Abstract
Starch and pullulan degrading enzymes are essential industrial biocatalysts. Pullulan-degrading enzymes are grouped into pullulanases (types I and type II) and pullulan hydrolase (types I, II and III). Generally, these enzymes hydrolyse the α-1,6 glucosidic bonds (and α-1,4 for certain enzyme groups) of substrates and form reducing sugars such as glucose, maltose, maltotriose, panose or isopanose. This review covers two main aspects: (i) bibliometric analysis of publications and patents related to pullulan-degrading enzymes and (ii) biological aspects of free and immobilised pullulan-degrading enzymes and protein engineering. The collective data suggest that most publications involved researchers within the same institution or country in the past and current practice. Multi-national interaction shall be improved, especially in tapping the enzymes from unculturable prokaryotes. While the understanding of pullulanases may reach a certain extend of saturation, the discovery of pullulan hydrolases is still limited. In this report, we suggest readers consider using the next-generation sequencing technique to fill the gaps of finding more new sequences encoding pullulan-degrading enzymes to expand the knowledge body of this topic.
1. Introduction
Pullulan is a biopolymer containing repeating units of maltotriose joined by α-1,6 glucosidic bonds and a small number of α-1,4 linked maltotetraose units. Pullulan powders are white, odourless, tasteless, non-toxic, non-pathogenic, non-mutagenic and water-soluble. Fungi (i.e., Aureobasidium, Cytaria and Cryphonectria), yeasts (i.e., Rhodototula) and lichen are pullulan producers [1]. Due to its high production yield and consistency in its composition from batch to batch, Aureobasidium pullulans is widely used to generate pullulan on an industrial scale [2,3,4]. Pullulan exhibits various prospects in maltotriose syrup production, food preservation, medicinal capsule, wound dressing and other industrial applications [5,6,7,8,9,10].
Pullulan-degrading enzymes can hydrolyse glucosidic bonds of polysaccharides (i.e., pullulan and starch), thus forming reducing sugars such as maltotriose, maltose, glucose, panose or isopanose [11]. Based on their substrate or product specificity, pullulan-degrading enzymes are divided into two major groups: (i) pullulanases (types I and type II pullulanases) and (ii) pullulan hydrolases (types I, II and III).
Type I pullulanase is required as one of the essential enzymes involved in producing glucose and maltose from pullulan and starch [12]. Generally, starch hydrolysis on the industrial scale can be divided into two major processes: liquefaction and saccharification. The liquefaction process involves three primary steps, which are: (i) the dissolution of starch granules at a high temperature between 90−110 °C; (ii) pH adjustment of the starch slurry; and (iii) the addition of thermostable α-amylase to reduce the viscosity of the starch slurry. Following that, the saccharification process is conducted by using several thermostable glycoside hydrolases (GH) enzymes (i.e., type I pullulanase, isoamylase, β-amylase and glucoamylase) under certain conditions (i.e., 60 °C and pH 4.5) to degrade the starch slurry in order to produce sugar syrups [13]. Type I pullulanase is also used in maltotriose syrup production from pullulan [14,15].
Apart from that, type II pullulanases are excellent biocatalysts in the starch processing industries. This enzyme group exhibits hydrolytic activity on both α-1,4 and α-1,6 glucosidic bonds. Because of this, the application of a single type II pullulanase could avoid or reduce the quantity of other GH enzymes (i.e., α-amylase, isoamylase, β-amylase and glucoamylase) during the starch liquefaction and saccharification processes [16]. Other than that, pullulanases (types I and II) are applied as anti-staling agents to improve the shelf life of baked products [12]. Staling is an undesirable reaction that causes bread to dry up, lose its crispiness and change its flavour [17]. In addition, alkali-tolerant pullulanases have great potential as additives in laundry and dishwashing detergents [18,19,20].
Close to a dozen research articles on pullulan-degrading enzymes (pullulanase and pullulan hydrolase) were published annually from 1966–1980, and the cumulative publication reached 20–65 articles yearly in the last decade. For interested readers, we have tabulated hundreds of characterised pullulan-degrading enzymes from microbial sources in Tables S1–S3. Several excellent reviews have covered fundamental science and the potential applications of pullulanase, and readers can refer to these prior references [11,21,22,23,24,25,26,27]. We systematically prepared this review to cover aspects that may have been overlooked in the earlier reviews. The current study includes (i) a bibliometric analysis related to the research publications and patents of pullulan-degrading enzymes; and (ii) an update on pullulan-degrading enzymes classification, structural and biochemical studies, immobilisation approaches and protein engineering studies. In addition, we also commented on the impact of next-generation metagenome sequencing on the new enzyme discovery. Bibliometric analysis is a statistical method for analysing scientific reports, as it allows a bird’s eye view of the footprints of a particular topic. None of the earlier reviews has covered bibliometric information on pullulan-degrading enzymes to the best of our knowledge. We hope this review will shed light on the emerging areas in this field after consolidating the bibliometric and the earlier scientific findings.
2. Bibliometric Analysis Related to the Publications and Patents of Pullulan-Degrading Enzymes
2.1. Publication Bibliometric Analysis
The Scopus database was used to retrieve documents published in 1966–2021. We used the Boolean expression ‘pullulanase or neopullulanase or isopullulanase or pullulan hydrolase’ to collect information from 1480 relevant documents (Oct 2021) (Figure 1a). 1390 hits are scientific articles, while the remaining are either review or book chapters. Web of Science (WOS) contained 1461 records using the identical Boolean expression. Unless specified, the information stated hereafter refers to the data retrieved from Scopus.
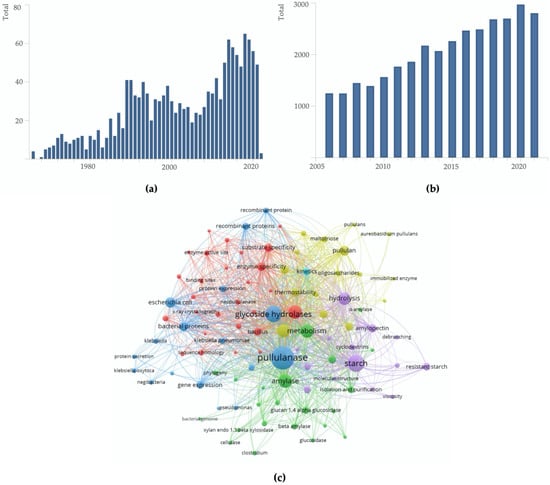
Figure 1.
Bibliometric analysis of pullulan-degrading enzymes. (a) Total publication from year 1966–2021; (b) Total citation in the past 15 years; and (c) Co-occurrence of keywords.
China, Japan, the United States, Germany, France, India, the United Kingdom and South Korea contributed 20.7%, 17.1%, 12.3%, 7.7%, 7.2%, 6.6%, 5.2% and 3.7% of the total documents in Scopus, respectively. A significant portion of knowledge derived from the scientific community are explained further in the third section, ‘Scientific review of pullulan-degrading enzymes’. Fewer than 10% of the documents are related to pullulan hydrolases (type I, neopullulanase; type II, isopullulanase; or pullulan hydrolase type III) [11,23,28,29,30,31,32,33,34,35,36,37]. The cumulative data indicate that this group of enzymes is getting less attention within the scientific community than the type I and type II pullulanases. So far, authors from Japan (Tokyo University of Agriculture and Technology, Osaka University and Ezaki Glico Co. Ltd) and South Korea (Seoul- and Incheon-National University) are the major research contributors to neopullulanase, isopullulanase or pullulan hydrolase type III. Examples of representative articles are listed in these references, and will be discussed in more detail in the subsequent parts [37,38,39,40,41].
Network analysis was calculated using VOSViewer bibliometric software v.1.6.17 [42], using information exported from Scopus. Figure 1c shows the co-occurring keywords in publications, and a threshold was set to 20 for the minimum number of occurrences. Out of 8436 keywords, 249 meet the requirements and we manually review the keywords. Furthermore, almost all these keywords are standard terms related to microbiology, biochemistry and enzymology. In the subsequent Section 3.6, we remarked that metagenome-driven biocatalyst would be a future trend. However, the latter is more complex, and international collaboration may be required.
The main research categories (Scopus) for the publications were Biochemistry, Genetics and Molecular Biology, Agricultural and Biological Sciences and Immunology and Microbiology (Table 1). The WOS database classified the publications into Biochemistry, Molecular Biology, Biotechnology, Applied Microbiology, Food Science Technology and Microbiology. A list of prominent authors that contributed to the body of knowledge of pullulan-degrading enzymes is provided in Table 2. Table 3 summarises a list of journals that researchers preferred to publish their findings. Section 3 will elaborate further on the scientific aspects of the enzymes.

Table 1.
Major research categories (Scopus) related to pullulan-degrading enzymes.

Table 2.
Prominent authors that have published more than 15 documents related to pullulan-degrading enzymes.

Table 3.
Name of journals with more than 15 publications related to pullulan-degrading enzymes.
2.2. Patents Bibliometric Analysis
Pullulanases are commercially available. Examples of these include Promozyme® D2 (Novozyme) and Optimax® L-1000 (DuPont Genencor® Science), which are the recombinant type I pullulanases originated from Bacillus acidopullulyticus and Bacillus deramificans, respectively. Other enzymes available in the market include PUL2 from Bacillus subtilis (Sunsonzymes) and PU-799 from Bacillus licheniformis (Boli Bioproducts); both manufacturers are in China. Patent protection and licensing are crucial for the manufacturer. The bibliometric information of patents related to pullulan-degrading enzymes was obtained using Lens.org (https://www.lens.org/). The database found 17,494 documents on pullulanases and pullulan hydrolases. Table 4 shows the top 10 owners, while Figure 2 summarises other patent bibliometric analyses for pullulan-degrading enzymes.

Table 4.
Numbers of patent documents owned by ten main companies.
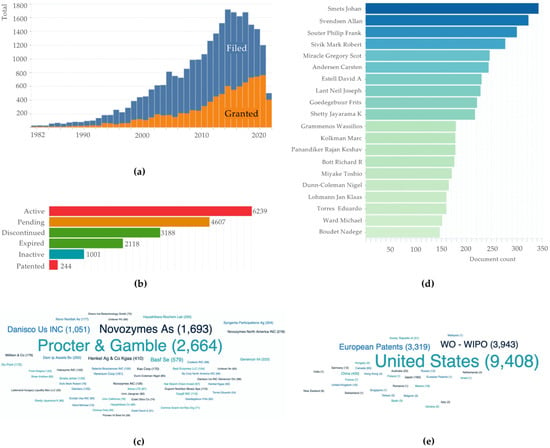
Figure 2.
Bibliometric analysis of pullulan-degrading enzymes: (a) Patent documents by filed and granted date; (b) Patent documents by legal status; (c) Top 50 applicants; (d) Top 20 inventors; and (e) Patent documents by jurisdiction.
Readers unfamiliar with patent law should be aware that patents do not last forever. In the United States, United Kingdom, Japan and many other countries, the patent term is up to 20 years from the filing date if the maintenance fees are paid on time. An example of an expired patent in 2010 is EP0415397A1, filed in 1990 by the Japanese Kao Corporation. It is forbidden to patent a native microorganism that produces biocatalysts. Claims of a patent, often located on the last page of the patent document, are the most critical section. A patent claim sets the boundaries of the invention. In biotechnology, the inventors can claim (i) genes, amino acid sequences and variants sequences (protein engineered or chimaera) of pullulan-degrading enzymes. The majority of the related patents protect gene-protein sequences. Once granted, the inventors of the gene patent dictate how the gene–enzyme can be used commercially. Examples of such patents include US8703465B2 (filed in 2010 by Novozymes AS and Novozymes North America Inc.) and patent US20170051265A1 (current assignee: Novozymes AS) that was filed in 2015. Both patents have anticipated an expiration in 2030 and 2035, respectively.
Hereafter is a statement taken from patent US20170051265A1. “The invention claimed is a polypeptide having pullulanase activity, selected from the group consisting of a polypeptide having at least 90% sequence identity to the mature polypeptide of SEQ ID NO: 9 or a polypeptide having at least 93% sequence identity to the mature polypeptide of SEQ ID NO: 11.” Where SEQ ID NO: 9 and NO: 11 referred to the protected protein sequences. It is common to state a claim with broader protection where patent US20170051265A1 set a 90% or 93% threshold. In the example given above, patent offices can exclude any other inventions with the primary sequence, that is, for instance, 94% identity to SEQ ID NO: 9 and NO: 11. Other patent files may adopt a lower threshold, such as patent WO 2014/150195Al, CN105960457A and EP2042593A2.
A patent can also claim (ii) a process. For example, inventors for patent US20170159090A1 claimed a composition comprising α-amylase, pullulanase and glucoamylase; and used that composition to make glucose syrup from liquefied starch. The current assignee for US20170159090A1 is Novozymes AS, and the filling was granted in 2019. In patent US4560651, the inventors claimed gene–protein as the independent claim and the process of using the enzyme for carbohydrate debranching as the dependent claim. In a different example, an application CN103388019A submitted by a Chinese company intends to protect the synthetic method (the process) of making ursolic acid glucose ester by using a mixture of lipase and amylase or pullulanase as biocatalysts for building-up reactions.
3. Scientific Review of Pullulan-Degrading Enzymes
3.1. Classification and Action of Pullulan-Degrading Enzymes
Pullulan-degrading enzymes are classified into two major groups: pullulanases (type I and type II) and pullulan hydrolases (types I, II and III) (Figure 3). Type I pullulanase (EC 3.2.1.41) acts on α-1,6 glucosidic bonds in pullulan to produce maltotriose (Figure 3a). The enzyme is also active against short branches (G1–G7) of starch, amylopectin, glycogen and β-limit dextrin, producing reducing sugars (i.e., glucose, maltose and maltotriose). Moreover, this enzyme group does not hydrolyse α-1,4 glucosidic bonds in the polysaccharides mentioned earlier [16], except the enzyme from Bacillus sp. AN-7 and Lactococcus lactis IBB 500 that possess the additional ability to hydrolyse α-1,4 glucosidic bonds in pullulan and form maltose [43,44]. Type I pullulanase is widely distributed in bacteria such as psychrophiles (i.e., Paenibacillus and Shewanella spp.), mesophiles (i.e., Bacillus, Exiguobacterium and Klebsiella spp.), thermophiles (i.e., Anoxybacillus, Geobacillus and Thermus spp.) and hyperthermophiles (i.e., Fervidobacterium and Thermotoga spp.) (Table S1) [45,46]. The optimum temperatures of type I pullulanases varied. For instance, the type I pullulanases from psychrophiles (i.e., Paenibacillus and Shewanella spp.) and mesophiles (i.e., Bacillus and Exiguobacterium spp.) are active at low temperatures of 35–50 °C. In contrast, those from thermophiles (i.e., Anoxybacillus, Geobacillus and Thermus spp.) and hyperthermophiles (i.e., Fervidobacterium and Thermotoga spp.) exhibit a higher optimum temperature in the range of 60–90 °C. From the industrial perspective, thermostable type I pullulanases are more feasible than heat-sensitive ones.
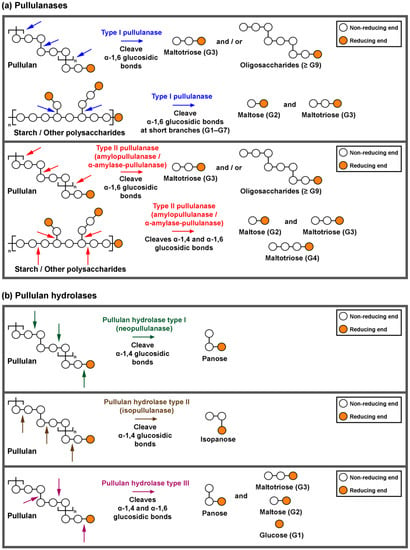
Figure 3.
The action pattern of (a) pullulanases (types I and II) on pullulan, starch or other polysaccharides and (b) pullulan hydrolases (types I, II and III) on pullulan.
Type II pullulanase (EC 3.2.1.1/41) can be divided into two subgroups: (i) amylopullulanase and (ii) α-amylase-pullulanase. These enzymes hydrolyse α-1,6 glucosidic bonds in pullulan. In addition, type II pullulanase can act on both α-1,4 and α-1,6 glucosidic bonds in starch and other polysaccharides [21]. Amylopullulanases from the following genera have been reported: thermophilic bacteria (i.e., Alkalilimnicola, Anoxybacillus, Geobacillus, Thermoanaerobacter, Thermoanaerobacterium and Thermus spp.) and hyperthermophilic archaea (i.e., Caldivirga, Desulfurococcus, Pyrococcus, Staphylothermus, Sulfolobus, Thermococcus and Thermofilum spp.) (Table S2) [47,48,49]. Other than that, a small number of currently known enzymes originated from mesophilic bacteria (i.e., Bacillus, Cohnella, Lactobacillus and Streptomyces spp.) and halo-mesophilic archaeon (i.e., Halorubrum sp.) [50,51]. Amylopullulanases listed in Table S2 are mainly recombinant enzymes; however, none of these enzymes have been crystallised. The molecular mass of amylopullulanases falls in 45–184 kDa. Some amylopullulanases exhibit high molecular mass (>200 kDa), such as those from Anoxybacillus sp. SK3-4 (225 kDa) that our group studied earlier [48]. Other examples are enzymes from Bacillus circulans F-2 (220 kDa), Bacillus sp. XAL601 (224 kDa), Geobacillus stearothermophilus TS-23 (220 kDa) and Thermoanaerobacterium thermosulfurigenes EM1 (205 kDa). Most of the currently known amylopullulanases are active at high temperatures (60–105 °C) and acidic (pH 3.0–6.5).
Another subgroup of type II pullulanase is α-amylase-pullulanase. At present, only six of these enzymes have been characterised. They all originated from mesophilic bacteria (i.e., Bacillus sp. KSM-1378, Bifidobacterium breve UCC2003, Bifidobacterium adolescentis P2P3, Lactobacillus plantarum L137, Streptococcus suis P1/7 and Alkalibacterium sp. SL3 [18,52,53,54,55]. This subgroup of enzymes exhibited a high molecular mass of approximately 200 kDa, and is active at 40–50 °C (Table S2). The action of α-amylase-pullulanases on polysaccharides (i.e., pullulan, starch, amylopectin, amylose and glycogen) generate reducing sugars of various lengths (G1–G6).
Pullulan hydrolases are further classified into three groups (Figure 3). Type I (neopullulanase, EC 3.2.1.135) hydrolyses α-1,4 glucosidic bonds of pullulan to form panose. In contrast, pullulan hydrolase type II (isopullulanase, EC 3.2.1.57) cleaves α-1,4 glucosidic bonds in pullulan and produces isopanose [23]. Pullulan hydrolase type III attacks α-1,4 and α-1,6 glucosidic bonds in pullulan and forms panose, maltotriose, maltose and glucose [30]. Furthermore, pullulan hydrolases can degrade starch, liberating reducing sugars such as glucose, maltose and maltotriose [21]. Table S3 tabulated the characterised pullulan hydrolases (neopullulanase, isopullulanase and pullulan hydrolase type III). Researchers have described neopullulanases from Paenibacillus, Bacillus, Bacteroides, Lactobacillus, Alicyclobacillus, Anoxybacillus and Geobacillus spp. The biochemical properties for isopullulanases from Bacillus sp. US149 and Aspergillus brasiliensis ATCC 9642T were reported in these articles [38,39,40,41,56]. Apart from that, only two pullulan hydrolases type III from Thermococcus spp. have been characterised [30,32].
Almost all pullulan hydrolases listed in Table S3 are recombinant enzymes, and exhibit a molecular mass of 53–200 kDa. The optimum temperature of pullulan hydrolases falls in the range of 37–70 °C, except for pullulan hydrolase type III, which they are active at 95 °C. These pullulan hydrolases favour acidic condition of pH 3.5–6.0. However, the neopullulanases from Bacillus sp. KSM-1876 and Micrococcus halobius OR-1 function at a slightly alkaline pH of 7.5 and 8.0, respectively [20,57].
Carbohydrate-Active enZYmes (CAZy) database classified pullulan-degrading enzymes in glycoside hydrolase (GH) families GH13, GH49 or GH57 [58]. Type I pullulanase, type II pullulanase, neopullulanase and pullulan hydrolase type III are members of GH13. Members within this family have consensus key features such as (i) they cleave the α-glucosidic bonds, (ii) the amino acid sequences contain four conserved regions, (iii) the enzymes possess a TIM barrel structure and (iv) Asp, Glu and Asp are the enzymes catalytic residues [59]. Among the type II pullulanases, some of the characterised enzymes from hyperthermophilic archaea (i.e., Caldivirga, Pyrococcus, Thermococcus, Sulfolobus and Staphylothermus spp.) have been classified in family GH57 [34]. Isopullulanases are categorised in family GH49 [11].
3.2. Domains, Structures and Properties of Pullulan-Degrading Enzymes
Pullulan-degrading enzymes are multi-domain proteins. Generally, these enzymes contain several domains such as carbohydrate-binding module (CBM), catalytic domain, C-terminal domain (i.e., domain C or amyC domain) and fibronectin type III (FnIII) domain [11]. CBM, a non-catalytic ancillary domain, assists protein attachment to polysaccharide’s surface (i.e., pullulan or starch). CBM also facilitates the degradation process by distorting the conformation or the packing of the polysaccharides. Based on the CAZy database (November 2021), CBMs are currently grouped into 88 primary sequence-based families [58]. CBMs with affinity for starch (CBM20, CBM21, CBM25, CBM26, CBM34, CBM41, CBM45, CBM48, CBM53, CBM58, CBM68, CBM69, CBM82 and CBM83) are commonly known as starch-binding domains (SBDs) [60]. Figure 4 shows the comparison of domains and structure architecture for each group of pullulan-degrading enzymes.
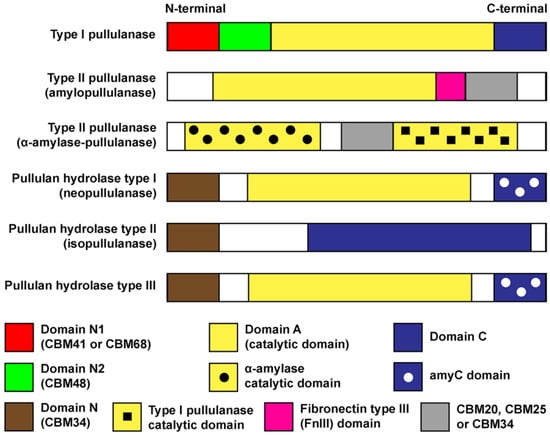
Figure 4.
Schematic representation of conserved domains in pullulan-degrading enzymes.
Initially, all type II pullulanases were named amylopullulanase. However, after more sequences were available, the differences between amylopullulanase and α-amylase-pullulanase were noticed. Now, amylopullulanase refers to a group of enzymes with a single catalytic domain (domain A). In contrast, α-amylase-pullulanase referred to enzymes with dual catalytic domains: (i) an α-amylase catalytic domain for α-1,4 bonds and (ii) type I pullulanase catalytic domain for α-1,6 glucosidic bonds (Figure 4) [11,18,52,53,54,55]. A recent article reported the role of CBM and the dual catalytic domains [61]. α-amylase-pullulanase (denoted as PulP) from Bifidobacterium adolescentis P2P3 underwent truncation study. Deletion of either catalytic domain affected the catalytic activity and thermostability. The removal of CBM41 and CBM25, located somewhere in the middle of the protein sequence, led to the loss of binding affinity and hydrolytic capability on raw and soluble starches. In a separate recent publication, the author preferred to name it pullulanase PulSL3 instead of α-amylase-pullulanase PulSL3 [55]. PulSL3 originated from Alkalibacterium sp. SL3 contains the typical dual catalytic domains explained earlier. Interestingly, the recombinant enzyme exhibited pullulanase activity, but lacked α-amylase activity. Huang et al. [55] created domain-truncated mutants and examined the function of each domain. For some reason, probably due to certain unfavourable residues, the N-terminal α-amylase domain is missing the α-1,4 hydrolytic ability.
As illustrated in Figure 4, pullulan hydrolases exhibit a less complicated architecture. Neopullulanase (pullulan hydrolase type I) and pullulan hydrolase type III have three domains (Domains N, A and C) [39]. Domains N and A function as starch-binding domain (CBM34) and catalytic domain, respectively. The C-terminal of these enzymes is composed of the amyC domain that orientates the substrate’s glucose chains so the hydrolytic action can take place [62]. Isopullulanase (pullulan hydrolase type II) possess only two domains, where domains N and C act as CBM34 and catalytic domain, respectively [39].
3.3. Single Immobilisation of Pullulan-Degrading Enzyme
In laboratories and industrial setups, enzymes immobilisation can increase enzyme activity or improve other catalytic features such as stability and specificity (selectivity); facilitate the recovery or the reusability of biocatalysts and enhance biochemical properties [63,64,65]. The methods for protein immobilisation are divided into three main categories: (i) the Entrapment (encapsulation) approach, a method based on the inclusion of the enzyme in a polymer network such as calcium alginate, polyacrylamide, silica, hollow fibre membrane or microcapsule [63], which often requires synthesis of the polymeric matrix together with the enzyme [66]; (ii) second, the cross-linked immobilisation strategy, which often employs linker reagents (i.e., glutaraldehyde) to produce carrier-free immobilised enzymes particles denoted as cross-linking enzymes crystals (CLECs) and cross-linking enzymes aggregates (CLEAs) [67,68]; and (iii) the binding approach, where protein is immobilised to the pre-fabricated solid support (carrier) via physical adsorption, ionic bonding or covalent attachment [69].
Some conventional immobilisation techniques, such as calcium alginate, could be an excellent example for students’ training; however, it is not feasible on an industrial scale. Choosing the ideal support for the enzyme is essential for a successful immobilisation process. One shall carefully examine the properties of the carriers; among the many, these are the critical selection criteria: particle size, pore size, surface area and hydrophilicity/hydrophobicity of the particle surface. Readers that would like to learn more on these topics may refer to several excellent review articles [63,68,70,71,72,73,74,75]. To date, many ready-to-use polymer supports are available at an affordable price. These include the epoxide or amino-epoxide-activated supports such as Sepabeads (Resindion S.R.L.), ReliZyme (Resindion S.R.L.), Immobead (ChiralVision) and Purolite (Purolite). Acrylic-based macroporous supports offer high mechanical stability, high resistance towards microbial contamination and low swelling effects in water, making them suitable for laboratory-, pilot- or industrial applications [76,77]. Furthermore, the material used in producing such supports is generally safe in food processing.
Unlike many immobilisation research on GH13 enzymes (i.e., α-amylase and α-glucosidase), studies on immobilisation of pullulan-degrading enzymes are rather scarce. Table 5 summarises the collective findings on single immobilised pullulan-degrading enzymes. Most of the immobilised enzymes were type I pullulanases that originated from several mesophilic bacteria such as Bacillus, Klebsiella and Fontibacillus spp. A small number of immobilisation studies used other types of pullulan-degrading enzymes. Examples include type II pullulanase (amylopullulanase) and neopullulanase from Anoxybacillus sp. WB42 and Geobacillus stearothermophilus TRS40, respectively [78,79].
Covalent attachment on a solid carrier is a favoured approach (Table 5). Lenders and Crichton were among the earliest groups that immobilised type I pullulanase using the covalent attachment on aldehyde-activated amylose conjugate [80]. The immobilisation construct increased the enzymes optimum temperature and thermostability. Other favourable covalent attachments materials for type I pullulanases include polyacrylate, agar, silica, calcium alginate, Duolite XAD761 and magnetic chitosan beads. Interestingly in one report, immobilised enzyme on carboxyl-activated calcium alginate beads enhanced thermostability by 50 times than its free enzyme counterpart at 60 °C [81].
Besides covalent attachment, pullulan-degrading enzymes were also immobilised using ionic binding. For example, the amylopullulanase from Anoxybacillus sp. WB42 was immobilised using ionic binding on magnetic carboxymethyl cellulose (CMC) nanoparticles [82]. The reusable immobilised enzyme was 27 cycles at 60 °C, and it had similar biochemical characteristics to the free enzyme. Entrapment (encapsulation), an aged immobilisation concept, can increase the enzymes’ thermostability. For instance, the type I pullulanase entrapped in silica-magnetic nanoparticles had a half-life activity of 5 h at 60 °C, while the free enzyme was only stable for an hour at an identical temperature [83] (Table 5).
On a pilot and industrial scale, immobilising using purified enzymes is costly. From a research (laboratory scale) point of view, immobilising using purified enzymes is better than crude proteins extracted from the whole cells. By doing so, we can eliminate the influences of non-targeted background proteins [67]. For instance, any changes to the enzymes biochemical characteristics after immobilisation is genuinely contributed by the effect of the immobilisation carrier or the chosen technique, rather than an uncertain reason such as complex interactions of the proteins and other factors [84]. However, a significant amount of enzymes immobilisation research on a lab-scale have been carried out using the crude enzyme [85].
Our lab investigated the effects of single enzyme-immobilisation on the product specificity of type I pullulanase (PulASK) from Anoxybacillus sp. SK3-4 [86] (Table 5). Four derivatives of PulASK were prepared by immobilising the purified enzyme using the covalent attachment on three epoxides (i.e., ReliZyme EP403/M, Immobead IB-150P and Immobead IB-150A) and an amino-epoxide (i.e., ReliZyme HFA403/M)-activated support. The collected data indicates that the free and immobilised PulASKs were optimally active at 60 °C and pH 6.0. However, the product specificities of free and immobilised enzymes were dissimilar [86]. Free PulASK was reactive towards pullulan and β-limit dextrin, but it could not hydrolyse the short branches in starch. Hence, no reducing sugars were formed. In contrast, the immobilised PulASKs were able to hydrolyse starch at both short and long branches producing reducing sugars (G3 and G2) and oligosaccharides (≥G8), respectively. Moreover, the ratio of reducing sugars and oligosaccharides produced by each PulASK derivative using pullulan, starch, β-limit dextrin, glycogen and amylopectin were dissimilar. These phenomena were hypothesised to be associated with changes in the enzyme binding pocket influenced by the carrier surface properties (hydrophobic or hydrophilic) and the spacer arms lengths between the supports and proteins.
3.4. Co-Immobilisation of Pullulan-Degrading Enzymes
Bioconversion processes using enzymes such as starch degradation industries require multi-enzymatic reactions in a cascade manner. For simplicity and cost-effective purpose, industrial players may prefer a one-pot reaction concept using a cocktail of several enzymes. In the same practice, researchers have developed the idea of co-immobilised enzymes [87], where various types of enzymes are immobilised together or separately but applied together [88]. In a report, Talekar et al. [89] co-immobilised type I pullulanase, glucoamylase and α-amylase by cross-linked enzyme aggregates (combi-CLEAs) method (Table 6). The one-pot starch hydrolysis shortened the process from 35 h (using free enzymes mixture) to 2 h (using co-immobilised enzymes). The co-immobilisation strategy exhibits multiple advantages [90]; however, some already known drawbacks are limited substrate diffusion and other issues [84].
Table 6 lists the cumulative studies on co-immobilisation of type I pullulanases with other CAZymes using various strategies, including the covalent attachment, ionic binding and enzymes combination of combi-CLEAs. All the co-immobilisation work summarised in Table 6 employed crude enzymes except for the enzymes from Anoxybacillus sp. SK3-4. Our lab has tried to understand the effects of co-immobilisation of type I pullulanase (PulASK) with an α-amylase (TASKA), both from Anoxybacillus sp. SK3-4 on the product specificity of enzymatic reaction [86] (Table 6). Both enzymes were individually purified to be at least 98% purity. ReliZyme HFA403/M support was chosen for co-immobilisation. Our experimental setups contained:
- (i)
- free enzymes mixture (PulASK+TASKA)
- (ii)
- individual immobilised enzymes mixture (HFA403/M–PulASK+ HFA403/M–TASKA)
- (iii)
- co-immobilised enzymes (PulASK–HFA403/M–TASKA).
The accumulated data from that setup indicated that the total amount and the generated spectrum of reducing sugars using (iii) co-immobilised enzymes were significantly different from either the (i) free enzymes mixture or (ii) the individual immobilised enzymes mixture [86]. We thought that structural changes in PulASK and TASKA may alter the substrate-enzyme-product dynamic interaction and thus drastically modify their product specificities. Nevertheless, due to random covalent attachment, that work could not pinpoint the exact residues bound to the carrier, or how these residues affect the product specificity of the bound enzyme.



Table 5.
List of single immobilised pullulan-degrading enzymes compared to their free enzyme counterpart.
Table 5.
List of single immobilised pullulan-degrading enzymes compared to their free enzyme counterpart.
| TE a | Source | Free Enzyme/Support | Topt (°C) | pHopt | Activity Half-Life | Reusability | Reaction Product b | Improvement after Immobilisation | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pullulan | Starch | |||||||||
| Binding to a carrier approach using covalent attachment | ||||||||||
| I c | Klebsiella pneumoniae | Free enzyme | 30 | 5.0 | 5 min at 50 °C | - | RD | ND | Thermostability | [80] |
| Amylose-aldehyde conjugate | 50 | 5.0 | >30 min at 50 °C | ND | RD | ND | ||||
| I | Bacillus acidopullulyticus | Free enzyme | 60 | 5.0 | 30 min at 60 °C | - | RD | ND | Thermostability | [91] |
| Chitosan-amino beads | 55 | 3.5 | 30 min at 55 °C | ND | RD | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 25 | 5.0 | 5 min at 50 °C | - | RD | ND | Thermostability and reusability | [92] |
| Polyacrylate -epoxide beads | 35 | 5.0 | 30 min at 50 °C | 8 cycles at 30 °C | RD | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 6.0 | 30 min at 45 °C | - | RD | ND | Thermostability | [93] |
| Agar-epoxide gel | 50 | 6.0 | 4 h at 45 °C | ND | RD | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 5.5 | 2 h at 70 °C | - | RD | ND | Thermostability and reusability | [94] |
| Magnetic chitosan-amino nanoparticles | 50 | 5.0 | 6 h at 70 °C | 10 cycles at 70 °C | RD | ND | ||||
| I c | Bacillus acidopullulyticus | Free enzyme | 50 | 5.0 | >6 h at 60 °C | - | RD | RD | Reusability | [95] |
| Duolite XAD761 beads | 60 | 5.5 | >6 h at 60 °C | 25 cycles at 60 °C | RD | RD | ||||
| I | Bacilius subtilis | Free enzyme | 50 | 4.4 | 5 h at 60 °C | - | RD | ND | Thermostability and reusability | [96] |
| Magnetic chitosan-amino nanoparticles | 50 | 3.5 | >5 h at 60 °C | 8 cycles at 50 °C | RD | ND | ||||
| I | Klebsiella pneumonia | Free enzyme | 60 | 5.5 | 1 h at 60 °C | - | RD | ND | Thermostability | [81] |
| Calcium alginate-carboxyl | 60 | 5.5 | 50 h at 60 °C | ND | RD | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 60 | 4.4 | ND | - | RD | ND | Reusability | [97] |
| Magnetic chitosan-amino nanoparticles | 60 | 4.4 | ND | 10 cycles at 50 °C | RD | ND | ||||
| I | Fontibacillus sp. DSHK 107 | Free enzyme | 35 | 5.0 | 5 h at 35 °C | - | RD | ND | Thermostability and reusability | [98] |
| Silica-amino nanoparticles | 35 | 6.0 | 24 h at 35 °C | 10 cycles at 30 °C | RD | ND | ||||
| I c | Anoxybacillus sp. SK3-4 | Free enzyme | 60 | 6.0 | >4 h at 60 °C | - | G3 | - | Reusability and product specificity on starch | [86] |
| ReliZyme HFA403/M | 60 | 6.0 | 3 h at 60 °C | 12 cycles at 60 °C | G3 | G3 | ||||
| II | Anoxybacillus sp. WB42 | Free enzyme | 65 | 6.0 | 30 min at 70 °C | - | RD | ND | Reusability | [78] |
| Magnetic Ni-IDA nanoparticles | 65 | 5.5 | 30 min at 70 °C | 12 cycles at 60 °C | RD | ND | ||||
| PHI c | Geobacillus stearothermophilus TRS40 | Free enzyme | 60 | 6.0 | >1 h at 60 °C | - | G1, G2, P | ND | Thermostability | [79] |
| Zetaffinity™ 10 beads | 60 | 6.0 | 30 h at 60 °C | ND | G1, G2, P | ND | ||||
| Binding to a carrier approach using ionic bonding | ||||||||||
| I | Streptomyces flavochromogenes | Free enzyme | 55 | 5.5 | 30 min at 55 °C | - | RD | ND | Thermostability | [99] |
| TEAE-cellulose-amino beads | 55 | 6.0 | 30 min at 55 °C | ND | RD | ND | ||||
| II | Anoxybacillus sp. WB42 | Free enzyme | 65 | 6.0 | 30 min at 70 °C | - | RD | ND | Reusability | [82] |
| Magnetic carboxymethyl cellulose (CMC)-amino nanoparticles | 65 | 6.0 | 30 min at 63 °C | 27 cycles at 60 °C | RD | ND | ||||
| Binding to a carrier approach using physical adsorption | ||||||||||
| I | Paenibacillus polymyxa Nws-pp2 | Free enzyme | ND | ND | ND | ND | ND | ND | One-step purification and immobilization | [100] |
| Magnetic nanoparticles | ND | ND | ND | ND | ND | ND | ||||
| Entrapment (encapsulation) approach | ||||||||||
| I | Klebsiella aerogenes | Free enzyme | 25 | ND | ND | - | G3 | ND | None | [101] |
| Polyacrylamide gel | 25 | ND | ND | ND | G3 | ND | ||||
| I | Bacillus acidopullulyticus | Free enzyme | 55 | 5.0 | 1 h at 55 °C | - | RD | RD | Thermostability | [102] |
| Calcium alginate beads | 55 | 5.0 | 2 h at 55 °C | ND | RD | RD | ||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 4.4 | 1 h at 60 °C | - | RD | ND | Thermostability and reusability | [103] |
| Chitosan-encapsulated magnetic κ-carrageenan nanoparticles | 50 | 4.4 | 3 h at 60 °C | 10 cycles at 50 °C | RD | ND | ||||
| Entrapment (encapsulation) approach | ||||||||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 4.4 | 20 min at 62 °C | - | ND | ND | Thermostability and reusability | [104] |
| Chitosan (50 kDa)-encapsulated magnetic κ-carrageenan nanoparticles | 50 | 4.4 | 3 h at 62 °C | 6 cycles at 62 °C | ND | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 4.4 | 1 h at 60 °C | - | RD | ND | Thermostability and reusability | [83] |
| Sol-gel-encapsulated magnetic chitosan nanoparticles | 50 | 4.4 | 5 h at 60 °C | 6 cycles at 50 °C | RD | ND | ||||
| I | Geobacillus kaustophilus DSM 7263T | Free enzyme | 65 | 6.0 | 45 h at 65 °C | - | G3 | ND | Thermostability and reusability | [105] |
| Calcium alginate beads | 65 | 6.0 | >70 h at 65 °C | 4 cycles at 65 °C | RD | ND | ||||
| I | Klebsiella pneumoniae | Free enzyme | 50 | 4.4 | ND | - | RD | RD | Thermostability, reusability, and product specificity on resistant starch | [106] |
| Sol-gel-encapsulated magnetic chitosan nanoparticles | 50 | 4.4 | ND | 8 cycles at 50 °C | RD | RD | ||||
| I | Bacillus subtilis | Free enzyme | 50 | 4.4 | 3 h at 50 °C | - | ND | ND | Thermostability and reusability | [107] |
| Streptavidin-encapsulated magnetic nanoparticles | 60 | 4.4 | >6 h at 65 °C | 8 cycles at 60 °C | ND | ND | ||||
II: type I pullulanase; II: type II pullulanase; PHI: pullulan hydrolase type I (neopullulanase); G1: glucose; G2: maltose; G3: maltotriose; P: panose; TE: type of enzyme; Topt: optimum temperature; pHopt: optimum pH; ND: not determined; Ni-IDA: nickel-iminodiacetic acid; RD: substrate was reduced as inferred from enzymatic assay; not applicable; a: the immobilisation processes were done using crude enzyme; b: reaction products were determined using high-performance liquid chromatography, high performance size exclusion chromatography or thin-layer chromatography in the respective reference; c: the immobilisation process was done using purified enzyme.

Table 6.
List of co-immobilisations of pullulan-degrading enzymes with various glycoside hydrolases.
Table 6.
List of co-immobilisations of pullulan-degrading enzymes with various glycoside hydrolases.
| TE a | Source | Support | Topt (°C) | pHopt | Activity Half-Life | Reusability | Starch Degradation Process | Ref. | |
|---|---|---|---|---|---|---|---|---|---|
| Time | Product b | ||||||||
| Binding to a carrier approach using covalent attachment | |||||||||
| Type I pullulanase Glucoamylase | Klebsiella aerogenes Aspergillus niger | Free enzymes mixture | 40 | 5.0 | 2 days at 50 °C | - | > 90 h | G2, G3, G4 | [108] |
| Bovine collagen-azide membrane | 60 | 7.0 | 7 days at 50 °C | ND | 80 h | G2, G3, G4 | |||
| Type I pullulanase Glucoamylase | Bacillus acidopullulyticus Aspergillus niger | Free enzymes mixture | 23 | 6.0 | ND | - | 24 h | RD | [109] |
| Hypol® 2002 foam | 23 | 6.0 | ND | ND | 4 h | RD | |||
| Type I pullulanase β-amylase | Klebsiella pneumoniae Barley | Free enzymes mixture | 30 | 4.5 | 10 min at 50 °C | - | 24 h | G2 | [110] |
| Polyacrylate-epoxide beads | 30 | 4.5 | 60 min at 60 °C | 30 cycles at 30 °C | 24 h | G2 | |||
| Type I pullulanase α-amylase Glucoamylase | ND Bacillus licheniforms Aspergillus niger | Free enzymes mixture | 70 | 6.0 | 2 h at 90 °C | - | 150 min | RD | [111] |
| Magnetic-amino nanoparticles | 80 | 7.0 | 4 h at 90 °C | 8 cycles at 70 °C | 90 min | RD | |||
| Type I pullulanase c α-amylase c | Anoxybacillus sp. SK3-4 Anoxybacillus sp. SK3-4 | Free enzymes mixture | 60 | 6.0 | ND | ND | 48 h | G1, G2, G4 | [86] |
| ReliZyme HFA403/M | 60 | 6.0 | ND | ND | 48 h | G1, G2, G3, G4, G5 | |||
| Binding to a carrier approach using ionic bonding | |||||||||
| Type I pullulanase β-amylase | Klebsiella pneumoniae Soybean | Free enzymes mixture | 60 | 6.0 | 6 days at 60 °C | - | 20 days | G2 | [112] |
| Chitopearl® BCW 3505 beads | 65 | 7.0 | 6 days at 60 °C | ND | 20 days | G2 | |||
| Type I pullulanase Maltotetraose- forming amylase | Klebsiella pneumoniae Pseudomonas stutzeri | Free enzymes mixture | 50 | 6.0 | 8 days at 55 °C | - | 25 days | G4 | [113] |
| Chitopearl® BCW 3505 beads | 50 | 6.0 | 8 days at 55 °C | ND | 25 days | G4 | |||
| Type I pullulanase Glucoamylase Xylose isomerase | Bacillus acidopullulyticus Aspergillus niger Streptomyces rubiginosus | Free enzymes mixture | 55 | 7.5 | ND | - | 5 h | F, G1 | [114] |
| BIOBONE™ beads | 55 | 7.0 | ND | ND | 5 h | F, G1 | |||
| Type I pullulanase β-amylase | Bacillus brevis Sweet potato | Free enzymes mixture | 50 | 6.0 | 30 min at 50 °C | - | 14 days | G2, G3 | [115] |
| Chitopearl® BCW 3505 beads | 70 | 6.0 | 30 min at 60 °C | ND | 14 days | G2, G3 | |||
| Cross-linking enzyme aggregates (CLEAs) approach | |||||||||
| Type I pullulanase Glucoamylase | ND ND | Free enzymes mixture | 60 | 5.0 | 1 h at 65 °C | - | 4 h | RD | [116] |
| CLEAs using glutaraldehyde | 70 | 7.0 | >4 h at 65 °C | 8 cycles at 70 °C | 3 h | RD | |||
| Type I pullulanase α-amylase Glucoamylase | ND ND ND | Free enzymes mixture | 65 | 6.0 | 1 h at 75 °C | - | 35 h | RD | [116] |
| CLEAs using glutaraldehyde | 75 | 7.0 | >3 h at 75 °C | 6 cycles at 75 °C | 2 h | RD | |||
F: fructose; G1: glucose; G2: maltose; G3: maltotriose; G4: maltotetraose; G5: maltopentaose; TE: type of enzyme; Topt,: optimum temperature; pHopt: optimum pH; ND: not determined; RD: substrate was reduced as inferred from enzymatic assay; not applicable; a: the immobilisation processes were done using crude enzyme; b: reaction products were determined using high performance liquid chromatography or thin-layer chromatography in the respective reference; c: the immobilisation process was done using purified enzyme.
3.5. Protein Engineering for Improving the Performance of Pullulan-Degrading Enzymes
Protein engineering, also known as protein mutagenesis, is an approach that alters or improves enzyme biochemical features (i.e., substrate specificity, product specificity, specific activity, reducing inhibition effects or thermostability). The strategies include (i) directed evolution (error-prone PCR, staggered extension PCR, DNA shuffling, etc.) and (ii) rational design or site-directed mutagenesis (mainly mega primer PCR and overlap extension PCR methods) [117]. Table 7 lists selected publications on this subject matter. As expected, there are relatively fewer attempts for pullulan hydrolases engineering [37,118,119].

Table 7.
Selected publications related to the engineering of pullulan-degrading enzymes (year 2000 onwards).
One central mutagenesis theme for pullulan-degrading enzymes is understanding the role of a residue at a particular position, specific amino acids stretch, motif or domain. For instance, Kim et al. [61] and Li et al. [126] performed CBM truncation to understand starch saccharification function. In another work, Kahar et al. elucidated that a short loop of the native protein prevents the native enzyme from generating reducing sugars from short linear or branched oligosaccharides [135]. In one particular study, Chen et al. shifted optimum pH from 5.0 to 4.0, and the mutant pullulanase exhibited an increased tolerance against acid denaturation [136]. Using pullulanase as a role model, the concept of ECSM (evolutionary coupling saturation mutagenesis) seems promising in cutting the time in identifying the amino acid positions for mutations [137]. The ECSM is an extension concept of an open-source software EVcouplings that perform computational predictions based on evolutionary sequence covariation (https://evcouplings.org).
Improving activity half-life or structural stability is another central theme for pullulanase. For instance, among the six constructed variant proteins, pullulanase mutant G692M from Geobacillus thermoleovorans had a two-fold improvement in the activity half-life [127]. Other outstanding thermostability improvements were performed by these studies [138,139] and others are listed in Table 7. In the past decade, several computation predictors were created to assist researchers in doing in silico protein structure stabilisation analysis before starting to construct variants on the lab bench (Table 8). Most of these predictors calculate ΔΔG (changes in the Gibbs free energy) between in silico native and variant proteins. In early times, these predictors applied the thermodynamic information recorded in the ProTherm database [140]. Approximately 20 years ago, ProTherm contained less than 6000 data entries and in 2013, the database updated to >25,000 thermodynamic data. Structure-based predictors, including FoldX, I-Mutant series, CUPSAT, PoPMuSiC, STRUM and etc., may outperform sequence-based predictors [141]. For the mutant pullulanase G692M that we stated at the beginning of this paragraph, the researchers utilised FoldX, I-Mutant 3.0 and dDFIRE [127]. From structural biologists’ perspective, X-ray protein structure is more accurate than homology-modelling predicted models. One may face drawbacks if the input files for structure-based predictors are the protein models instead of high-resolution X-ray structures. Crystallography is costly and the state-of-the-art AlphaFold may be an option to generate a better model structure.

Table 8.
List of software or programs for simulating protein macromolecules for thermostability mutagenesis.
More recent predictors apply protein structural datasets and evolutional information to train machine learning (Table 8). A few years ago, a research team noticed several problems with ProTherm, and they later cleaned the database before developing the PON-tstab predictor [148]. To overcome other limitations of ProTherm, the developers for DDGun and DDGun3D used a method that combines anti-symmetric features for predicting the ΔΔG upon variation, and claimed that this untrained and straightforward method has excellent performance. So far, only a few predictors address the anti-symmetric issue, and examples of these programs include PROTS-RF, INPS, SDM, ProTstab and a few others [149]. Predictor DeepDDG, a neural network approach, was trained using ProTherm, and manually curated literature data. The developer suggested that DeepDDG outperformed the other eleven commonly used predictors [146]. The identical research group that created PON-tstab later proposed a new platform, ProTstab, which used a >3500 proteolysis and mass spectrometry (LiP-MS) dataset to train the gradient-boosting based machine learning platform. Different algorithms may suggest contradictory suggestions, and it is time-consuming to try one predictor at a time. One may consider iStable 2.0, an integrated platform consisting of eleven structure- and sequence-based tools [147]. Readers can refer to this excellent review that compares the limitations and challenges of each predictor from the angle of future applications in precision medicine [149]. We observed that not many pullulanase protein engineers use the tools mentioned above; therefore, keywords stated in this subsection will not appear in bibliometric records (Figure 1c).
3.6. Influence of Next-Generation Metagenome Sequencing on Novel Pullulan-Degrading Enzymes Discovery
Next-generation sequencing (NGS) is a powerful tool in natural science and biotechnology in the current era. Metagenome sequencing with computation pipelines can generate a tremendous amount of novel macromolecule sequences. Interested readers in this topic can refer to these publications for more insights [150,151,152,153,154]. We searched using the keyword ‘metagenome and X’ (where X is cellulase, lipase, amylase or pullulanase), the hit of research articles in the Scopus database is 180, 137, 60, 5, respectively. The articles related to pullulanase are listed in the references [155,156,157,158,159]. No relevant documents for pullulan hydrolase types I, II and III were found.
A type I pullulanase derived from Reshi hot spring metagenome dataset was reported in 2021 [157]. The enzyme, named PulM, has very low sequence similarity to currently characterised pullulanases. For instance, PulM shares only 42.9% identity to a type I pullulanase from Geobacillus stearothermophilus. PulM had an optimum catalytic temperature at 40 °C. Interestingly, the enzyme remained 50% active at 4 °C, a relatively higher activity than other pullulanases obtained from isolated bacteria sampled from hot springs [157]. In another recent article, a novel type I pullulanase (PersiPul1) was established from cow rumen metagenomic data [156]. The enzyme is active from 30–80 °C and retained close to 60% of its original activity at 80 °C. In the same study, the researchers blended a cocktail of type I pullulanase (PersiPul1) and α-amylase PersiAmy2 (also derived from metagenome strategy, [160]). They used the enzyme cocktail to improve bread physical and sensory properties [156].
Amylopullulanase PulSS4 was a type II pullulanase derived via the metagenome approach [159]. The gene encoding this enzyme was identified in a gut metagenome (clonal functional library approach) of Hermetia illucens (black soldier fly). PulSS4 closest counterpart (51%) is a protein from the anaerobic bacterium Amphibacillus xylanus. The authors claimed that PulSS4 had the maximum activity at pH 9.0 and exhibited a broad pH tolerance [159].
Our laboratory recently performed Illumina shotgun sequencing using environmental DNA extracted from biofilms attaching to plant litters socked in a local hot spring. Using the assembled contigs (Figure 5c), we have mined >10,000 sequences related to various CAZymes and identified close to 40 sequences for pullulan-degrading enzymes. All of them are type I or type II pullulanases, except one was detected as neopullulanase. This may suggest that pullulan hydrolases are rarely encoded in the genomes of thermophiles. Nevertheless, this approach enables us to find novel pullulanase sequences from rare prokaryotes, for instance, Candidatus Sericytochromatia and Candidatus Roseilinea. Our team is currently analysing the high-quality genomes generated by metagenome-assembled genomes (MAGs) using short-reads from Illumina. Besides, Oxford Nanopore and HiFi PacBio long-reads sequencing will soon become a common practice for MAGS. The MAGs strategy might extend our ability to mine novel pullulan-degrading enzymes.
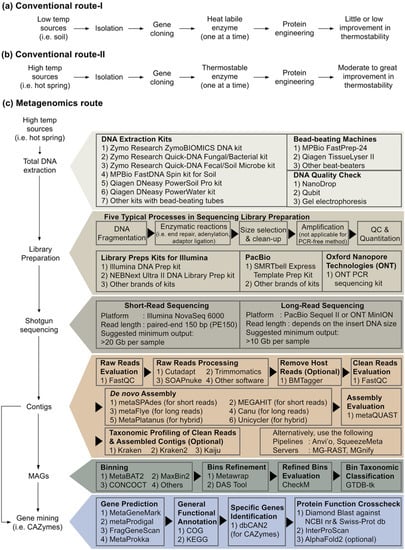
Figure 5.
Research flow chart to find novel active and thermostable pullulan-degrading enzymes: (a) Conventional route using a low or moderate temperature initial resources (i.e., soil or water). Low throughput approach. (b) Conventional path using a high-temperature initial resource (i.e., hot spring, vent, etc.). Low throughput approach but may obtain excellent thermostable enzymes. (c) Metagenomic-driven route using high-temperature initial resources. High throughput, more costly and computationally intensive methodology.
Suppose searching for active and thermostable pullulan-degrading enzymes is the aim of our research. Despite the feasibility of protein engineering approach to design enzymes with improved thermostability and longer half-life, especially among the thermolabile origins, the engineered enzymes might be far from the temperature threshold required by the starch- and pullulan processing industries (Figure 5a). Harness native biocatalyst from a large pool of metagenome-derived enzymes, particularly from samples such as high-temperature hot springs, is perhaps a better route than the protein engineering approach (Figure 5c). Biologists may be unacquainted with the advanced bioinformatic tools summarised in Figure 5c. In addition, from our own experience and the limited examples of pullulan-degrading enzymes [155,156,157,158,159], metagenome-derived enzymes may exhibit low sequence similarity to patented sequences; therefore, it is of great advantage from a legal perspective when it comes to commercialisation (refer to Section 2.2).
4. Conclusions
This current review contained two main parts, which are: (1) bibliometric analyses on publications and patents; and (2) the scientific review of pullulan-degrading enzymes. We believe that the understanding of type I and type II pullulanases is reaching saturation, since enzymology research on these groups of enzymes was initiated decades ago. We deduced from the bibliometric data that some earlier prominent researchers are losing their interest in this topic. The recommendation of the following articles may be part of the complete solutions to make another breakthrough [150,161]. Studies on pullulan hydrolase types I, II and III have not reached any significant discoveries lately. The number of reported genes encoding pullulan hydrolases is more petite in total quantity, probably because microorganisms prefer to conserve the type I and type II pullulanases. We also made a review on single- and co-enzymes immobilisations. In short, the numbers of immobilisation studies is relatively low compared to their counterparts, such as glucoamylase and α-amylase. Co-immobilization of pullulan-degrading enzymes with other CAZymes is still a considerable challenge, particularly in maintaining a suitable enzyme uniformity and increasing the enzyme activities, stabilities or product specificities. We believe that fellow scientists and engineers’ dedication to developing a more affordable, reliable and reusable metal-organic frameworks (MOFs)-based carrier [162] will shed new light on single- and co-immobilisation research of pullulan-degrading enzymes. NGS-based amplicon sequencing (metataxonomic) has become a standard tool in understanding the microbial population. Another NGS approach, whole metagenomic sequencing, is relatively more costly than metataxonomic. However, with a significant reduction of sequencing price, outsourcing metagenomic sequencing is now a common practice in moderate to rich laboratories. Although this current review did not include metagenome-assembled genomes (MAGs) in detail, this tool allows us to peep the genomes of many unculturable microorganisms and possibly mine their genes, such as pullulan-degrading enzymes. The publication on MAGs is growing; however, only a handful of researchers apply it to discover pullulan-, starch- or lignocellulose-hydrolytic enzymes, thus placing little focus on bibliometric records. Shotgun metagenomic pairing with MAGs requires extensive bioinformatics effort. From the co-occurrence bibliometric network map described earlier, publications are missing keywords related to NGS, MAGs and bioinformatics pipelines. We believe most biologists or enzymologists may not be well-versed in bioinformatics analyses involving command lines. International collaboration is highly advised to overcome this limitation; we anticipate that the paradigm will change soon.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/catal12020143/s1. Table S1: List of sources and biochemical characteristics of purified microbial type I pullulanases, Table S2: List of sources and biochemical characteristics of purified microbial type II pullulanases and Table S3: List of sources and biochemical characteristics of purified microbial pullulan hydrolases.
Author Contributions
Conceptualization and writing original draft preparation, U.M.K. and K.M.G.; review and editing, K.J.L., N.A.L. and S.I.A.; methodology, K.J.L., U.M.K., K.M.G. and S.I.A.; software, N.A.L. and K.M.G.; validation, U.M.K.; and data curation and figures preparation, U.M.K., K.J.L. and K.M.G. All authors have read and agreed to the published version of the manuscript.
Funding
U.M.K. is grateful for funding received from Malaysia FRGS grant (FRGS/1/2020/WAB11/MESTECC/02/1) and National Institutes of Biotechnology Malaysia (NIBM) Seed Fund Grant 2021 (NIBM/SF/2021-13). This research was funded by Malaysia Fundamental Research Grant Scheme (FRGS) 5F241 and 5F245 with the respective reference code FRGS/1/2019/STG03/UTM/02/1 and FRGS/1/2019/STG04/UTM/02/4.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Singh, R.S.; Kaur, N.; Rana, V.; Kennedy, J.F. Pullulan: A novel molecule for biomedical applications. Carbohydr. Polym. 2017, 171, 102–121. [Google Scholar] [CrossRef]
- Singh, R.S.; Saini, G.K.; Kennedy, J.F. Pullulan: Microbial sources, production and applications. Carbohydr. Polym. 2008, 73, 515–531. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chi, Z.; Wang, G.Y.; Wang, Z.P.; Liu, G.L.; Lee, C.F.; Ma, Z.C.; Chi, Z.M. Taxonomy of Aureobasidium spp. and biosynthesis and regulation of their extracellular polymers. Crit. Rev. Microbiol. 2015, 41, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Sugumaran, K.R.; Ponnusami, V. Review on production, downstream processing and characterization of microbial pullulan. Carbohydr. Polym. 2017, 173, 573–591. [Google Scholar]
- Prajapati, V.D.; Jani, G.K.; Khanda, S.M. Pullulan: An exopolysaccharide and its various applications. Carbohydr. Polym. 2013, 95, 540–549. [Google Scholar] [CrossRef]
- Wu, S.J.; Chen, J. Preparation of maltotriose from fermentation broth by hydrolysis of pullulan using pullulanase. Carbohydr. Polym. 2014, 107, 94–97. [Google Scholar] [CrossRef]
- Klein, M.N.; Kupper, K.C. Biofilm production by Aureobasidium pullulans improves biocontrol against sour rot in citrus. Food Microbiol. 2018, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kimoto, T.; Shibuya, T.; Shiobara, S. Safety studies of a novel starch, pullulan: Chronic toxicity in rats and bacterial mutagenicity. Food Chem. Toxicol. 1997, 35, 323–329. [Google Scholar] [CrossRef]
- Cheng, K.C.; Demirci, A.; Catchmark, J.M. Pullulan: Biosynthesis, production, and applications. Appl. Microbiol. Biotechnol. 2011, 92, 29–44. [Google Scholar] [CrossRef]
- Singh, R.S.; Kaur, N.; Rana, V.; Kennedy, J.F. Recent insights on applications of pullulan in tissue engineering. Carbohydr. Polym. 2016, 153, 455–462. [Google Scholar] [CrossRef]
- Nisha, M.; Satyanarayana, T. Characteristics, protein engineering and applications of microbial thermostable pullulanases and pullulan hydrolases. Appl. Microbiol. Biotechnol. 2016, 100, 5661–5679. [Google Scholar] [CrossRef] [PubMed]
- Patel, A.K.; Singhania, R.R.; Pandey, A. Novel enzymatic processes applied to the food industry. Curr. Opin. Food Sci. 2016, 7, 64–72. [Google Scholar] [CrossRef]
- Buchholz, K.; Seibel, J. Industrial carbohydrate biotransformations. Carbohydr. Res. 2008, 343, 1966–1979. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Saini, G.K.; Kennedy, J.F. Maltotriose syrup preparation from pullulan using pullulanase. Carbohydr. Polym. 2010, 80, 401–407. [Google Scholar] [CrossRef]
- Mishra, B.; Manikanta, A.; Zamare, D. Preparation of maltotriose syrup from microbial pullulan by using pullulanase enzyme. Biosci. Biotechnol. Res. Asia 2016, 13, 481–485. [Google Scholar] [CrossRef]
- Bertoldo, C.; Antranikian, G. Starch-hydrolyzing enzymes from thermophilic archaea and bacteria. Curr. Opin. Chem. Biol. 2002, 6, 151–160. [Google Scholar] [CrossRef]
- Gupta, R.; Gigras, P.; Mohapatra, H.; Goswami, V.K.; Chauhan, B. Microbial α-amylases: A biotechnological perspective. Process Biochem. 2003, 38, 1599–1616. [Google Scholar] [CrossRef]
- Ara, K.; Saeki, K.; Igarashi, K.; Takaiwa, M.; Uemura, T.; Hagihara, H.; Kawai, S.; Ito, S. Purification and characterization of an alkaline amylopullulanase with both α-1,4 and α-1,6 hydrolytic activity from alkalophilic Bacillus sp. KSM-1378. BBA-Gen. Subj. 1995, 1243, 315–324. [Google Scholar] [CrossRef]
- Rajaei, S.; Noghabi, K.A.; Sadeghizadeh, M.; Zahiri, H.S. Characterization of a pH and detergent-tolerant, cold-adapted type I pullulanase from Exiguobacterium sp. SH3. Extremophiles 2015, 19, 1145–1155. [Google Scholar] [CrossRef]
- Rajdevi, K.P.; Yogeeswaran, G. Cooperativity and substrate specificity of an alkaline amylase and neopullulanase complex of Micrococcus halobius OR-1. Appl. Biochem. Biotechnol.-Part A Enzym. Eng. Biotechnol. 2001, 90, 233–250. [Google Scholar] [CrossRef]
- Domań-Pytka, M.; Bardowski, J. Pullulan degrading enzymes of bacterial origin. Crit. Rev. Microbiol. 2004, 30, 107–121. [Google Scholar] [CrossRef]
- Haki, G.D.; Rakshit, S.K. Developments in industrially important thermostable enzymes: A review. Bioresour. Technol. 2003, 89, 17–34. [Google Scholar] [CrossRef]
- Hii, S.L.; Tan, J.S.; Ling, T.C.; Ariff, A. Bin Pullulanase: Role in starch hydrolysis and potential industrial applications. Enzym. Res. 2012, 2012, 921362. [Google Scholar]
- Xu, P.; Zhang, S.Y.; Luo, Z.G.; Zong, M.H.; Li, X.X.; Lou, W.Y. Biotechnology and bioengineering of pullulanase: State of the art and perspectives. World J. Microbiol. Biotechnol. 2021, 37, 1–10. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, S.; Lv, M. Structural and functional analysis of GH57 family thermostable amylopullulanase—A review. Wei Sheng Wu Xue Bao 2011, 51, 21–28. [Google Scholar] [PubMed]
- Xia, W.; Zhang, K.; Su, L.; Wu, J. Microbial starch debranching enzymes: Developments and applications. Biotechnol. Adv. 2021, 50, 107786. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Nie, Y.; Xu, Y. Industrially produced pullulanases with thermostability: Discovery, engineering, and heterologous expression. Bioresour. Technol. 2019, 278, 360–371. [Google Scholar] [CrossRef]
- Guo, J.; Coker, A.R.; Wood, S.P.; Cooper, J.B.; Keegan, R.M.; Ahmad, N.; Muhamma, M.A.; Rashid, N.; Akhtar, M. Structure and function of the type III pullulan hydrolase from Thermococcus kodakarensis. Acta Crystallogr. Sect. D Struct. Biol. 2018, 74, 305–314. [Google Scholar] [CrossRef]
- Ryan, S.M.; Fitzgerald, G.F.; Van Sinderen, D. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl. Environ. Microbiol. 2006, 72, 5289–5296. [Google Scholar] [CrossRef]
- Ahmad, N.; Rashid, N.; Haider, M.S.; Akram, M.; Akhtar, M. Novel maltotriose-hydrolyzing thermoacidophilic type III pullulan hydrolase from Thermococcus kodakarensisxs. Appl. Environ. Microbiol. 2014, 80, 1108–1115. [Google Scholar] [CrossRef]
- Zeng, J.; He, C.; Guo, J.; Yuan, L. Effect of truncation of N-Terminal structural modules on enzymatic properties of thermoacidiphilic type III pullulan hydrolase TK-PUL. Shipin Kexue/Food Sci. 2021, 42, 201–208. [Google Scholar]
- Niehaus, F.; Peters, A.; Groudieva, T.; Antranikian, G. Cloning, expression and biochemical characterisation of a unique thermostable pullulan-hydrolysing enzyme from the hyperthermophilic archaeon Thermococcus aggregans. FEMS Microbiol. Lett. 2000, 190, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Rashid, N.; Shehzad, A.; Ahmad, N.; Hussain, Z.; Haider, M.S. Valorization of waste foods using pullulan hydrolase from Thermococcus kodakarensis. Amylase 2019, 2, 39–43. [Google Scholar] [CrossRef]
- Møller, M.S.; Henriksen, A.; Svensson, B. Structure and function of α-glucan debranching enzymes. Cell. Mol. Life Sci. 2016, 73, 2619–2641. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.; Imai, T. Bacillus stearothermophilus KP 1064 pullulan hydrolase—Its assignment to a unique type of maltogenic α-amylase but to neither pullulanase nor isopullulanase. Appl. Microbiol. Biotechnol. 1985, 21, 20–26. [Google Scholar]
- Toor, K.J.; Ahmad, N.; Muhammad, M.A.; Rashid, N. TK-PUL, a pullulan hydrolase type III from Thermococcus kodakarensis, a potential candidate for simultaneous liquefaction and saccharification of starch. Amylase 2021, 4, 45–55. [Google Scholar] [CrossRef]
- Akeboshi, H.; Tonozuka, T.; Furukawa, T.; Ichikawa, K.; Aoki, H.; Shimonishi, A.; Nishikawa, A.; Sakano, Y. Insights into the reaction mechanism of glycosyl hydrolase family 49: Site-directed mutagenesis and substrate preference of isopullulanase. Eur. J. Biochem. 2004, 271, 4420–4427. [Google Scholar] [CrossRef]
- Aoki, H.; Yopi; Sakano, Y. Molecular cloning and heterologous expression of the isopullulanase gene from Aspergillus niger A.T.C.C. 9642. Biochem. J. 1997, 323, 757–764. [Google Scholar] [CrossRef][Green Version]
- Mizuno, M.; Koide, A.; Yamamura, A.; Akeboshi, H.; Yoshida, H.; Kamitori, S.; Sakano, Y.; Nishikawa, A.; Tonozuka, T. Crystal Structure of Aspergillus niger Isopullulanase, a Member of Glycoside Hydrolase Family 49. J. Mol. Biol. 2008, 376, 210–220. [Google Scholar] [CrossRef]
- Kamasaka, H.; Sugimoto, K.; Takata, H.; Nishimura, T.; Kuriki, T. Bacillus stearothermophilus neopullulanase selective hydrolysis of amylose to maltose in the presence of amylopectin. Appl. Environ. Microbiol. 2002, 68, 1658–1664. [Google Scholar] [CrossRef][Green Version]
- Jung, J.H.; An, Y.K.; Son, S.Y.; Jeong, S.Y.; Seo, D.H.; Kim, M.K.; Park, C.S. Characterization of a novel extracellular α-amylase from Ruminococcus bromii ATCC 27255 with neopullulanase-like activity. Int. J. Biol. Macromol. 2019, 130, 605–614. [Google Scholar] [CrossRef] [PubMed]
- van Eck, N.J.; Waltman, L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics 2010, 84, 523–538. [Google Scholar] [CrossRef] [PubMed]
- Waśko, A.; Polak-Berecka, M.; Targoński, Z. Purification and characterization of pullulanase from Lactococcus lactis. Prep. Biochem. Biotechnol. 2011, 41, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Kunamneni, A.; Singh, S. Improved high thermal stability of pullulanase from a newly isolated thermophilic Bacillus sp. AN-7. Enzym. Microb. Technol. 2006, 39, 1399–1404. [Google Scholar] [CrossRef]
- Xu, J.; Ren, F.; Huang, C.H.; Zheng, Y.; Zhen, J.; Sun, H.; Ko, T.P.; He, M.; Chen, C.C.; Chan, H.C.; et al. Functional and structural studies of pullulanase from Anoxybacillus sp. LM18-11. Proteins Struct. Funct. Bioinform. 2014, 82, 1685–1693. [Google Scholar] [CrossRef]
- Yang, S.; Yan, Q.; Bao, Q.; Liu, J.; Jiang, Z. Expression and biochemical characterization of a novel type I pullulanase from Bacillus megaterium. Biotechnol. Lett. 2017, 39, 397–405. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Fu, J.; Pan, Y.; Li, D. Sequence analysis and biochemical properties of an acidophilic and hyperthermophilic amylopullulanase from Thermofilum pendens. Int. J. Biol. Macromol. 2018, 114, 235–243. [Google Scholar] [CrossRef]
- Kahar, U.M.; Chan, K.-G.; Salleh, M.M.; Hii, S.M.; Goh, K.M. A High Molecular-Mass Anoxybacillus sp. SK3-4 Amylopullulanase: Characterization and Its Relationship in Carbohydrate Utilization. Int. J. Mol. Sci. 2013, 14, 11302–11318. [Google Scholar] [CrossRef]
- Mesbah, N.M.; Wiegel, J. Biochemical characterization of halophilic, alkalithermophilic amylopullulanase PulD7 and truncated amylopullulanases PulD7ΔN and PulD7ΔC. Int. J. Biol. Macromol. 2018, 111, 632–638. [Google Scholar] [CrossRef]
- Roodi, F.Z.; Aminzadeh, S.; Farrokhi, N.; Karkhane, A.A.; Haghbeen, K. Cohnella amylopullulanases: Biochemical characterization of two recombinant thermophilic enzymes. PLoS ONE 2017, 12, e0175013. [Google Scholar]
- Vishnu, C.; Naveena, B.J.; Altaf, M.; Venkateshwar, M.; Reddy, G. Amylopullulanase—A novel enzyme of L. amylophilus GV6 in direct fermentation of starch to L(+) lactic acid. Enzym. Microb. Technol. 2006, 38, 545–550. [Google Scholar] [CrossRef]
- Motherway, M.O.C.; Fitzgerald, G.F.; Neirynck, S.; Ryan, S.; Steidler, L.; Van Sinderen, D. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2008, 74, 6271–6279. [Google Scholar] [CrossRef] [PubMed]
- Ferrando, M.L.; Fuentes, S.; De Greeff, A.; Smith, H.; Wells, J.M. ApuA, a multifunctional α-glucan-degrading enzyme of Streptococcus suis, mediates adhesion to porcine epithelium and mucus. Microbiology 2010, 156, 2818–2828. [Google Scholar] [CrossRef]
- Kim, J.H.; Sunako, M.; Ono, H.; Murooka, Y.; Fukusaki, E.; Yamashita, M. Characterization of gene encoding amylopullulanase from plant-originated lactic acid bacterium, Lactobacillus plantarum L137. J. Biosci. Bioeng. 2008, 106, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Lin, Y.; Wang, G.; Lin, J. Gene cloning, expression and biochemical characterization of a new multi-domain, halotolerant and SDS-resistant alkaline pullulanase from Alkalibacterium sp. SL3. Process Biochem. 2020, 96, 1–10. [Google Scholar] [CrossRef]
- Roy, A.; Messaoud, E.B.; Bejar, S. Isolation and purification of an acidic pullulanase type II from newly isolated Bacillus sp. US149. Enzym. Microb. Technol. 2003, 33, 720–724. [Google Scholar] [CrossRef]
- Igarashi, K.; Ara, K.; Saeki, K.; Ozaki, K.; Kawai, S.; Ito, S. Nucleotide sequence of the gene that encodes a neopullulanase from an alkalophilic Bacillus. Biosci. Biotechnol. Biochem. 1992, 56, 514–516. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Van Der Maarel, M.J.E.C.; Van Der Veen, B.; Uitdehaag, J.C.M.; Leemhuis, H.; Dijkhuizen, L. Properties and applications of starch-converting enzymes of the α-amylase family. J. Biotechnol. 2002, 94, 137–155. [Google Scholar] [CrossRef]
- Christiansen, C.; Abou Hachem, M.; Janeček, Š.; Viksø-Nielsen, A.; Blennow, A.; Svensson, B. The carbohydrate-binding module family 20—Diversity, structure, and function. FEBS J. 2009, 276, 5006–5029. [Google Scholar] [CrossRef]
- Kim, S.-Y.; Kim, H.; Kim, Y.-J.; Jung, D.-H.; Seo, D.-H.; Jung, J.-H.; Park, C.-S. Enzymatic analysis of truncation mutants of a type II pullulanase from Bifidobacterium adolescentis P2P3, a resistant starch-degrading gut bacterium. Int. J. Biol. Macromol. 2021, in press. [Google Scholar] [CrossRef]
- Robert, X.; Haser, R.; Gottschalk, T.E.; Ratajczak, F.; Driguez, H.; Svensson, B.; Aghajari, N. The structure of barley α-amylase isozyme 1 reveals a novel role of domain C in substrate recognition and binding: A pair of sugar tongs. Structure 2003, 11, 973–984. [Google Scholar] [CrossRef]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A. Characteristic features and biotechnological applications of cross-linked enzyme aggregates (CLEAs). Appl. Microbiol. Biotechnol. 2011, 92, 467–477. [Google Scholar] [CrossRef]
- Eş, I.; Vieira, J.D.G.; Amaral, A.C. Principles, techniques, and applications of biocatalyst immobilization for industrial application. Appl. Microbiol. Biotechnol. 2015, 99, 2065–2082. [Google Scholar] [CrossRef] [PubMed]
- Vaghari, H.; Jafarizadeh-Malmiri, H.; Mohammadlou, M.; Berenjian, A.; Anarjan, N.; Jafari, N.; Nasiri, S. Application of magnetic nanoparticles in smart enzyme immobilization. Biotechnol. Lett. 2016, 38, 223–233. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Galan, C.; Berenguer-Murcia, Á.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Sheldon, R.A. Cross-linked enzyme aggregates (CLEA®s): Stable and recyclable biocatalysts. Biochem. Soc. Trans. 2007, 35, 1583–1587. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Basso, A.; Brady, D. New frontiers in enzyme immobilisation: Robust biocatalysts for a circular bio-based economy. Chem. Soc. Rev. 2021, 50, 5850–5862. [Google Scholar] [CrossRef]
- Bernal, C.; Rodríguez, K.; Martínez, R. Integrating enzyme immobilization and protein engineering: An alternative path for the development of novel and improved industrial biocatalysts. Biotechnol. Adv. 2018, 36, 1470–1480. [Google Scholar] [CrossRef]
- Mylkie, K.; Nowak, P.; Rybczynski, P.; Ziegler-Borowska, M. Polymer-coated magnetite nanoparticles for protein immobilization. Materials 2021, 14, 248. [Google Scholar] [CrossRef] [PubMed]
- Polaina, J.; MacCabe, A.P. Industrial Enzymes: Structure, Function and Applications; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Meryam Sardar, R.A. Enzyme Immobilization: An Overview on Nanoparticles as Immobilization Matrix. Biochem. Anal. Biochem. 2015, 4, 1. [Google Scholar] [CrossRef]
- Hwang, E.T.; Lee, S. Multienzymatic Cascade Reactions via Enzyme Complex by Immobilization. ACS Catal. 2019, 9, 4402–4425. [Google Scholar] [CrossRef]
- Gennari, A.; Führ, A.J.; Volpato, G.; Volken de Souza, C.F. Magnetic cellulose: Versatile support for enzyme immobilization—A review. Carbohydr. Polym. 2020, 246, 116646. [Google Scholar] [CrossRef]
- Mateo, C.; Grazú, V.; Pessela, B.C.C.; Montes, T.; Palomo, J.M.; Torres, R.; López-Gallego, F.; Fernández-Lafuente, R.; Guisán, J.M. Advances in the design of new epoxy supports for enzyme immobilization-stabilization. Biochem. Soc. Trans. 2007, 35, 1593–1601. [Google Scholar] [CrossRef]
- Contesini, F.J.; Figueira, J.d.A.; Kawaguti, H.Y.; Fernandes, P.C.d.B.; Carvalho, P.d.O.; Nascimento, M.d.G.; Sato, H.H. Potential applications of carbohydrases immobilization in the food industry. Int. J. Mol. Sci. 2013, 14, 1335–1369. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Zhou, Z. Regulation of the catalytic behavior of pullulanases chelated onto nickel (II)-modified magnetic nanoparticles. Enzym. Microb. Technol. 2017, 101, 9–16. [Google Scholar] [CrossRef]
- Kuriki, T.; Tsuda, M.; Imanaka, T. Continuous production of panose by immobilized neopullulanase. J. Ferment. Bioeng. 1992, 73, 198–202. [Google Scholar] [CrossRef]
- Lenders, J.-P.; Crichton, R.R. Thermal stabilization of amylolytic enzymes by covalent coupling to soluble polysaccharides. Biotechnol. Bioeng. 1984, 26, 1343–1351. [Google Scholar] [CrossRef]
- Ali, G.; Dulong, V.; Gasmi, S.N.; Rihouey, C.; Picton, L.; Le Cerf, D. Covalent immobilization of pullulanase on alginate and study of its hydrolysis of pullulan. Biotechnol. Prog. 2015, 31, 883–889. [Google Scholar] [CrossRef]
- Wang, J.; Liu, Z.; Zhou, Z. Improving pullulanase catalysis via reversible immobilization on modified Fe3O4@polydopamine nanoparticles. Appl. Biochem. Biotechnol. 2017, 182, 1467–1477. [Google Scholar] [CrossRef]
- Long, J.; Li, X.; Zhan, X.; Xu, X.; Tian, Y.; Xie, Z.; Jin, Z. Sol–gel encapsulation of pullulanase in the presence of hybrid magnetic (Fe3O4–chitosan) nanoparticles improves thermal and operational stability. Bioprocess Biosyst. Eng. 2017, 40, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef] [PubMed]
- Cowan, D.A.; Fernandez-Lafuente, R. Enhancing the functional properties of thermophilic enzymes by chemical modification and immobilization. Enzym. Microb. Technol. 2011, 49, 326–346. [Google Scholar] [CrossRef] [PubMed]
- Kahar, U.M.; Chan, K.G.; Sani, M.H.; Mohd Noh, N.I.; Goh, K.M. Effects of single and co-immobilization on the product specificity of type I pullulanase from Anoxybacillus sp. SK3-4. Int. J. Biol. Macromol. 2017, 104, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Ji, Q.; Wang, B.; Tan, J.; Zhu, L.; Li, L. Immobilized multienzymatic systems for catalysis of cascade reactions. Process Biochem. 2016, 51, 1193–1203. [Google Scholar] [CrossRef]
- Schoffelen, S.; Van Hest, J.C.M. Chemical approaches for the construction of multi-enzyme reaction systems. Curr. Opin. Struct. Biol. 2013, 23, 613–621. [Google Scholar] [CrossRef]
- Talekar, S.; Pandharbale, A.; Ladole, M.; Nadar, S.; Mulla, M.; Japhalekar, K.; Pattankude, K.; Arage, D. Carrier free co-immobilization of alpha amylase, glucoamylase and pullulanase as combined cross-linked enzyme aggregates (combi-CLEAS): A tri-enzyme biocatalyst with one pot starch hydrolytic activity. Bioresour. Technol. 2013, 147, 269–275. [Google Scholar] [CrossRef]
- Xue, R.; Woodley, J.M. Process technology for multi-enzymatic reaction systems. Bioresour. Technol. 2012, 115, 183–195. [Google Scholar] [CrossRef]
- Kusano, S.; Shiraishi, T.; Takahashi, S.I.; Fujimoto, D.; Sakano, Y. Immobilization of Bacillus acidopullulyticus pullulanase and properties of the immobilized pullulanases. J. Ferment. Bioeng. 1989, 68, 233–237. [Google Scholar] [CrossRef]
- Dessouki, A.M.; Issa, G.I.; Atia, K.S. Pullulanase immobilization on natural and synthetic polymers. J. Chem. Technol. Biotechnol. 2011, 76, 700–706. [Google Scholar] [CrossRef]
- Kuroiwa, T.; Shoda, H.; Ichikawa, S.; Sato, S.; Mukataka, S. Immobilization and stabilization of pullulanase from Klebsiella pneumoniae by a multipoint attachment method using activated agar gel supports. Process Biochem. 2005, 40, 2637–2642. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, X.; Zheng, S.; Sun, H. Photochemical preparation of magnetic chitosan beads for immobilization of pullulanase. Biochem. Eng. J. 2009, 46, 83–87. [Google Scholar] [CrossRef]
- Singh, R.S.; Saini, G.K.; Kennedy, J.F. Covalent immobilization and thermodynamic characterization of pullulanase for the hydrolysis of pullulan in batch system. Carbohydr. Polym. 2010, 81, 252–259. [Google Scholar] [CrossRef]
- Long, J.; Jiao, A.; Wei, B.; Wu, Z.; Zhang, Y.; Xu, X.; Jin, Z. A novel method for pullulanase immobilized onto magnetic chitosan/Fe3O4 composite nanoparticles by in situ preparation and evaluation of the enzyme stability. J. Mol. Catal. B Enzym. 2014, 109, 53–61. [Google Scholar] [CrossRef]
- Long, J.; Li, X.; Wu, Z.; Xu, E.; Xu, X.; Jin, Z.; Jiao, A. Immobilization of pullulanase onto activated magnetic chitosan/Fe3O4 nanoparticles prepared by in situ mineralization and effect of surface functional groups on the stability. Colloids Surf. A Physicochem. Eng. Asp. 2015, 472, 69–77. [Google Scholar] [CrossRef]
- Alagöz, D.; Yildirim, D.; Güvenmez, H.K.; Sihay, D.; Tükel, S.S. Covalent immobilization and characterization of a novel pullulanase from Fontibacillus sp. strain DSHK 107 onto Florisil® and nano-silica for pullulan hydrolysis. Appl. Biochem. Biotechnol. 2016, 179, 1262–1274. [Google Scholar] [CrossRef]
- Ohba, R.; Chaen, H.; Hayashi, S.; Ueda, S. Immobilization of Streptomyces flavochromogenes pullulanase on tannic acid and TEAE–cellulose. Biotechnol. Bioeng. 1978, 20, 665–676. [Google Scholar] [CrossRef]
- Jiang, M.; Zhang, L.; Wang, F.; Zhang, J.; Liu, G.; Gao, B.; Wei, D. Novel application of magnetic protein: Convenient one-step purification and immobilization of proteins. Sci. Rep. 2017, 7, 13329. [Google Scholar] [CrossRef]
- Ohtani, N.; Ishidao, T.; Iwai, Y.; Arai, Y. Hydrolysis of starch or pullulan by glucoamylase or pullulanase immobilized on poly(N-isopropylacrylamide) gel. Colloid Polym. Sci. 1999, 277, 197–202. [Google Scholar] [CrossRef]
- Roy, I.; Gupta, M.N. Hydrolysis of starch by a mixture of glucoamylase and pullulanase entrapped individually in calcium alginate beads. Enzym. Microb. Technol. 2004, 34, 26–32. [Google Scholar] [CrossRef]
- Long, J.; Wu, Z.; Li, X.; Xu, E.; Xu, X.; Jin, Z.; Jiao, A. New method for the immobilization of pullulanase onto hybrid magnetic (Fe3O4-κ-carrageenan) nanoparticles by electrostatic coupling with pullulanase/chitosan complex. J. Agric. Food Chem. 2015, 63, 3534–3542. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Ma, X.; Han, Y.; Wei, Y.; Guo, Q.; Yang, S.; Zhang, Y.; Liao, W.; Gao, Y. Effect of chitosan molecular weight on zein-chitosan nanocomplexes: Formation, characterization, and the delivery of quercetagetin. Int. J. Biol. Macromol. 2020, 164, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, F.; Lin, L.; He, D.; Chen, J.; Wei, W.; Wei, D. Biochemical characterization of a novel thermostable type I pullulanase produced recombinantly in Bacillus subtilis. Starch/Staerke 2018, 70, 1700179. [Google Scholar] [CrossRef]
- Long, J.; Zhang, B.; Li, X.; Zhan, X.; Xu, X.; Xie, Z.; Jin, Z. Effective production of resistant starch using pullulanase immobilized onto magnetic chitosan/Fe3O4 nanoparticles. Food Chem. 2018, 239, 276–286. [Google Scholar] [CrossRef]
- Long, J.; Li, X.; Liu, X.; Jin, Z.; Xie, Z.; Xu, X.; Lu, C. Preparation of streptavidin-coated magnetic nanoparticles for specific immobilization of enzymes with high activity and enhanced stability. Ind. Eng. Chem. Res. 2021, 60, 1542–1552. [Google Scholar] [CrossRef]
- Ram, K.A.; Venkatasubramanian, K. Enhancement of starch conversion efficiency with free and immobilized pullulanase and α-1,4-glucosidase. Biotechnol. Bioeng. 1982, 24, 355–369. [Google Scholar] [CrossRef]
- Chakrabarti, A.C.; Storey, K.B. Co-immobilization of amyloglucosidase and pullulanase for enhanced starch hydrolysis. Appl. Microbiol. Biotechnol. 1990, 33, 48–50. [Google Scholar] [CrossRef]
- Atia, K.S.; Ismail, S.A.; El-Arnaouty, M.B.; Dessouki, A.M. Use of Co-immobilized β-amylase and pullulanase in reduction of saccharification time of starch and increase in maltose yield. Biotechnol. Prog. 2003, 19, 853–857. [Google Scholar] [CrossRef]
- Talekar, S.; Joshi, A.; Kambale, S.; Jadhav, S.; Nadar, S.; Ladole, M. A tri-enzyme magnetic nanobiocatalyst with one pot starch hydrolytic activity. Chem. Eng. J. 2017, 325, 80–90. [Google Scholar] [CrossRef]
- Yoshida, M.; Oishi, K.; Kimura, T.; Ogata, M.; Nakakuki, T. Continuous production of maltose using a dual immobilized enzyme system. Agric. Biol. Chem. 1989, 53, 3139–3142. [Google Scholar] [CrossRef]
- Kimura, T.; Ogata, M.; Kobayashi, H.; Yoshida, M.; Oishi, K.; Nakakuki, T. Continuous production of maltotetraose using a dual immobilized enzyme system of maltotetraose-forming amylase and pullulanase. Biotechnol. Bioeng. 1990, 36, 790–796. [Google Scholar] [CrossRef] [PubMed]
- Schafhagser, D.Y.; Storey, K.B. Fructose production: Coimmobilized amyloglucosidase, pullulanase, and glucose isomerase on BIOBONETM. Appl. Biochem. Biotechnol. Part A Enzym. Eng. Biotechnol. 1992, 36, 63–74. [Google Scholar] [CrossRef]
- Noda, T.; Furuta, S.; Suda, I. Sweet potato β-amylase immobilized on chitosan beads and its application in the semi-continuous production of maltose. Carbohydr. Polym. 2001, 44, 189–195. [Google Scholar] [CrossRef]
- Talekar, S.; Desai, S.; Pillai, M.; Nagavekar, N.; Ambarkar, S.; Surnis, S.; Ladole, M.; Nadar, S.; Mulla, M. Carrier free co-immobilization of glucoamylase and pullulanase as combi-cross linked enzyme aggregates (combi-CLEAs). RSC Adv. 2013, 3, 2265–2271. [Google Scholar] [CrossRef]
- Goh, K.M.; Hong, G.P.; Chyi, N.H.; Ng, P.; Piaw, C.K.; Rahman, R.N.Z.R.A. Trends and tips in protein engineering, a review. J. Teknol. 2012, 59, 21–31. [Google Scholar]
- Ece, S.; Evran, S.; Janda, J.O.; Merkl, R.; Sterner, R. Improving thermal and detergent stability of Bacillus stearothermophilus neopullulanase by rational enzyme design. Protein Eng. Des. Sel. 2014, 28, 147–151. [Google Scholar] [CrossRef]
- Miyazaki, T.; Yashiro, H.; Nishikawa, A.; Tonozuka, T. The side chain of a glycosylated asparagine residue is important for the stability of isopullulanase. J. Biochem. 2015, 157, 225–234. [Google Scholar] [CrossRef]
- Zeng, Y.; Zheng, H.; Shen, Y.; Xu, J.; Tan, M.; Liu, F.; Song, H. Identification and analysis of binding residues in the CBM68 of pullulanase PulA from Anoxybacillus sp. LM18-11. J. Biosci. Bioeng. 2019, 127, 8–15. [Google Scholar] [CrossRef]
- Li, S.F.; Xu, J.Y.; Bao, Y.J.; Zheng, H.C.; Song, H. Structure and sequence analysis-based engineering of pullulanase from Anoxybacillus sp. LM18-11 for improved thermostability. J. Biotechnol. 2015, 210, 8–14. [Google Scholar] [CrossRef]
- Duan, X.; Chen, J.; Wu, J. Improving the thermostability and catalytic efficiency of Bacillus deramificans pullulanase by site-directed mutagenesis. Appl. Environ. Microbiol. 2013, 79, 4072–4077. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Chu, X.; Lv, J.; Li, Q.; Tian, J.; Wu, N. Improving the thermostability of acidic pullulanase from bacillus naganoensis by rational design. PLoS ONE 2016, 11, e0165006. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.Y.; Xie, N.Z.; Du, Q.S.; Qin, Y.; Li, J.X.; Meng, J.Z.; Huang, R.B. Active hydrogen bond network (AHBN) and applications for improvement of thermal stability and pH-sensitivity of pullulanase from Bacillus naganoensis. PLoS ONE 2017, 12, e0169080. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Nie, Y.; Mu, X.; Xu, Y.; Xiao, R. Disorder prediction-based construct optimization improves activity and catalytic efficiency of Bacillus naganoensis pullulanase. Sci. Rep. 2016, 6, 24574. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dong, F.; Lin, L.; He, D.; Wei, W.; Wei, D. N-Terminal Domain Truncation and Domain Insertion-Based Engineering of a Novel Thermostable Type i Pullulanase from Geobacillus thermocatenulatus. J. Agric. Food Chem. 2018, 66, 10788–10798. [Google Scholar] [CrossRef]
- Bi, J.; Chen, S.; Zhao, X.; Nie, Y.; Xu, Y. Computation-aided engineering of starch-debranching pullulanase from Bacillus thermoleovorans for enhanced thermostability. Appl. Microbiol. Biotechnol. 2020, 104, 7551–7562. [Google Scholar] [CrossRef]
- Mu, G.C.; Nie, Y.; Mu, X.Q.; Xu, Y.; Xiao, R. Single Amino Acid Substitution in the Pullulanase of Klebsiella variicola for Enhancing Thermostability and Catalytic Efficiency. Appl. Biochem. Biotechnol. 2015, 176, 1736–1745. [Google Scholar] [CrossRef]
- Pang, B.; Zhou, L.; Cui, W.; Liu, Z.; Zhou, Z. Improvement of the Thermostability and Activity of Pullulanase from Anoxybacillus sp. WB42. Appl. Biochem. Biotechnol. 2020, 191, 942–954. [Google Scholar] [CrossRef]
- Nisha, M.; Satyanarayana, T. The role of N1 domain on the activity, stability, substrate specificity and raw starch binding of amylopullulanase of the extreme thermophile Geobacillus thermoleovorans. Appl. Microbiol. Biotechnol. 2015, 99, 5461–5474. [Google Scholar] [CrossRef]
- Nisha, M.; Satyanarayana, T. Characterization of recombinant amylopullulanase (gt-apu) and truncated amylopullulanase (gt-apuT) of the extreme thermophile Geobacillus thermoleovorans NP33 and their action in starch saccharification. Appl. Microbiol. Biotechnol. 2013, 97, 6279–6292. [Google Scholar] [CrossRef]
- Lin, F.P.; Ho, Y.H.; Lin, H.Y.; Lin, H.J. Effect of C-terminal truncation on enzyme properties of recombinant amylopullulanase from Thermoanaerobacter pseudoethanolicus. Extremophiles 2012, 16, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.Y.; Chuang, H.H.; Lin, F.P. Biochemical characterization of engineered amylopullulanase from Thermoanaerobacter ethanolicus 39E-implicating the non-necessity of its 100 C-terminal amino acid residues. Extremophiles 2008, 12, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Sunako, M.; Ono, H.; Murooka, Y.; Fukusaki, E.; Yamashita, M. Characterization of the C-terminal truncated form of amylopullulanase from Lactobacillus plantarum L137. J. Biosci. Bioeng. 2009, 107, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Kahar, U.M.; Ng, C.L.; Chan, K.-G.; Goh, K.M. Characterization of a type I pullulanase from Anoxybacillus sp. SK3-4 reveals an unusual substrate hydrolysis. Appl. Microbiol. Biotechnol. 2016, 100, 6291–6307. [Google Scholar] [CrossRef]
- Chen, A.; Xu, T.; Ge, Y.; Wang, L.; Tang, W.; Li, S. Hydrogen-bond-based protein engineering for the acidic adaptation of Bacillus acidopullulyticus pullulanase. Enzym. Microb. Technol. 2019, 124, 79–83. [Google Scholar] [CrossRef]
- Wang, X.; Jing, X.; Deng, Y.; Nie, Y.; Xu, F.; Xu, Y.; Zhao, Y.L.; Hunt, J.F.; Montelione, G.T.; Szyperski, T. Evolutionary coupling saturation mutagenesis: Coevolution-guided identification of distant sites influencing Bacillus naganoensis pullulanase activity. FEBS Lett. 2020, 594, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Bi, J.; Jing, X.; Wu, L.; Zhou, X.; Gu, J.; Nie, Y.; Xu, Y. Computational design of noncanonical amino acid-based thioether staples at N/C-terminal domains of multi-modular pullulanase for thermostabilization in enzyme catalysis. Comput. Struct. Biotechnol. J. 2021, 19, 577–585. [Google Scholar] [CrossRef]
- Wang, X.; Nie, Y.; Xu, Y. Improvement of the Activity and Stability of Starch-Debranching Pullulanase from Bacillus naganoensis via Tailoring of the Active Sites Lining the Catalytic Pocket. J. Agric. Food Chem. 2018, 66, 13236–13242. [Google Scholar] [CrossRef]
- Bava, K.A.; Gromiha, M.M.; Uedaira, H.; Kitajima, K.; Sarai, A. ProTherm, version 4.0: Thermodynamic database for proteins and mutants. Nucleic Acids Res. 2004, 32, D120–D121. [Google Scholar] [CrossRef]
- Montanucci, L.; Capriotti, E.; Frank, Y.; Ben-Tal, N.; Fariselli, P. DDGun: An untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinform. 2019, 20, 335. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Quan, L.; Lv, Q.; Zhang, Y. STRUM: Structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics 2016, 32, 2936–2946. [Google Scholar] [CrossRef]
- Pandurangan, A.P.; Ochoa-Montaño, B.; Ascher, D.B.; Blundell, T.L. SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017, 45, W229–W235. [Google Scholar] [CrossRef]
- Yang, Y.; Ding, X.; Zhu, G.; Niroula, A.; Lv, Q.; Vihinen, M. ProTstab—Predictor for cellular protein stability. BMC Genom. 2019, 20, 804–809. [Google Scholar] [CrossRef]
- Cao, H.; Wang, J.; He, L.; Qi, Y.; Zhang, J.Z. DeepDDG: Predicting the Stability Change of Protein Point Mutations Using Neural Networks. J. Chem. Inf. Model. 2019, 59, 1508–1514. [Google Scholar] [CrossRef]
- Chen, C.W.; Lin, M.H.; Liao, C.C.; Chang, H.P.; Chu, Y.W. iStable 2.0: Predicting protein thermal stability changes by integrating various characteristic modules. Comput. Struct. Biotechnol. J. 2020, 18, 622–630. [Google Scholar] [CrossRef]
- Yang, Y.; Urolagin, S.; Niroula, A.; Ding, X.; Shen, B.; Vihinen, M. Pon-tstab: Protein variant stability predictor. Importance of training data quality. Int. J. Mol. Sci. 2018, 19, 1009. [Google Scholar] [CrossRef]
- Sanavia, T.; Birolo, G.; Montanucci, L.; Turina, P.; Capriotti, E.; Fariselli, P. Limitations and challenges in protein stability prediction upon genome variations: Towards future applications in precision medicine. Comput. Struct. Biotechnol. J. 2020, 18, 1968–1979. [Google Scholar] [CrossRef]
- Goh, K.M.; Shahar, S.; Chan, K.-G.; Chong, C.S.; Amran, S.I.; Sani, M.H.; Zakaria, I.I.; Kahar, U.M. Current status and potential applications of underexplored prokaryotes. Microorganisms 2019, 7, 468. [Google Scholar] [CrossRef]
- Urbieta, M.S.; Donati, E.R.; Chan, K.-G.; Shahar, S.; Sin, L.L.; Goh, K.M. Thermophiles in the genomic era: Biodiversity, science, and applications. Biotechnol. Adv. 2015, 33, 633–647. [Google Scholar] [CrossRef]
- Kaushal, G.; Kumar, J.; Sangwan, R.S.; Singh, S.P. Metagenomic analysis of geothermal water reservoir sites exploring carbohydrate-related thermozymes. Int. J. Biol. Macromol. 2018, 119, 882–895. [Google Scholar] [CrossRef]
- Reichart, N.J.; Bowers, R.M.; Woyke, T. High potential for biomass-degrading enzymes revealed by hot spring metagenomics. Front. Microbiol. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Liew, K.J.; Lim, C.C.; Chan, C.S.; Wei, K.Y.; Salleh, M.M.; Sani, R.K.; Chan, K.G.; Goh, K.M. Direct Cellulase Gene Amplification from Hot Spring Using the Guidance of 16S rRNA Amplicon Metagenomics; Academic Press: London, UK, 2018; ISBN 9780128134030. [Google Scholar]
- Perez-Mon, C.; Qi, W.; Vikram, S.; Frossard, A.; Makhalanyane, T.; Cowan, D.; Frey, B. Shotgun metagenomics reveals distinct functional diversity and metabolic capabilities between 12,000-year-old permafrost and active layers on Muot da Barba Peider (Swiss Alps). Microb. Genom. 2021, 7, 000558. [Google Scholar]
- Sadeghian Motahar, S.F.; Salami, M.; Ariaeenejad, S.; Emam-Djomeh, Z.; Sheykh Abdollahzadeh Mamaghani, A.; Kavousi, K.; Moghadam, M.; Hosseini Salekdeh, G. Synergistic effect of metagenome-derived starch-degrading enzymes on quality of functional bread with antioxidant activity. Starch/Staerke 2021, 74, 1–2. [Google Scholar] [CrossRef]
- Thakur, M.; Sharma, N.; Rai, A.K.; Singh, S.P. A novel cold-active type I pullulanase from a hot-spring metagenome for effective debranching and production of resistant starch. Bioresour. Technol. 2021, 320, 124288. [Google Scholar] [CrossRef]
- Yarygin, K.; Tyakht, A.; Larin, A.; Kostryukova, E.; Kolchenko, S.; Bitner, V.; Alexeev, D. Abundance profiling of specific gene groups using precomputed gut metagenomes yields novel biological hypotheses. PLoS ONE 2017, 12, e0176154. [Google Scholar] [CrossRef]
- Lee, Y.S.; Seo, S.H.; Yoon, S.H.; Kim, S.Y.; Hahn, B.S.; Sim, J.S.; Koo, B.S.; Lee, C.M. Identification of a novel alkaline amylopullulanase from a gut metagenome of Hermetia illucens. Int. J. Biol. Macromol. 2016, 82, 514–521. [Google Scholar] [CrossRef]
- Sadeghian Motahar, S.F.; Ariaeenejad, S.; Salami, M.; Emam-Djomeh, Z.; Sheykh Abdollahzadeh Mamaghani, A. Improving the quality of gluten-free bread by a novel acidic thermostable α-amylase from metagenomics data. Food Chem. 2021, 352, 129307. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Brady, D. The limits to biocatalysis: Pushing the envelope. Chem. Commun. 2018, 54, 6088–6104. [Google Scholar] [CrossRef]
- Dalapati, R.; Nandi, S.; Gogoi, C.; Shome, A.; Biswas, S. Metal–Organic framework (MOF) derived recyclable, superhydrophobic composite of cotton fabrics for the facile removal of oil spills. ACS Appl. Mater. Interfaces 2021, 13, 8563–8573. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).