Mohammadi, S. Biological Activity of Quercetin-3-O-Glucoside, a Known Plant Flavonoid. Russ. J. Bioorg. Chem. 2009, 35, 376–378. [Google Scholar] [CrossRef]
- Zhang, X.; Thuong, P.T.; Jin, W.; Su, N.D.; Sok, D.E.; Bae, K.; Kang, S.S. Antioxidant Activity of Anthraquinones and Flavonoids from Flower of Reynoutria sachalinensis. Arch. Pharm. Res. 2005, 28, 22–27. [Google Scholar] [CrossRef]
- Csepregi, K.; Neugart, S.; Schreiner, M.; Hideg, É. Comparative Evaluation of Total Antioxidant Capacities of Plant Polyphenols. Molecules 2016, 21, 208. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Rossello, A.; Stura, E.A.; Orlandini, E. Synthesis and Structural Analysis of Halogen Substituted Fibril Formation Inhibitors of Human Transthyretin (TTR). J. Enzym. Inhib. Med. Chem. 2016, 31, 40–51. [Google Scholar] [CrossRef][Green Version]
- Oza, V.B.; Smith, C.; Raman, P.; Koepf, E.K.; Lashuel, H.A.; Petrassi, H.M.; Chiang, K.P.; Powers, E.T.; Sachettinni, J.; Kelly, J.W. Synthesis, Structure, and Activity of Diclofenac Analogues as Transthyretin Amyloid Fibril Formation Inhibitors. J. Med. Chem. 2002, 45, 321–332. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Tonali, N.; Fruchart-Gaillard, C.; Shepard, W.; Nuti, E.; Camodeca, C.; Rossello, A.; Orlandini, E. Monoaryl Derivatives as Transthyretin Fibril Formation Inhibitors: Design, Synthesis, Biological Evaluation and Structural Analysis. Bioorganic Med. Chem. 2020, 28, 115673. [Google Scholar] [CrossRef]
- Yokoyama, T.; Kosaka, Y.; Mizuguchi, M. Inhibitory Activities of Propolis and Its Promising Component, Caffeic Acid Phenethyl Ester, against Amyloidogenesis of Human Transthyretin. J. Med. Chem. 2014, 57, 8928–8935. [Google Scholar] [CrossRef]
- Ciccone, L.; Nencetti, S.; Rossello, A.; Tepshi, L.; Stura, E.A.; Orlandini, E. X-Ray Crystal Structure and Activity of Fluorenyl-Based Compounds as Transthyretin Fibrillogenesis Inhibitors. J. Enzym. Inhib. Med. Chem. 2016, 31, 824–833. [Google Scholar] [CrossRef]
- Ciccone, L.; Vera, L.; Tepshi, L.; Rosalia, L.; Rossello, A.; Stura, E.A. Multicomponent Mixtures for Cryoprotection and Ligand Solubilization. Biotechnol. Rep. 2015, 7, 120–127. [Google Scholar] [CrossRef]
- Ciccone, L.; Tepshi, L.; Nencetti, S.; Stura, E.A. Transthyretin Complexes with Curcumin and Bromo-Estradiol: Evaluation of Solubilizing Multicomponent Mixtures. New Biotechnol. 2015, 32, 54–64. [Google Scholar] [CrossRef]
- Polsinelli, I.; Savko, M.; Rouanet-Mehouas, C.; Ciccone, L.; Nencetti, S.; Orlandini, E.; Stura, E.A.; Shepard, W. Comparison of Helical Scan and Standard Rotation Methods in Single-Crystal X-Ray Data Collection Strategies. J. Synchrotron Radiat. 2017, 24, 42–52. [Google Scholar] [CrossRef]
- Polsinelli, I.; Nencetti, S.; Shepard, W.; Ciccone, L.; Orlandini, E.; Stura, E.A. A New Crystal Form of Human Transthyretin Obtained with a Curcumin Derived Ligand. J. Struct. Biol. 2016, 194, 8–17. [Google Scholar] [CrossRef]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W. PHENIX: A Comprehensive Python-Based System for Macromolecular Structure Solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and Development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Moon, J.-K.; Shibamoto, T. Antioxidant Assays for Plant and Food Components. J. Agric. Food Chem. 2009, 57, 1655–1666. [Google Scholar] [CrossRef]
- Ciccone, L.; Petrarolo, G.; Barsuglia, F.; Fruchart-Gaillard, C.; Cassar Lajeunesse, E.; Adewumi, A.T.; Soliman, M.E.S.; La Motta, C.; Orlandini, E.; Nencetti, S. Nature-Inspired O-Benzyl Oxime-Based Derivatives as New Dual-Acting Agents Targeting Aldose Reductase and Oxidative Stress. Biomolecules 2022, 12, 448. [Google Scholar] [CrossRef]
- Montaño, M.; Cocco, E.; Guignard, C.; Marsh, G.; Hoffmann, L.; Bergman, Å.; Gutleb, A.C.; Murk, A.J. New Approaches to Assess the Transthyretin Binding Capacity of Bioactivated Thyroid Hormone Disruptors. Toxicol. Sci. 2012, 130, 94–105. [Google Scholar] [CrossRef]
- Ciccone, L.; Tonali, N.; Shepard, W.; Nencetti, S.; Orlandini, E. Physiological Metals Can Induce Conformational Changes in Transthyretin Structure: Neuroprotection or Misfolding Induction? Crystals 2021, 11, 354. [Google Scholar] [CrossRef]
- The GIMP Development Team GIMP; 2019.
- Iacopini, P.; Baldi, M.; Storchi, P.; Sebastiani, L. Catechin, Epicatechin, Quercetin, Rutin and Resveratrol in Red Grape: Content, in Vitro Antioxidant Activity and Interactions. J. Food Compos. Anal. 2008, 21, 589–598. [Google Scholar] [CrossRef]
- Dhouafli, Z.; Cuanalo-Contreras, K.; Hayouni, E.A.; Mays, C.E.; Soto, C.; Moreno-Gonzalez, I. Inhibition of Protein Misfolding and Aggregation by Natural Phenolic Compounds. Cell. Mol. Life Sci. 2018, 75, 3521–3538. [Google Scholar] [CrossRef]
- Amato, A.; Terzo, S.; Mulè, F. Natural Compounds as Beneficial Antioxidant Agents in Neurodegenerative Disorders: A Focus on Alzheimer’s Disease. Antioxidants 2019, 8, 608. [Google Scholar] [CrossRef]

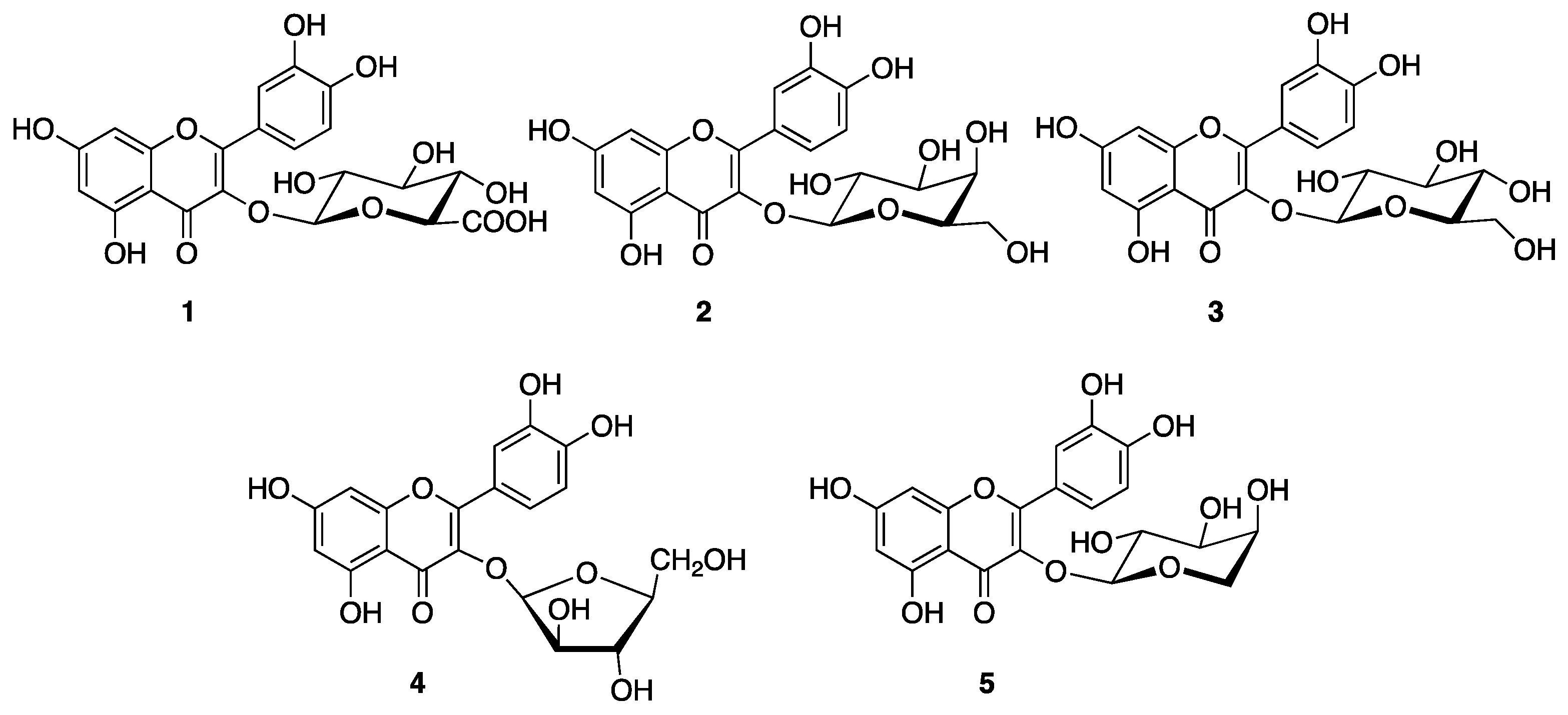


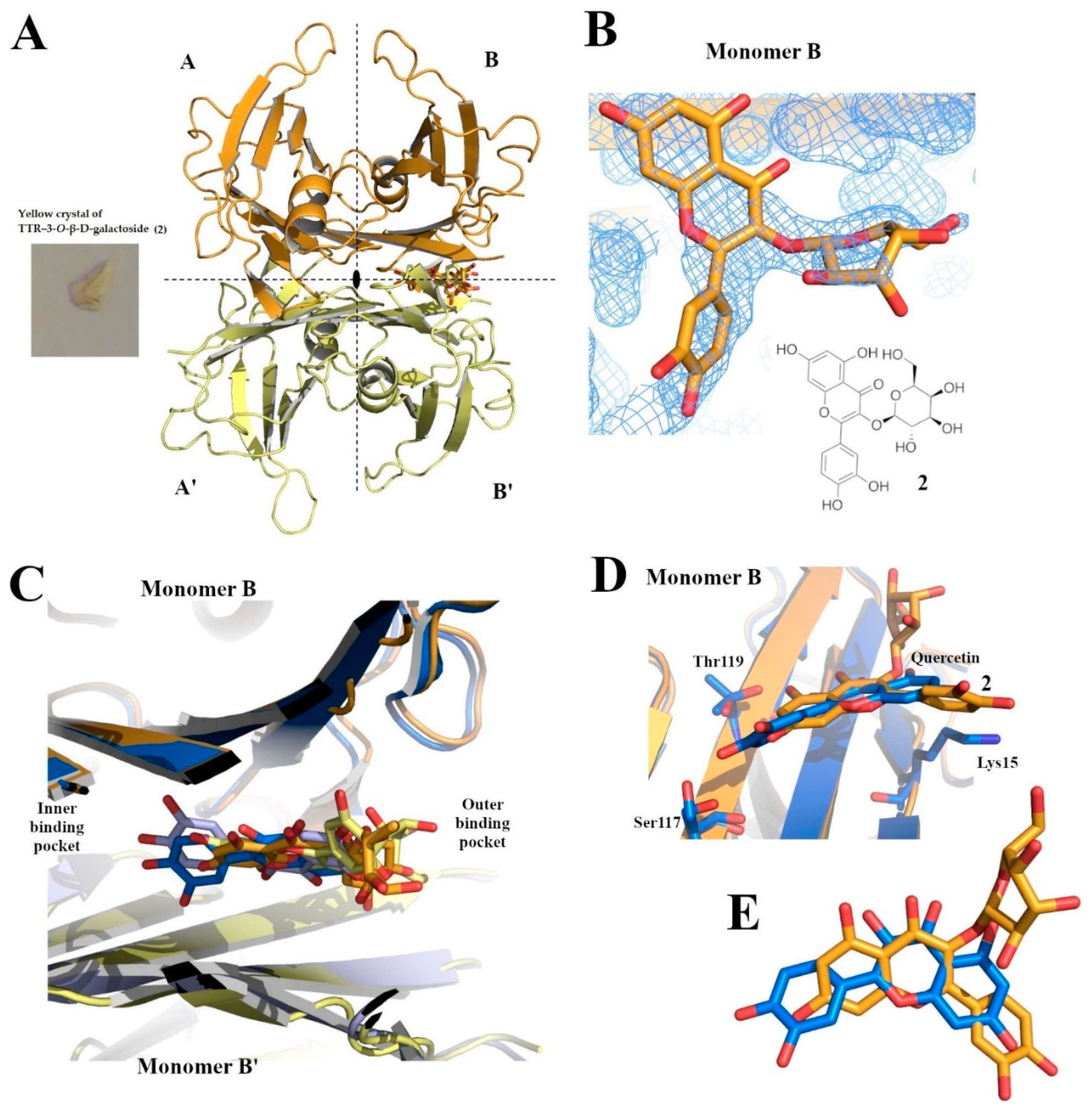
| Structure | TTR–QUE 3-O-β-d-Galactopyranoside Crystal Complex |
|---|---|
| PDB code | 7Z60 |
| Crystallization | 80% (30% PEG 4000, 0.2 M Imidazole Malate pH 6.0) and 15% (12% MPEG 5000, 0.1 Sodium Acetate pH 5.5) |
| Data Collection | |
| Source | SOLEIL PROXIMA2A |
| Wavelength (A°) | 0.980 |
| Space group | P21212 |
| Unit-cell (A°) | a = 43.230, b = 85.600, c = 64.430; α = β = γ = 90° |
| Molec./asym. | 2 |
| Resolution (A°) | 42.80–1.40/1.43–1.40 |
| CC1/2 (%) | 99.9/39.6 |
| 〈I/σ(I)〉 | 18.10/0.93 |
| No. of reflections | 569968 |
| No. of unique reflections | 45603 |
| Completeness (%) | 99.7/93.0 |
| Multiplicity | 13.3/13.3 |
| Refinement | Refmac |
| Resolution (A°) | 42.80–1.40/1.43–1.40 |
| No. of reflections | 45603 |
| R-work | 18.23 |
| R-free | 21.9 |
| RMSD Bond lengths (A°) | 0.01 |
| RMSD Bond angles (°) | 1.75 |
| Ramachandran favored | 96.60% |
| Ramachandran outliers | 0 |
| Compounds | Radical Scavenging Activity a RC50 (µM) | R2 |
|---|---|---|
| Quercetin | 5.4 ± 1.0 [66] | 0.989 |
| 1 | 4.78 ± 1.07 | 0.997 |
| 2 | 24.96 ± 1.09 | 0.988 |
| 3 | 4.47 ± 1.09 | 0.978 |
| 4 | 29.40 ± 1.08 | 0.987 |
| 5 | 5.62 ± 1.08 | 0.995 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciccone, L.; Tonali, N.; Fruchart-Gaillard, C.; Barlettani, L.; Rossello, A.; Braca, A.; Orlandini, E.; Nencetti, S. Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization. Crystals 2022, 12, 638. https://doi.org/10.3390/cryst12050638
Ciccone L, Tonali N, Fruchart-Gaillard C, Barlettani L, Rossello A, Braca A, Orlandini E, Nencetti S. Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization. Crystals. 2022; 12(5):638. https://doi.org/10.3390/cryst12050638
Chicago/Turabian StyleCiccone, Lidia, Nicolò Tonali, Carole Fruchart-Gaillard, Lucia Barlettani, Armando Rossello, Alessandra Braca, Elisabetta Orlandini, and Susanna Nencetti. 2022. "Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization" Crystals 12, no. 5: 638. https://doi.org/10.3390/cryst12050638
APA StyleCiccone, L., Tonali, N., Fruchart-Gaillard, C., Barlettani, L., Rossello, A., Braca, A., Orlandini, E., & Nencetti, S. (2022). Antioxidant Quercetin 3-O-Glycosylated Plant Flavonols Contribute to Transthyretin Stabilization. Crystals, 12(5), 638. https://doi.org/10.3390/cryst12050638










