Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method
Abstract
1. Introduction
2. Results
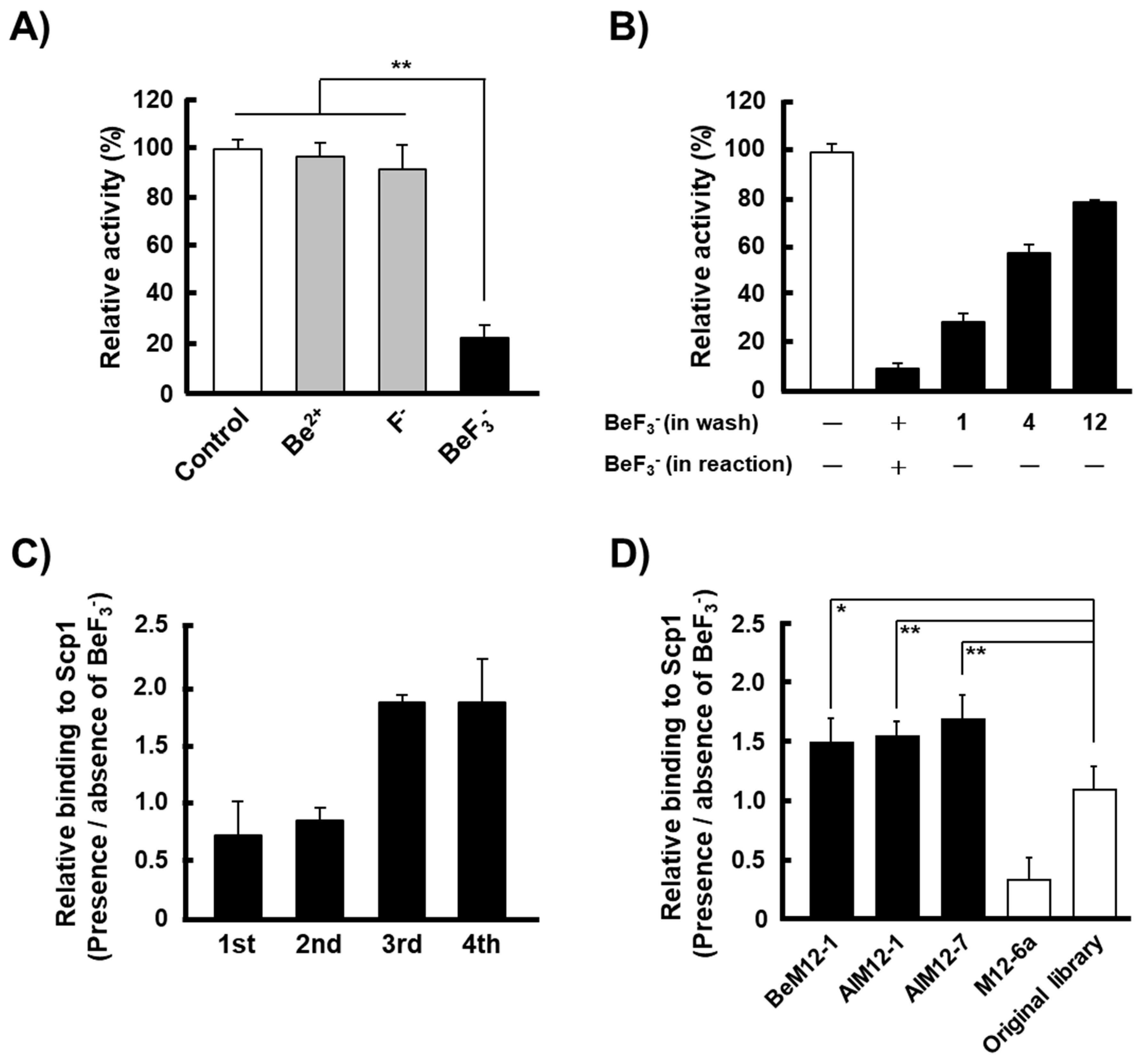
2.1. Specific Inhibition of rScp1 by BeF3−
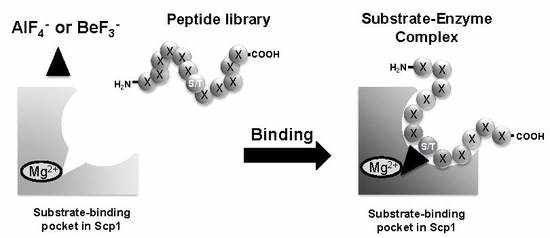
2.2. Screening of Scp1 Substrate Candidates Using the PMPD Method with BeF3−
2.3. Ion-Specific Binding of Isolated Peptide-Displayed Phages to rScp1
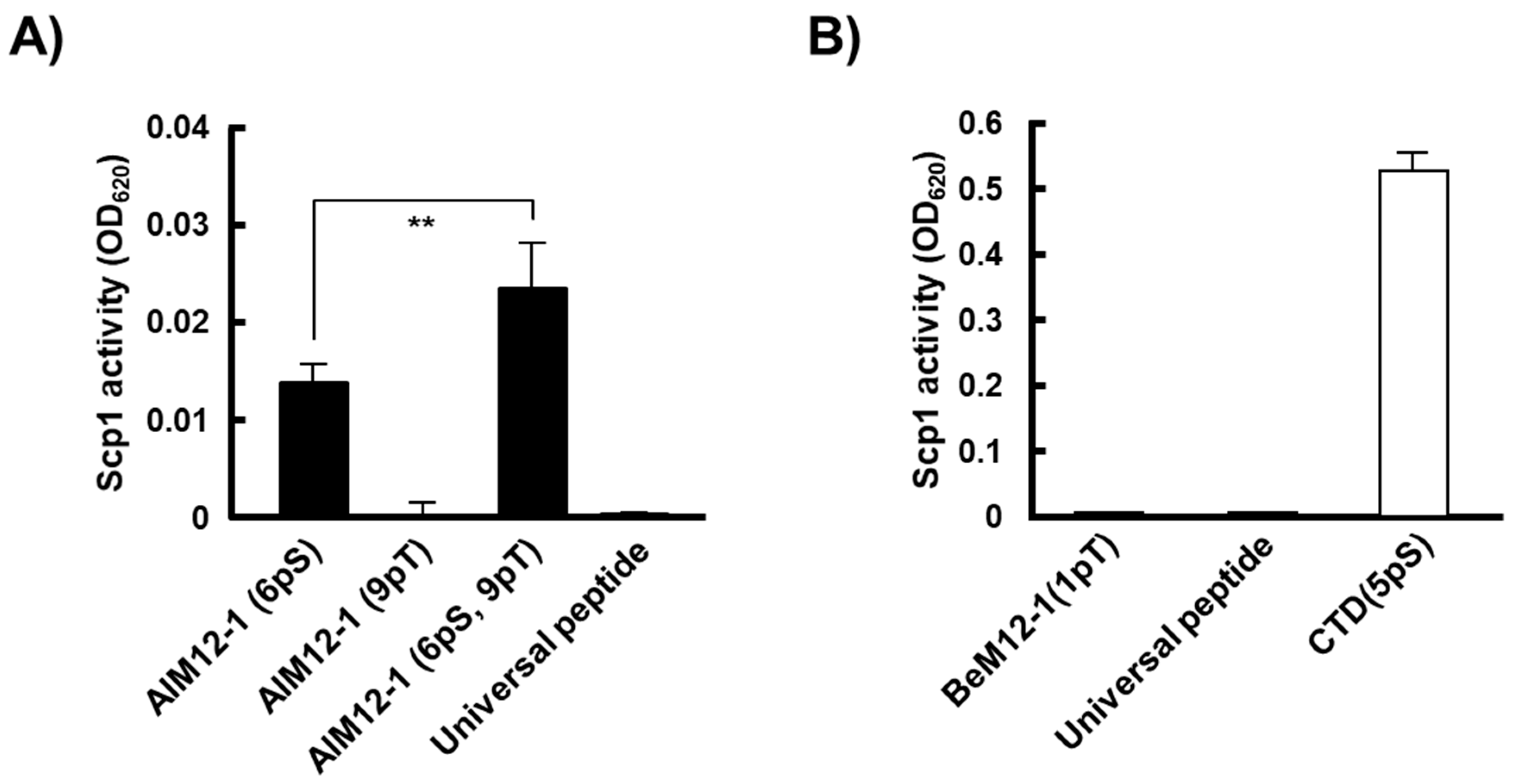
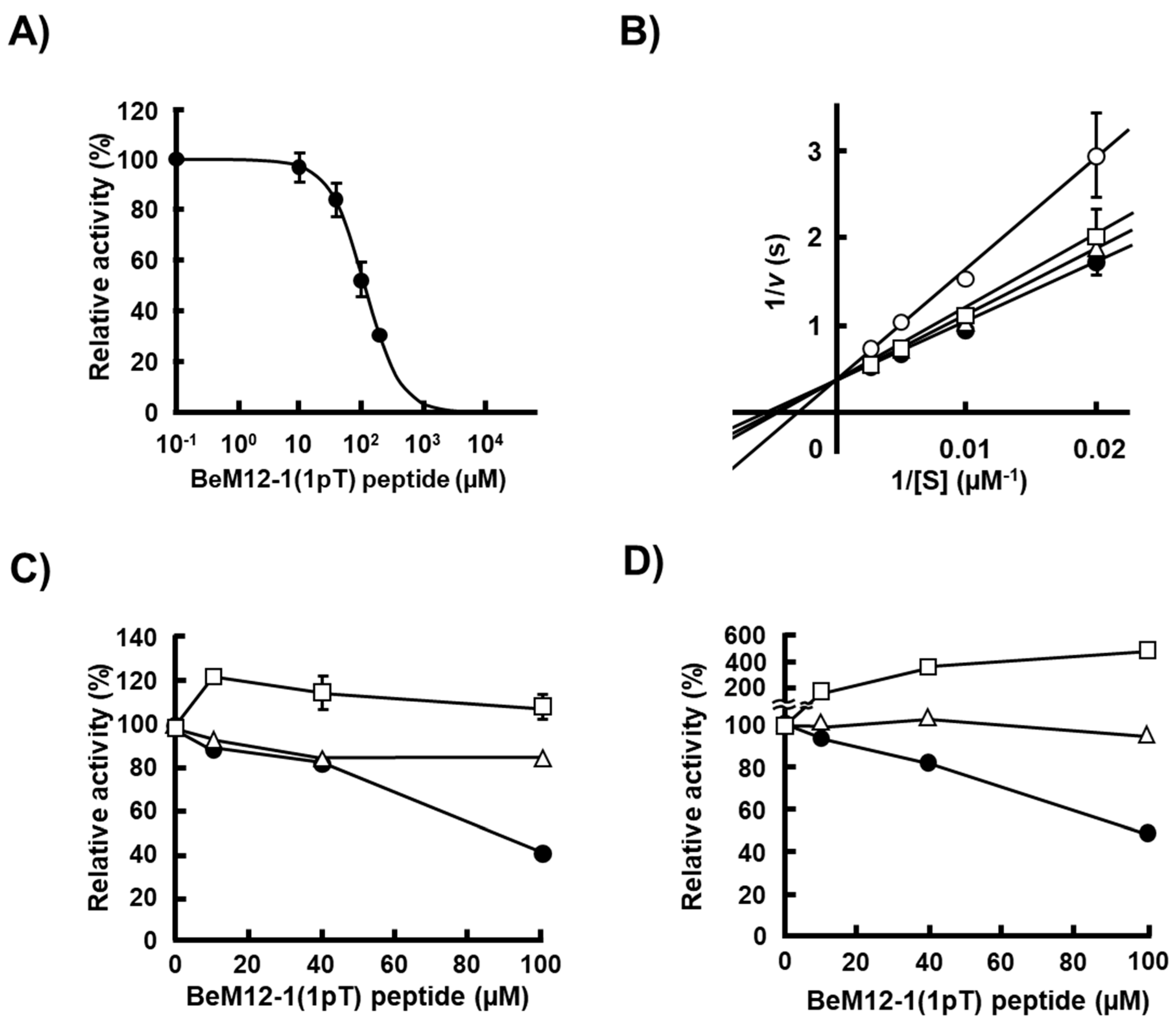
2.4. Phosphatase Activity of rScp1 against Phosphorylated Peptides Identified by the PMPD Method
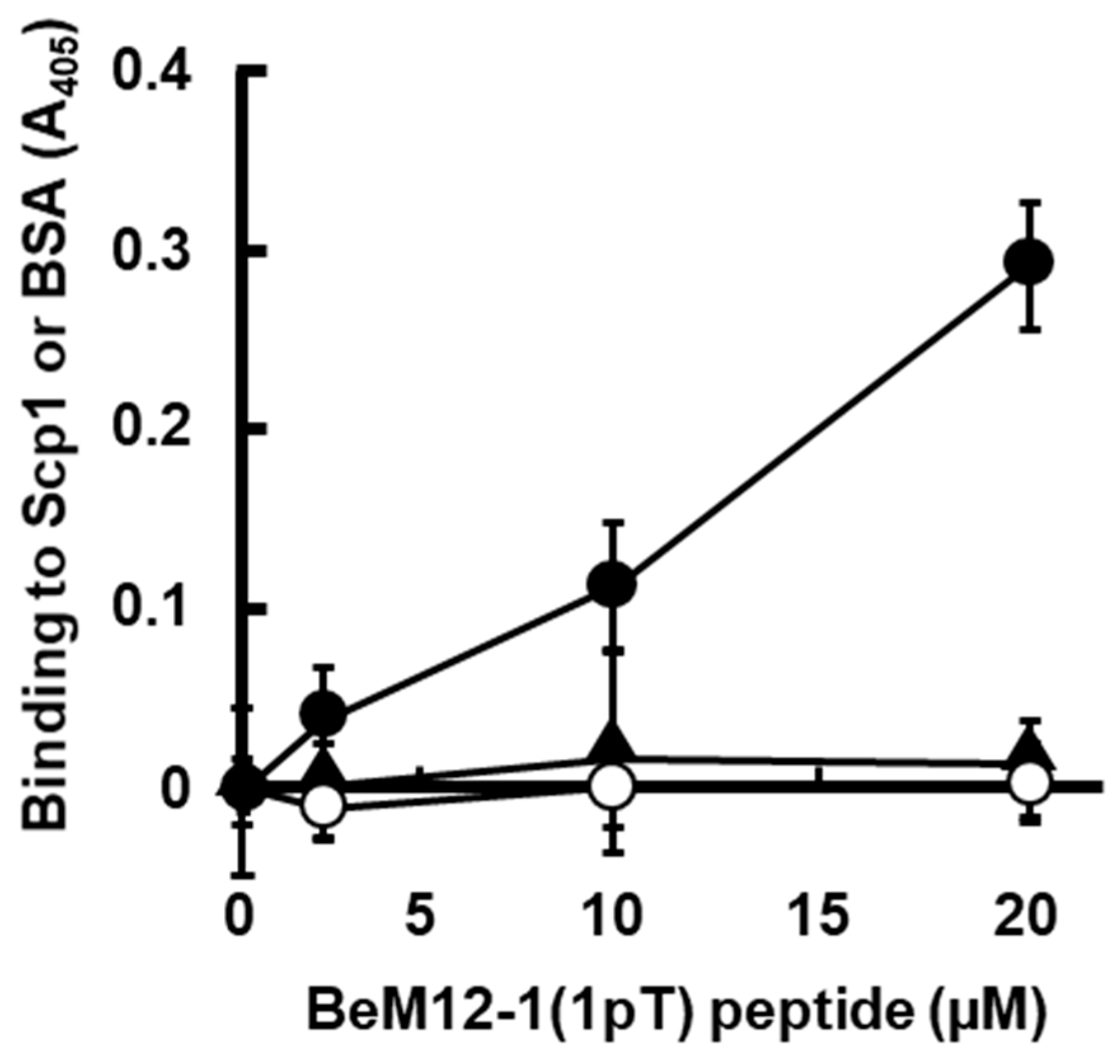
2.5. Inhibition of BeM12-1(1pT) Binding to rScp1 by BeF3−
2.6. Inhibitory Activity of BeM12-1(1pT) against rScp1
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Synthesis of Scp1 Substrate Candidate Peptides
4.3. In Vitro Phosphatase Assay of rScp1
4.4. Inhibition Assay of rScp1 by Inhibitors including BeF3−
4.5. BeF3− Inhibition and Reactivity Assay on ELISA Plate
4.6. Screening of Phage Displayed Combinatorial Peptide Library against rScp1 with Mg2+ and BeF3−
4.7. Binding Analysis of Isolated Phage Clones to rScp1 with Ions in ELISA
4.8. Binding Analysis of Biotinylated BeM12-1(1pT) Peptide to rScp1
5. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Olsen, J.V.; Blagoev, B.; Gnad, F.; Macek, B.; Kumar, C.; Mortensen, P.; Mann, M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006, 127, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Allen, K.N.; Dunaway-Mariano, D. Phosphoryl group transfer: Evolution of a catalytic scaffold. Trends Biochem. Sci. 2004, 29, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kim, Y.; Genoud, N.; Gao, J.; Kelly, J.W.; Pfaff, S.L.; Gill, G.N.; Dixon, J.E.; Noel, J.P. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol. Cell 2006, 24, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.O.; Campagnoni, C.W.; Kampf, K.; Feng, J.M.; Handley, V.W.; Schonmann, V.; Bongarzone, E.R.; Reyes, S.; Campagnoni, A.T. Identification of a protein that interacts with the golli-myelin basic protein and with nuclear LIM interactor in the nervous system. J. Neurosci. Res. 2004, 75, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Wright, J.; Dou, S.; Olsen, P.; Teeter, L.; Adams, G.; Graviss, E. Ethnic divergence and linkage disequilibrium of novel SNPs in the human NLI-IF gene: Evidence of human origin and lack of association with tuberculosis susceptibility. J. Hum. Genet. 2002, 47, 140–145. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, W.H.; Su, Y.H.; Hsu, W.H.; Wang, C.C.; Arbiser, J.L.; Yang, M.H. Imipramine blue halts head and neck cancer invasion through promoting F-box and leucine-rich repeat protein 14-mediated Twist1 degradation. Oncogene 2016, 35, 2287–2298. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Fu, J.; Shen, T.; Lin, X.; Liao, L.; Feng, X.H.; Xu, J. The Small C-terminal Domain Phosphatase 1 Inhibits Cancer Cell Migration and Invasion by Dephosphorylating Ser(P)68-Twist1 to Accelerate Twist1 Protein Degradation. J. Biol. Chem. 2016, 291, 11518–11528. [Google Scholar] [CrossRef]
- Liao, P.; Wang, W.; Li, Y.; Wang, R.; Jin, J.; Pang, W.; Chen, Y.; Shen, M.; Wang, X.; Jiang, D.; et al. Palmitoylated SCP1 is targeted to the plasma membrane and negatively regulates angiogenesis. Elife 2017, 6, e22058. [Google Scholar] [CrossRef]
- Lin, Y.C.; Lu, L.T.; Chen, H.Y.; Duan, X.; Lin, X.; Feng, X.H.; Tang, M.J.; Chen, R.H. SCP phosphatases suppress renal cell carcinoma by stabilizing PML and inhibiting mTOR/HIF signaling. Cancer Res. 2014, 74, 6935–6946. [Google Scholar] [CrossRef]
- Knockaert, M.; Sapkota, G.; Alarcón, C.; Massagué, J.; Brivanlou, A.H. Unique players in the BMP pathway: Small C-terminal domain phosphatases dephosphorylate Smad1 to attenuate BMP signaling. Proc. Natl. Acad. Sci. USA 2006, 103, 11940–11945. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, G.; Knockaert, M.; Alarcón, C.; Montalvo, E.; Brivanlou, A.H.; Massagué, J. Dephosphorylation of the linker regions of Smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J. Biol. Chem. 2006, 281, 40412–40419. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Evers, B.M.; Zhou, B.P. Small C-terminal domain phosphatase enhances snail activity through dephosphorylation. J. Biol. Chem. 2009, 284, 640–648. [Google Scholar] [CrossRef] [PubMed]
- Ballas, N.; Grunseich, C.; Lu, D.D.; Speh, J.C.; Mandel, G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 2005, 121, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Yeo, M.; Lee, S.K.; Lee, B.; Ruiz, E.C.; Pfaff, S.L.; Gill, G.N. Small CTD phosphatases function in silencing neuronal gene expression. Science 2005, 307, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Burkholder, N.T.; Mayfield, J.E.; Yu, X.; Irani, S.; Arce, D.K.; Jiang, F.; Matthews, W.L.; Xue, Y.; Zhang, Y.J. Phosphatase activity of small C-terminal domain phosphatase 1 (SCP1) controls the stability of the key neuronal regulator RE1-silencing transcription factor (REST). J. Biol. Chem. 2018, 293, 16851–16861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Cho, E.J.; Burstein, G.; Siegel, D.; Zhang, Y. Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem. Biol. 2011, 6, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Park, H.; Lee, H.S.; Ku, B.; Lee, S.R.; Kim, S.J. Two-track virtual screening approach to identify both competitive and allosteric inhibitors of human small C-terminal domain phosphatase 1. J. Comput. Aided. Mol. Des. 2017, 31, 743–753. [Google Scholar] [CrossRef]
- Nozaka, A.; Nishiwaki, A.; Nagashima, Y.; Endo, S.; Kuroki, M.; Nakajima, M.; Narukawa, M.; Kamisuki, S.; Arazoe, T.; Taguchi, H.; et al. Chloramphenicol inhibits eukaryotic Ser/Thr phosphatase and infection-specific cell differentiation in the rice blast fungus. Sci. Rep. 2019, 9, 9283. [Google Scholar] [CrossRef]
- Williams, M.P.; Pounder, R.E. Review article: The pharmacology of rabeprazole. Aliment Pharmacol. Ther. 1999, 13, 3–10. [Google Scholar] [CrossRef]
- Barford, D. Molecular mechanisms of the protein serine/threonine phosphatases. Trends Biochem. Sci. 1996, 21, 407–412. [Google Scholar] [CrossRef]
- Wu, D.; De Wever, V.; Derua, R.; Winkler, C.; Beullens, M.; Van Eynde, A.; Bollen, M. A substrate-trapping strategy for protein phosphatase PP1 holoenzymes using hypoactive subunit fusions. J. Biol. Chem. 2018, 293, 15152–15162. [Google Scholar] [CrossRef] [PubMed]
- Otsubo, K.; Yoneda, T.; Kaneko, A.; Yagi, S.; Furukawa, K.; Chuman, Y. Development of a Substrate Identification Method for Human Scp1 Phosphatase Using Phosphorylation Mimic Phage Display. Protein Pept. Lett. 2018, 25, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Lassila, J.K.; Zalatan, J.G.; Herschlag, D. Biological phosphoryl-transfer reactions: Understanding mechanism and catalysis. Annu. Rev. Biochem. 2011, 80, 669–702. [Google Scholar] [CrossRef] [PubMed]
- Cleland, W.W.; Hengge, A.C. Enzymatic mechanisms of phosphate and sulfate transfer. Chem. Rev. 2006, 106, 3252–3278. [Google Scholar] [CrossRef] [PubMed]
- Kamenski, T.; Heilmeier, S.; Meinhart, A.; Cramer, P. Structure and mechanism of RNA polymerase II CTD phosphatases. Mol. Cell 2004, 15, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Shuman, S.; Lima, C.D. The structure of Fcp1, an essential RNA polymerase II CTD phosphatase. Mol. Cell 2008, 32, 478–490. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Wang, W.; Kim, R.; Yokota, H.; Damo, S.; Kim, S.H.; Wemmer, D.; Kustu, S.; Yan, D. BeF3- acts as a phosphate analog in proteins phosphorylated on aspartate: Structure of a BeF3− complex with phosphoserine phosphatase. Proc. Natl. Acad. Sci. USA 2001, 98, 8525–8530. [Google Scholar] [CrossRef] [PubMed]
- Yan, D.; Cho, H.S.; Hastings, C.A.; Igo, M.M.; Lee, S.Y.; Pelton, J.G.; Stewart, V.; Wemmer, D.E.; Kustu, S. Beryllofluoride mimics phosphorylation of NtrC and other bacterial response regulators. Proc. Natl. Acad. Sci. USA 1999, 96, 14789–14794. [Google Scholar] [CrossRef] [PubMed]
- Eick, D.; Geyer, M. The RNA polymerase II carboxy-terminal domain (CTD) code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Jasnovidova, O.; Stefl, R. The CTD code of RNA polymerase II: A structural view. Wiley Interdiscip. Rev. RNA 2013, 4, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Prevost, M.S.; Pinotsis, N.; Dumoux, M.; Hayward, R.D.; Waksman, G. The Legionella effector WipB is a translocated Ser/Thr phosphatase that targets the host lysosomal nutrient sensing machinery. Sci. Rep. 2017, 7, 9450. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Dayaram, T.; Gilmartin, A.G.; Ganji, G.; Pemmasani, S.K.; Van Der Key, H.; Shohet, J.M.; Donehower, L.A.; Kumar, R. WIP1 phosphatase as a potential therapeutic target in neuroblastoma. PLoS ONE 2015, 10, e0115635. [Google Scholar] [CrossRef] [PubMed]
- Nesti, E.; Corson, G.M.; McCleskey, M.; Oyer, J.A.; Mandel, G. C-terminal domain small phosphatase 1 and MAP kinase reciprocally control REST stability and neuronal differentiation. Proc. Natl. Acad. Sci. USA 2014, 111, 3929–3936. [Google Scholar] [CrossRef] [PubMed]
- Wrighton, K.H.; Willis, D.; Long, J.; Liu, F.; Lin, X.; Feng, X.H. Small C-terminal domain phosphatases dephosphorylate the regulatory linker regions of Smad2 and Smad3 to enhance transforming growth factor-beta signaling. J. Biol. Chem. 2006, 281, 38365–38375. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, P.; Shen, M.; Chen, T.; Chen, Y.; Li, Y.; Lin, X.; Ge, X.; Wang, P. SCP1 regulates c-Myc stability and functions through dephosphorylating c-Myc Ser62. Oncogene 2016, 35, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Elliott, A.M.; Simard, L.R.; Coghlan, G.; Chudley, A.E.; Chodirker, B.N.; Greenberg, C.R.; Burch, T.; Ly, V.; Hatch, G.M.; Zelinski, T. A novel mutation in KIAA0196: Identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J. Med. Genet. 2013, 50, 819–822. [Google Scholar] [CrossRef] [PubMed]
- Van Beuningen, S.F.B.; Will, L.; Harterink, M.; Chazeau, A.; van Battum, E.Y.; Frias, C.P.; Franker, M.A.M.; Katrukha, E.A.; Stucchi, R.; Vocking, K.; et al. TRIM46 Controls Neuronal Polarity and Axon Specification by Driving the Formation of Parallel Microtubule Arrays. Neuron 2015, 88, 1208–1226. [Google Scholar] [CrossRef] [PubMed]
- Debnath, S.; Kosek, D.; Tagad, H.D.; Durell, S.R.; Appella, D.H.; Acevedo, R.; Grishaev, A.; Dyda, F.; Appella, E.; Mazur, S.J. A trapped human PPM1A-phosphopeptide complex reveals structural features critical for regulation of PPM protein phosphatase activity. J. Biol. Chem. 2018, 293, 7993–8008. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Gong, Z.; Pan, C.; Xie, D.D.; Tang, J.Y.; Cui, M.; Xu, Y.F.; Yao, W.; Pang, Q.; Xu, Z.G.; et al. Metal-dependent protein phosphatase 1A functions as an extracellular signal-regulated kinase phosphatase. FEBS J. 2013, 280, 2700–2711. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.; Kaldis, P.; Solomon, M.J. Dephosphorylation of human cyclin-dependent kinases by protein phosphatase type 2C alpha and beta 2 isoforms. J. Biol. Chem. 2000, 275, 34744–34749. [Google Scholar] [CrossRef] [PubMed]
- Nash, P.; Tang, X.; Orlicky, S.; Chen, Q.; Gertler, F.B.; Mendenhall, M.D.; Sicheri, F.; Pawson, T.; Tyers, M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 2001, 414, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, N.; Xu, L.; Pasek, D.A.; Evans, K.E.; Chen, S.R.; Meissner, G. Calmodulin regulation and identification of calmodulin binding region of type-3 ryanodine receptor calcium release channel. Biochemistry 2005, 44, 15074–15081. [Google Scholar] [CrossRef] [PubMed]
- Hunter, T.; Sefton, B.M. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc. Natl. Acad. Sci. USA 1980, 77, 1311–1315. [Google Scholar] [CrossRef] [PubMed]
- Ubersax, J.A.; Ferrell, J.E., Jr. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Tong, P.; Zhao, W.; Li, Y.; Zhang, L.; Xia, Y.; Yu, Y. The REST gene signature predicts drug sensitivity in neuroblastoma cell lines and is significantly associated with neuroblastoma tumor stage. Int. J. Mol. Sci. 2014, 15, 11220–11233. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Bai, L.M.; Yang, Y.P.; Luo, W.F.; Hu, W.D.; Chen, J.P.; Mao, C.J.; Liu, C.F. Effects of IL-6 secreted from astrocytes on the survival of dopaminergic neurons in lipopolysaccharide-induced inflammation. Neurosci. Res. 2009, 65, 252–258. [Google Scholar] [CrossRef]
- Rigamonti, D.; Mutti, C.; Zuccato, C.; Cattaneo, E.; Contini, A. Turning REST/NRSF dysfunction in Huntington’s disease into a pharmaceutical target. Curr. Pharm. Des. 2009, 15, 3958–3967. [Google Scholar] [CrossRef]
- Chen, L.; Chen, Y.; Li, B. The efficacy and safety of proton-pump inhibitors in treating patients with non-erosive reflux disease: A network meta-analysis. Sci. Rep. 2016, 6, 32126. [Google Scholar] [CrossRef]
| Name | Sequence | Frequency |
|---|---|---|
| BeM12-1 | TAKYLPMRPGPL | 12 |
| AlM12-1 | DYHDPSLPTLRK | 5 |
| AlM12-7 | SALPWFWSMDPS | 3 |
| Others | 6 | |
| Total | 26 |
| Name | Sequence |
|---|---|
| BeM12-1(1pT) | Ac-(pT)AKYLPMRPGPL-NH2 |
| AlM12-1(6pS) | Ac-DYHDP(pS)LPTLRK-NH2 |
| AlM12-1(9pT) | Ac-DYHDPSLP(pT)LRK-NH2 |
| AlM12-1(6pS, 9pT) | Ac-DYHDP(pS)LP(pT)LRK-NH2 |
| Universal peptide | H-RRA(pT)VA-OH |
| CTD(5pS) | Ac-SPSYSPT(pS)PS-NH2 |
| p53(15P) | Ac-VEPPL(pS)QETFSDLW-NH2 |
| Bio-BeM12-1(1pT) | bio-ε-(pT)AKYLPMRPGPL-NH2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoshida, T.; Yamazaki, K.; Imai, S.; Banno, A.; Kaneko, A.; Furukawa, K.; Chuman, Y. Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method. Catalysts 2019, 9, 842. https://doi.org/10.3390/catal9100842
Yoshida T, Yamazaki K, Imai S, Banno A, Kaneko A, Furukawa K, Chuman Y. Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method. Catalysts. 2019; 9(10):842. https://doi.org/10.3390/catal9100842
Chicago/Turabian StyleYoshida, Takuya, Kazuki Yamazaki, Shunta Imai, Akinori Banno, Atsushi Kaneko, Kazuhiro Furukawa, and Yoshiro Chuman. 2019. "Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method" Catalysts 9, no. 10: 842. https://doi.org/10.3390/catal9100842
APA StyleYoshida, T., Yamazaki, K., Imai, S., Banno, A., Kaneko, A., Furukawa, K., & Chuman, Y. (2019). Identification of a Specific Inhibitor of Human Scp1 Phosphatase Using the Phosphorylation Mimic Phage Display Method. Catalysts, 9(10), 842. https://doi.org/10.3390/catal9100842