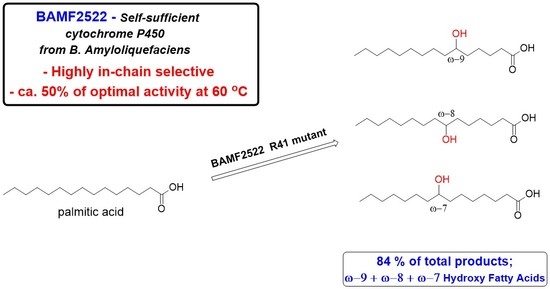
Optimization and Engineering of a Self-Sufficient CYP102 Enzyme from Bacillus amyloliquefaciens towards Synthesis of In-Chain Hydroxy Fatty Acids
Abstract
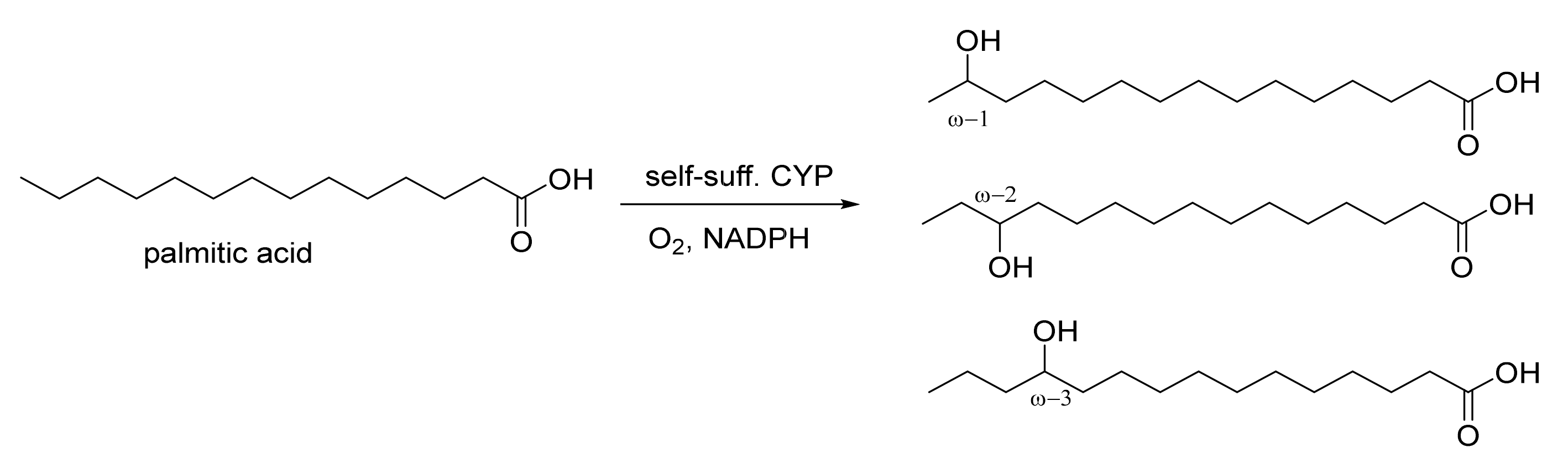
1. Introduction
2. Results and Discussion
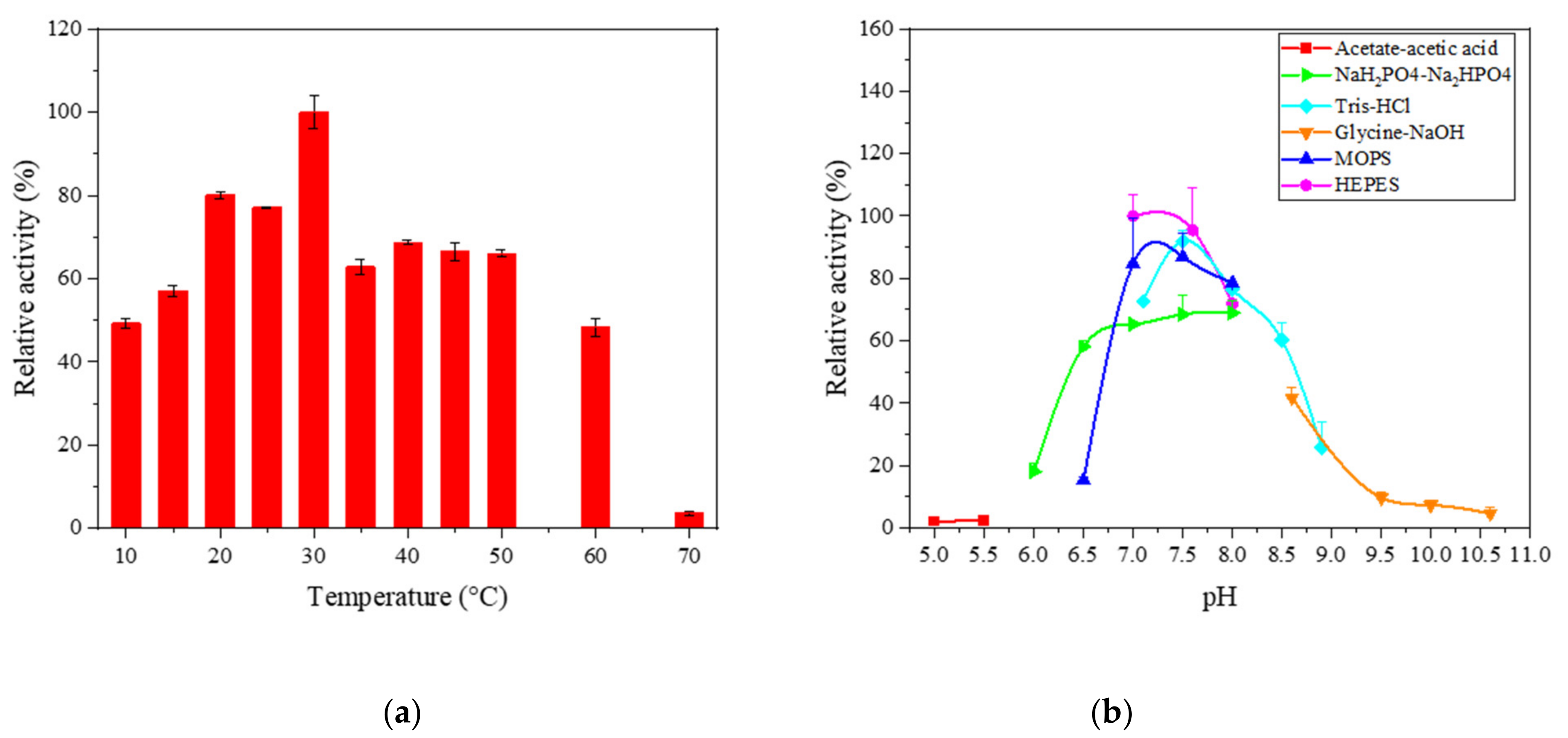
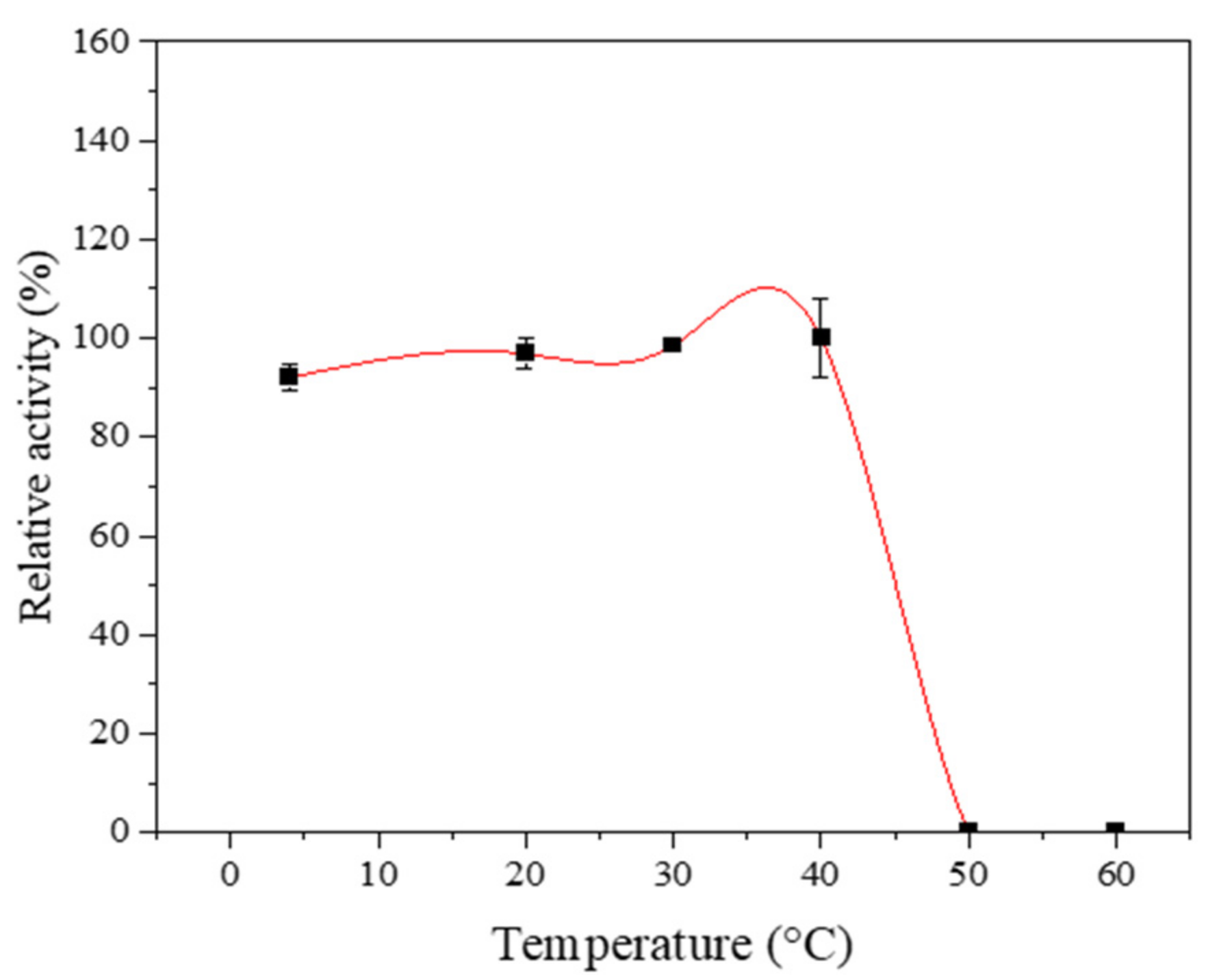
2.1. Temperature, pH and Thermostability Analysis
2.2. Active Site Mutagenesis of BAMF2522 to Enhance Formation of In-Chain HFAs
2.3. Site-Saturation Mutagenesis of BAMF2522 to Enhance Selective Formation of In-Chain HFAs with Lauric Acid
3. Materials and Methods
3.1. Chemicals
3.2. Site-Directed and Site Saturation Mutagenesis
3.3. Protein Expression and Purification
3.4. Activity Assays for Determination of Optimum Conditions
3.5. Whole Cell Assays for Regioselectivity Analysis
3.6. Product Analysis by GC-FID and GC-MS
3.7. Structural Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Groves, J.T. Cytochrome P450 enzymes: Understanding the biochemical hieroglyphs. F1000Research 2015, 4. [Google Scholar] [CrossRef]
- Guengerich, F.P. Mechanisms of Cytochrome P450-Catalyzed Oxidations. ACS Catal. 2018, 8, 10964–10976. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, C.J.; Bell, S.G.; Wong, L.L. P450(BM3) (CYP102A1): Connecting the dots. Chem. Soc. Rev. 2012, 41, 1218–1260. [Google Scholar] [CrossRef] [PubMed]
- Dennig, A.; Kuhn, M.; Tassoti, S.; Thiessenhusen, A.; Gilch, S.; Bulter, T.; Haas, T.; Hall, M.; Faber, K. Oxidative Decarboxylation of Short-Chain Fatty Acids to 1-Alkenes. Angew. Chem. Int. Ed. Engl. 2015, 54, 8819–8822. [Google Scholar] [CrossRef] [PubMed]
- Oliw, E.H.; Bylund, J.; Herman, C. Bisallylic hydroxylation and epoxidation of polyunsaturated fatty acids by cytochrome P450. Lipids 1996, 31, 1003–1021. [Google Scholar] [CrossRef] [PubMed]
- Ahr, H.; King, L.; Nastainczyk, W.; Ullrich, V. The mechanism of reductive dehalogenation of halothane by liver cytochrome P450. Biochem. Pharmacol. 1982, 31, 383–390. [Google Scholar] [CrossRef]
- Rahimtula, A.D.; O’Brien, P.J. Hydroperoxide dependent O-dealkylation reactions catalyzed by liver microsomal cytochrome P450. Biochem. Biophys. Res. Commun. 1975, 62, 268–275. [Google Scholar] [CrossRef]
- Volz, T.J.; Rock, D.A.; Jones, J.P. Evidence for two different active oxygen species in cytochrome P450 BM3 mediated sulfoxidation and N-dealkylation reactions. J. Am. Chem. Soc. 2002, 124, 9724–9725. [Google Scholar] [CrossRef]
- Hammer, S.C.; Kubik, G.; Watkins, E.; Huang, S.; Minges, H.; Arnold, F.H. Anti-Markovnikov alkene oxidation by metal-oxo—mediated enzyme catalysis. Science 2017, 358, 215–218. [Google Scholar] [CrossRef]
- Denisov, I.G. Structure and Chemistry of Cytochrome P450. Chem. Rev. 2005, 105, 2253–2277. [Google Scholar] [CrossRef]
- Eser, B.E.; Zhang, Y.; Zong, L.; Guo, Z. Self-sufficient Cytochrome P450s and Their Potential Applications in Biotechnology. Chin. J. Chem. Eng. 2020. [Google Scholar] [CrossRef]
- Jung, S.T.; Lauchli, R.; Arnold, F.H. Cytochrome P450: Taming a wild type enzyme. Curr. Opin. Biotechnol. 2011, 22, 809–817. [Google Scholar] [CrossRef]
- Fasan, R. Tuning P450 Enzymes as Oxidation Catalysts. ACS Catal. 2012, 2, 647–666. [Google Scholar] [CrossRef]
- Urlacher, V.B.; Girhard, M. Cytochrome P450 Monooxygenases in Biotechnology and Synthetic Biology. Trends Biotechnol. 2019, 37, 882–897. [Google Scholar] [CrossRef]
- Kaluzna, I.; Schmitges, T.; Straatman, H.; van Tegelen, D.; Müller, M.; Schürmann, M.; Mink, D. Enabling Selective and Sustainable P450 Oxygenation Technology. Production of 4-Hydroxy-α-isophorone on Kilogram Scale. Org. Process. Res. Dev. 2016, 20, 814–819. [Google Scholar] [CrossRef]
- Pflug, S.; Richter, S.M.; Urlacher, V.B. Development of a fed-batch process for the production of the cytochrome P450 monooxygenase CYP102A1 from Bacillus megaterium in E. coli. J. Biotechnol. 2007, 129, 481–488. [Google Scholar] [CrossRef]
- Ciaramella, A.; Minerdi, D.; Gilardi, G. Catalytically self-sufficient cytochromes P450 for green production of fine chemicals. Rendiconti Lincei 2016, 28, 169–181. [Google Scholar] [CrossRef]
- Hammerer, L.; Winkler, C.K.; Kroutil, W. Regioselective Biocatalytic Hydroxylation of Fatty Acids by Cytochrome P450s. Catal. Lett. 2018, 148, 787–812. [Google Scholar] [CrossRef]
- Metzger, J.O.; Bornscheuer, U. Lipids as renewable resources: Current state of chemical and biotechnological conversion and diversification. Appl. Microbiol. Biotechnol. 2006, 71, 13–22. [Google Scholar] [CrossRef]
- Lu, W.; Ness, J.E.; Xie, W.; Zhang, X.; Minshull, J.; Gross, R.A. Biosynthesis of monomers for plastics from renewable oils. J. Am. Chem. Soc. 2010, 132, 15451–15455. [Google Scholar] [CrossRef]
- Durairaj, P.; Malla, S.; Nadarajan, S.P.; Lee, P.G.; Jung, E.; Park, H.H.; Kim, B.G.; Yun, H. Fungal cytochrome P450 monooxygenases of Fusarium oxysporum for the synthesis of omega-hydroxy fatty acids in engineered Saccharomyces cerevisiae. Microb. Cell Fact. 2015, 14, 45. [Google Scholar] [CrossRef]
- Zhang, Y.; Eser, B.E.; Kristensen, P.; Guo, Z. Fatty acid hydratase for value-added biotransformation: A review. Chin. J. Chem. Eng. 2020, 28, 2051–2063. [Google Scholar] [CrossRef]
- Kim, K.R.; Oh, D.K. Production of hydroxy fatty acids by microbial fatty acid-hydroxylation enzymes. Biotechnol. Adv. 2013, 31, 1473–1485. [Google Scholar] [CrossRef]
- Ogawa, J. New lipid science in our inner ecosystem. Eur. J. Lipid Sci. Technol. 2015, 117, 577–578. [Google Scholar] [CrossRef]
- Moraes-Vieira, P.M.; Saghatelian, A.; Kahn, B.B. GLUT4 expression in adipocytes regulates de novo lipogenesis and levels of a novel class of lipids with antidiabetic and anti-inflammatory effects. Diabetes 2016, 65, 1808–1815. [Google Scholar] [CrossRef]
- Syed, I.; Lee, J.; Peroni, O.D.; Yore, M.M.; Moraes-Vieira, P.M.; Santoro, A.; Wellenstein, K.; Smith, U.; McGraw, T.E.; Saghatelian, A.; et al. Methodological Issues in Studying PAHSA Biology: Masking PAHSA Effects. Cell Metab. 2018, 28, 543–546. [Google Scholar] [CrossRef]
- Yore, M.M.; Syed, I.; Moraes-Vieira, P.M.; Zhang, T.; Herman, M.A.; Homan, E.A.; Patel, R.T.; Lee, J.; Chen, S.; Peroni, O.D.; et al. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 2014, 159, 318–332. [Google Scholar] [CrossRef]
- Zhou, P.; Santoro, A.; Peroni, O.D.; Nelson, A.T.; Saghatelian, A.; Siegel, D.; Kahn, B.B. PAHSAs enhance hepatic and systemic insulin sensitivity through direct and indirect mechanisms. J. Clin. Investig. 2019, 129, 4138–4150. [Google Scholar] [CrossRef]
- Serhan, C.N. Pro-resolving lipid mediators are leads for resolution physiology. Nature 2014, 510, 92–101. [Google Scholar] [CrossRef]
- Fredman, G.; Spite, M. Specialized pro-resolving mediators in cardiovascular diseases. Mol. Asp. Med. 2017, 58, 65–71. [Google Scholar] [CrossRef]
- Spite, M.; Claria, J.; Serhan, C.N. Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2014, 19, 21–36. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Groves, J.T. Beyond ferryl-mediated hydroxylation: 40 years of the rebound mechanism and C-H activation. J. Biol. Inorg. Chem. 2017, 22, 185–207. [Google Scholar] [CrossRef] [PubMed]
- Buergler, M.B.; Dennig, A.; Nidetzky, B. Process intensification for cytochrome P450 BM3-catalyzed oxy-functionalization of dodecanoic acid. Biotechnol. Bioeng. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lundemo, M.T.; Woodley, J.M. Guidelines for development and implementation of biocatalytic P450 processes. Appl. Microbiol. Biotechnol. 2015, 99, 2465–2483. [Google Scholar] [CrossRef]
- Maseme, M.J.; Pennec, A.; van Marwijk, J.; Opperman, D.J.; Smit, M.S. CYP505E3: A Novel Self-Sufficient ω-7 In-Chain Hydroxylase. Angew. Chem. Int. Ed. 2020, 59, 10359–10362. [Google Scholar] [CrossRef]
- Zong, L.; Gao, R.; Guo, Z.; Shao, Z.; Wang, Y.; Eser, B.E. Characterization and modification of two self-sufficient CYP102 family enzymes from Bacillus amyloliquefaciens DSM 7 with distinct regioselectivity towards fatty acid hydroxylation. Biochem. Eng. J. 2021, 166, 107871. [Google Scholar] [CrossRef]
- Manning, J.; Tavanti, M.; Porter, J.L.; Kress, N.; De Visser, S.P.; Turner, N.J.; Flitsch, S.L. Regio- and Enantio-selective Chemo-enzymatic C-H-Lactonization of Decanoic Acid to (S)-delta-Decalactone. Angew. Chem. Int. Ed. Engl. 2019, 58, 5668–5671. [Google Scholar] [CrossRef]
- Saab-Rincón, G.; Alwaseem, H.; Guzmán-Luna, V.; Olvera, L.; Fasan, R. Stabilization of the Reductase Domain in the Catalytically Self-Sufficient Cytochrome P450BM3 by Consensus-Guided Mutagenesis. ChemBioChem 2018, 19, 622–632. [Google Scholar] [CrossRef]
- Ruettinger, R.T.; Fulco, A. Epoxidation of unsaturated fatty acids by a soluble cytochrome P-450-dependent system from Bacillus megaterium. J. Biol. Chem. 1981, 256, 5728–5734. [Google Scholar] [CrossRef]
- Fürst, M.J.L.J.; Kerschbaumer, B.; Rinnofner, C.; Migglautsch, A.K.; Winkler, M.; Fraaije, M.W. Exploring the Biocatalytic Potential of a Self-Sufficient Cytochrome P450 from Thermothelomyces thermophila. Adv. Synth. Catal. 2019, 361, 2487–2496. [Google Scholar] [CrossRef]
- Baker, G.J.; Girvan, H.M.; Matthews, S.; McLean, K.J.; Golovanova, M.; Waltham, T.N.; Rigby, S.E.J.; Nelson, D.R.; Blankley, R.T.; Munro, A.W. Expression, Purification, and Biochemical Characterization of the Flavocytochrome P450 CYP505A30 from Myceliophthora thermophila. ACS Omega 2017, 2, 4705–4724. [Google Scholar] [CrossRef]
- Eiben, S.; Bartelmäs, H.; Urlacher, V.B. Construction of a thermostable cytochrome P450 chimera derived from self-sufficient mesophilic parents. Appl. Microbiol. Biotechnol. 2007, 75, 1055–1061. [Google Scholar] [CrossRef]
- Salazar, O.; Cirino, P.C.; Arnold, F.H. Thermostabilization of a cytochrome P450 peroxygenase. ChemBioChem 2003, 4, 891–893. [Google Scholar] [CrossRef]
- Zorn, K.; Oroz-Guinea, I.; Brundiek, H.; Bornscheuer, U.T. Engineering and application of enzymes for lipid modification, an update. Prog. Lipid Res. 2016, 63, 153–164. [Google Scholar] [CrossRef]
- Dietrich, M.; Do, T.A.; Schmid, R.D.; Pleiss, J.; Urlacher, V.B. Altering the regioselectivity of the subterminal fatty acid hydroxylase P450 BM-3 towards gamma- and delta-positions. J. Biotechnol. 2009, 139, 115–117. [Google Scholar] [CrossRef]
- Cirino, P.C.; Arnold, F.H. Regioselectivity and activity of cytochrome P450 BM-3 and mutant F87A in reactions driven by hydrogen peroxide. Adv. Synth. Catal. 2002, 344, 932–937. [Google Scholar] [CrossRef]
- Brühlmann, F.; Fourage, L.; Ullmann, C.; Haefliger, O.P.; Jeckelmann, N.; Dubois, C.; Wahler, D. Engineering cytochrome P450 BM3 of Bacillus megaterium for terminal oxidation of palmitic acid. J. Biotechnol. 2014, 184, 17–26. [Google Scholar] [CrossRef]
- Haines, D.C.; Hegde, A.; Chen, B.; Zhao, W.; Bondlela, M.; Humphreys, J.M.; Mullin, D.A.; Tomchick, D.R.; Machius, M.; Peterson, J.A. A Single Active-Site Mutation of P450BM-3 Dramatically Enhances Substrate Binding and Rate of Product Formation. Biochemistry 2011, 50, 8333–8341. [Google Scholar] [CrossRef]
- Di Nardo, G.; Gilardi, G. Optimization of the bacterial cytochrome P450 BM3 system for the production of human drug metabolites. Int. J. Mol. Sci. 2012, 13, 15901–15924. [Google Scholar] [CrossRef]
- Seifert, A.; Vomund, S.; Grohmann, K.; Kriening, S.; Urlacher, V.B.; Laschat, S.; Pleiss, J. Rational Design of a Minimal and Highly Enriched CYP102A1 Mutant Library with Improved Regio-, Stereo- and Chemoselectivity. ChemBioChem 2009, 10, 853–861. [Google Scholar] [CrossRef]
- Munday, S.D.; Maddigan, N.K.; Young, R.J.; Bell, S.G. Characterisation of two self-sufficient CYP102 family monooxygenases from Ktedonobacter racemifer DSM44963 which have new fatty acid alcohol product profiles. Biochim. Biophys. Acta 2016, 1860, 1149–1162. [Google Scholar] [CrossRef]
- Maga, J.A.; Katz, I. Lactones in foods. Crit. Rev. Food Sci. Nutr. 1976, 8, 1–56. [Google Scholar] [CrossRef]
- An, J.-U.; Oh, D.-K. Increased production of γ-lactones from hydroxy fatty acids by whole Waltomyces lipofer cells induced with oleic acid. Appl. Microbiol. Biotechnol. 2013, 97, 8265–8272. [Google Scholar] [CrossRef]
- Barr, I.; Guo, F. Pyridine Hemochromagen Assay for Determining the Concentration of Heme in Purified Protein Solutions. Bio-protocol 2015, 5, e1594. [Google Scholar] [CrossRef]
- Eser, B.E.; Poborsky, M.; Dai, R.; Kishino, S.; Ljubic, A.; Takeuchi, M.; Jacobsen, C.; Ogawa, J.; Kristensen, P.; Guo, Z. Rational Engineering of Hydratase from Lactobacillus acidophilus Reveals Critical Residues Directing Substrate Specificity and Regioselectivity. ChemBioChem 2020, 21, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.; Sali, A. Comparative Protein Structure MODELING Using MODELLER. Curr. Protoc. Bioinform. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—Visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]


| Entry | Subst. | Variant | Total Conv. (%) 1 | Product Distribution (%) 1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ω-9 | ω-8 | ω-7 | ω-6 | ω-5 | ω-4 | ω-3 | ω-2 | ω-1 | ||||
| 1 | C12:0 | Wild-type | 10 | - | - | - | - | <2 | <2 | 40 | 40 | 17 |
| 2 | C12:0 | F89V | 5 | - | - | - | - | 12 | 8 | 49 | 21 | 9 |
| 3 | C12:0 | F89I/A331V | 4 | - | - | - | - | 7 | 13 | 36 | 18 | 27 |
| 4 | C12:0 | RY5 3 | 1.2 | - | - | - | - | 10 | 14 ± 7 | 34 | 15 | 27 ± 6 |
| 5 | C12:0 | R31 4 | 3 | - | - | - | - | 8 ± 2 | 18 | 22 | 40 ± 10 | 12 ± 5 |
| 6 | C12:0 | R41 5 | 2 | - | - | - | - | 18 ± 4 | 19 | 36 | 13 | 13 ± 3 |
| 7 | C14:0 | Wild-type | 28 | - | - | <2 | <1 | 2 ± 1 | 9 | 46 | 33 | 8 |
| 8 | C14:0 | F89V | 29 ± 7 | - | - | 35 | 10 | 19 | 15 | 12 | 7 | 3 ± 1 |
| 9 | C14:0 | F89I | 3 | - | - | 16 | 12 | 35 | 13 | 13 ± 4 | 6 ± 2 | 6 |
| 10 | C14:0 | F89I/A331V | 19 | - | - | 17 | 6 | 29 | 20 | 10 ± 3 | 4 ± 2 | 13 |
| 11 | C14:0 | RY5 3 | 4 ± 3 | - | - | 12 | 7 | 28 | 15 ± 3 | 14 ± 3 | 9 | 16 |
| 12 | C14:0 | R31 4 | 6 | - | - | 32 ± 8 | 12 | 26 | 20 ± 5 | 11 ± 3 | <1 | <1 |
| 13 | C14:0 | R41 5 | 9 | - | - | 36 | 10 ± 4 | 30 | 17 | 8 | <1 | <1 |
| 14 | C16:0 | Wild-type | 14 | - | - | 20 | 9 ± 3 | 8 | 10 | 19 | 24 | 10 |
| 15 | C16:0 | F89I | 14 ± 3 | 71 2 | 7 | 3 | 2 | 2 | 5 | 9 | ||
| 16 | C16:0 | A331V | 29 | - | - | 24 | 9 | 7 | 4 | 9 | 20 | 27 |
| 17 | C16:0 | F89I/A331V | 8 | 49 ± 2 2 | 5 | 3 | 2 | 3 | 9 | 29 | ||
| 18 | C16:0 | RY5 3 | 10 | 48 ± 1 2 | 6 | 4 ± 1 | 9 | 7 | 11 | 18 | ||
| 19 | C16:0 | R31 4 | 5 | 77 ± 2 2 | 7 ± 1 | 2 ± 1 | 2 | <2 | 6 | 5 | ||
| 20 | C16:0 | R41 5 | 7 | 84 ± 1 2 | 6 | 2 ± 1 | <2 | <2 | <2 | 4 | ||
| 21 | C20:0 | Wild-type | 3 | - | - | - | - | - | - | 49 | 39 | 13 |
| 22 | C20:0 | A331V | 8 ± 2 | - | - | - | - | 9 ± 3 | 11 | 21 | 27 | 31 |
| Entry | Variant | Total Conv. (%) 1 | Product Distribution (%) 1 | ||||
|---|---|---|---|---|---|---|---|
| ω-5 | ω-4 | ω-3 | ω-2 | ω-1 | |||
| 1 | Wild-type | 10 | <1 | 2 ± 1 | 40 | 40 | 17 |
| 2 | A(Ala) | 3 | 29 | 15 | 41 | 12 | 2 |
| 3 | L(Leu) | 3 ± 1 | <1 | 4 ± 1 | 36 | 45 | 14 |
| 4 | I(Ile) | < 1 | - | - | 50 | 22 ± 7 | 29 |
| 5 | V(Val) | 5 | 12 | 8 | 49 | 21 | 9 |
| 6 | M(Met) | 3 | 3 | 3 ± 1 | 52 | 31 | 10 |
| 7 | P(Pro) | 4 ± 1 | 31 | 19 | 38 | 10 | 2 |
| 8 | S(Ser) | 4 ± 1 | 17 | 11 | 50 | 18 | 5 |
| 9 | T(Thr) | 2 | 14 ± 3 | 12 ± 3 | 48 | 21 | 7 ± 2 |
| 10 | N(Asn) | 1 | - | - | 48 | 39 | 13 |
| 11 | H(His) | 6 ± 4 | 2 | 2 | 48 | 36 | 12 |
| Entry | Variant | Total Conv. (%) 1 | Product Distribution (%) 1 | ||||
|---|---|---|---|---|---|---|---|
| ω-5 | ω-4 | ω-3 | ω-2 | ω-1 | |||
| 1 | Wild-type | 10 | <2 | 2 ± 1 | 40 | 40 | 17 |
| 2 | G(Gly) | 4 | - | - | 33 | 38 | 29 |
| 3 | I(Ile) | 1.4 | - | - | 25 ± 8 | 51 | 25 |
| 4 | V(Val) | 6 | - | <2 | 29 | 39 | 30 |
| 5 | P(Pro) | 6 | - | - | 28 | 37 | 35 |
| 6 | S(Ser) | 6 | - | - | 42 | 45 | 14 |
| 7 | T(Thr) | 7 | - | - | 31 | 38 | 31 |
| 8 | C(Cys) | 6 | - | - | 43 | 31 | 26 |
| 9 | D(Asp) | 2 | - | - | 30 | 52 | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zong, L.; Zhang, Y.; Shao, Z.; Wang, Y.; Guo, Z.; Gao, R.; Eser, B.E. Optimization and Engineering of a Self-Sufficient CYP102 Enzyme from Bacillus amyloliquefaciens towards Synthesis of In-Chain Hydroxy Fatty Acids. Catalysts 2021, 11, 665. https://doi.org/10.3390/catal11060665
Zong L, Zhang Y, Shao Z, Wang Y, Guo Z, Gao R, Eser BE. Optimization and Engineering of a Self-Sufficient CYP102 Enzyme from Bacillus amyloliquefaciens towards Synthesis of In-Chain Hydroxy Fatty Acids. Catalysts. 2021; 11(6):665. https://doi.org/10.3390/catal11060665
Chicago/Turabian StyleZong, Li, Yan Zhang, Zhengkang Shao, Yingwu Wang, Zheng Guo, Renjun Gao, and Bekir Engin Eser. 2021. "Optimization and Engineering of a Self-Sufficient CYP102 Enzyme from Bacillus amyloliquefaciens towards Synthesis of In-Chain Hydroxy Fatty Acids" Catalysts 11, no. 6: 665. https://doi.org/10.3390/catal11060665
APA StyleZong, L., Zhang, Y., Shao, Z., Wang, Y., Guo, Z., Gao, R., & Eser, B. E. (2021). Optimization and Engineering of a Self-Sufficient CYP102 Enzyme from Bacillus amyloliquefaciens towards Synthesis of In-Chain Hydroxy Fatty Acids. Catalysts, 11(6), 665. https://doi.org/10.3390/catal11060665