1. Introduction
Cancers are among the leading causes of death worldwide and account for a significant number of fatalities [
1]. Lung cancer is considered one of the most critical forms of cancer because of its high incidence and mortality rates. Therefore, the early detection and appropriate treatment of lung cancer are crucial. Chest computed tomography (CT) plays a critical role in lung cancer diagnosis. Lung nodules are the most common radiological manifestations of lung cancer, and their detection and analysis are vital for early diagnosis. CT is useful for detecting lung nodules and revealing their morphological features, which are essential for distinguishing between benign and malignant nodules and determining treatment strategies. The National Lung Screening Trial (NLST) conducted in the United States demonstrated that low-dose CT screening significantly reduced lung-cancer-related mortality [
2]. However, differential diagnosis based on imaging requires the consideration of multiple factors, including tumor shape, internal characteristics, and relationships with surrounding tissues. Consequently, the diagnostic accuracy significantly depends on the experience and expertise of the physician. The classification of benign and malignant nodules can be challenging, particularly in cases involving small or subtle lesions. Recent studies have increasingly focused on automatic classification methods using artificial intelligence (AI), particularly deep-learning techniques [
3,
4], to address these issues. Numerous image classification models have been proposed to support lung cancer diagnoses [
5,
6,
7,
8,
9,
10]. For example, Wang et al. developed a method using TransUNet to classify benign and malignant nodules and achieved an accuracy of 84.62% and an area under the receiver operating characteristic curve (AUC) of 0.862 [
8]. Raunak et al. developed a three-dimensional DenseNet-based neural network called MoDenseNet and obtained a classification accuracy of 90.4% on the LIDC-IDRI dataset [
9]. Onishi et al. introduced a method combining convolutional neural networks (CNNs) and generative adversarial networks (GANs), attaining a classification accuracy of 81.7% and an AUC of 0.841 [
10]. These techniques enable high-precision feature extraction by learning from large-scale medical image datasets, thereby increasing the expectations for clinical applications. However, many current image classification models lack transparency in their decision-making processes, making it difficult for physicians to interpret their output. These findings highlight the need for explainability and transparency in clinical practice. Therefore, it is essential to achieve high classification performance and clarify the reasoning for the decisions of the model.
In response to this challenge, increasing attention has been paid to image-finding generation, a technique that produces natural language descriptions from images. In particular, vision–language models (VLMs), which integrate visual and textual information, represent a next-generation approach that enables natural language generation based on visual features. VLMs have advanced rapidly in computer vision and natural language processing, and their applications in the medical domain are expanding. In chest X-ray studies, for example, Wang et al. proposed a method for generating radiology reports using a CNN–recurrent neural network architecture. They obtained a bilingual evaluation understudy (BLEU)-1 score of 0.286 and a recall-oriented understudy for gisting evaluation (ROUGE)-L score of 0.226 on the ChestXRay-14 dataset [
11]. Hou et al. introduced a report generation approach that combined CNNs and Transformers [
12] using DenseNet121 for feature extraction and text generation through a transformer-based decoder. They obtained a BLEU-1 score of 0.232 and a ROUGE-L score of 0.240 [
13]. Teramoto et al. established a model that combined a CNN-based feature extractor with a Transformer text decoder to perform image classification and report generation for lung cytology images. Their CNN classifier achieved an accuracy of 95.8% in classifying benign and malignant nodules, and the generated text attained a BLEU-4 score of 0.828 and a semantic propositional image caption evaluation (SPICE) score of 0.832 [
14]. Recently, approaches that use bidirectional integration of image and language information have attracted attention. For example, some studies investigated the reverse direction, automatically generating cytological images of lung cancer from textual findings [
15]. In the case of chest CT imaging, Ates et al. proposed DCFormer, an efficient 3D VLM that utilizes decomposed convolutions to capture spatial dependencies in volumetric data while significantly reducing computational cost [
16]. In addition, Zhang et al. developed a large-scale grounded vision–language dataset for chest CT analysis, providing region-level text grounding, segmentation masks, and visual question–answering pairs to advance multimodal learning in medical imaging [
17]. However, studies focusing on image-finding generation for diagnostic decision-making in lung nodules are scarce. To the best of our knowledge, no study has reported a method for generating descriptive outputs based on CT images to classify benign and malignant nodules.
The aim of this study was to improve the explainability of image classification models for lung cancer diagnosis by developing a method to generate image findings from chest CT images using VLMs and classify benign and malignant nodules. This approach is intended to enhance the practicality of diagnostic support and assist physicians in clinical decision-making.
The main contributions of this study are as follows:
A Novel Multimodal Diagnostic Support Method for Diagnosing Chest CT: This is study proposes a novel multimodal diagnostic support approach that simultaneously generates image findings focusing on the nodule regions and classifies the malignancy of lung nodules using a VLM.
Enhanced Explainability through Report Generation: Unlike most previous studies that focused solely on binary classification, the proposed method integrates the generation of descriptive findings as a form of explicit diagnostic reasoning. This integration enhances explainability and transparency by enabling the model to provide a diagnostic rationale for its classification outputs.
Fine-Tuning with Limited Clinical Data: This study demonstrates that comparable accuracy to conventional classification models can be achieved by fine-tuning VLMs using a small number of clinical CT cases. As collecting large-scale annotated datasets in the medical domain is often difficult, this characteristic highlights the practical value and applicability of the proposed method.
Clinical Relevance and Decision Support: By generating image findings, the proposed method can complement physicians’ decision-making processes and potentially enhance trust and usability in clinical environments.
2. Materials and Methods
2.1. Outline
An overview of the proposed method is shown in
Figure 1. The nodule region was extracted from the chest CT image and fed into a VLM. The model generated a descriptive report that included both imaging findings and malignancy assessments. By analyzing the generated text, this method simultaneously performs image interpretation and malignancy classification.
In this study, the ROI was manually defined, although we plan to automate ROI extraction using nodule detection in future work. As previous studies have already demonstrated high accuracy in automatic nodule detection [
18], we manually set the ROI in this study for controlled evaluation.
2.2. Dataset
The dataset used in this study consisted of noncontrast chest CT images from 77 patients who underwent examinations at Fujita Health University Hospital. Malignancy in each case was confirmed via pathological diagnosis, comprising 26 benign and 51 malignant cases. All CT scans were acquired using an Aquilion ONE scanner (Canon Medical Systems) and reconstructed under optimized conditions for a clear visualization of the lung fields. The scan conditions are shown in
Table 1.
This study was approved by the Institutional Ethics Committee (approval number: HM25-018), and informed consent was obtained from all patients under the condition that the data were anonymized.
2.3. Image Preprocessing
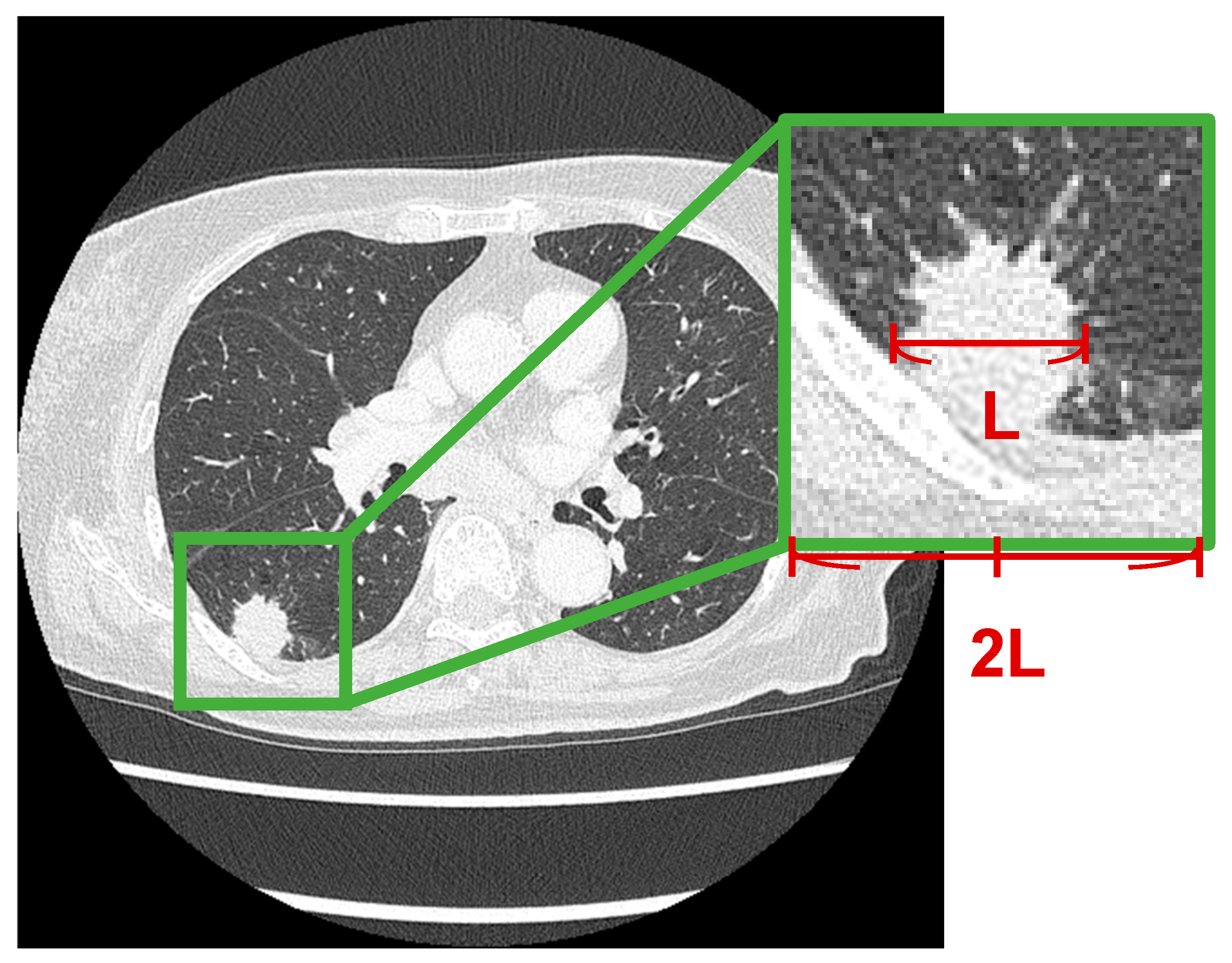
In this study, regions of interest (ROIs) around lung nodules were extracted from CT images as shown in
Figure 2. In each case, an ROI centered on the tumor was extracted. The longest diameter of the lung nodule was measured, and a square ROI with a side length twice the measured diameter was defined based on the center of the nodule. This square region was cropped from the CT images.
For image preprocessing, the grayscale values of the CT images—originally stored in a 16-bit format—were converted to 8-bit images by applying a window width (WW) of 1600 and a window level (WL) of −600.
2.4. Text Preprocessing
Figure 3 shows a representative example of the dataset constructed in this study. Imaging findings were described in Japanese for each nodule region using the terminology commonly used by respiratory physicians in routine clinical practice. As all the VLMs used in this study are specific to the English language, these descriptions were translated into English for input into the image captioning model. In addition, a statement indicating if the results were benign or malignant was added at the end of each report to allow for the simultaneous generation of imaging findings and malignancy classifications.
The translation process aimed to preserve the medical meaning of the original descriptions while maintaining consistency in terminology. The wording of technical terms was refined based on advice from respiratory physicians, ensuring consistent phrasing and eliminating lexical variations, thereby facilitating stable learning by the model.
2.5. Vision–Language Models
VLMs can be broadly categorized into two paradigms. One focuses on learning visual–textual alignment through contrastive learning, as exemplified by contrastive language–image pretraining (CLIP), which is primarily used for classification and retrieval. The other focuses on direct text generation from visual input, as represented by bootstrapping language–image pretraining (BLIP) and the generative image-to-text transformer (GiT). Given that the goal of this study was not only to classify lung nodules but also to generate image findings, we adopted the latter type of model.
Both BLIP and GiT are pretrained on large-scale image–text paired datasets and are widely used in image-captioning tasks, in which visual information is translated into natural language. Each VLM typically consists of an image encoder that extracts visual features and a text decoder that generates textual descriptions. In this study, we applied these VLMs to chest CT images to generate descriptive reports on lung nodules and to perform malignancy classification based on the generated content.
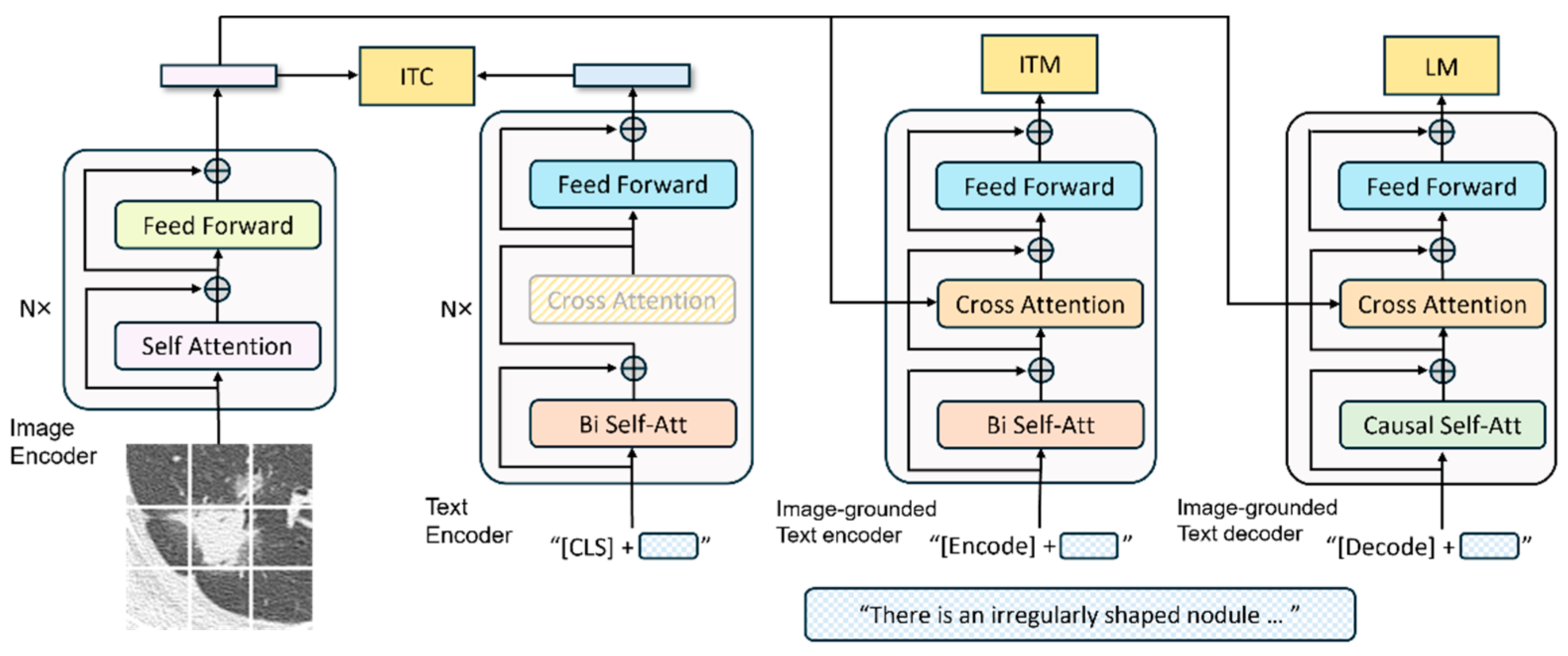
BLIP [
19] is a multimodal model that leverages both visual and linguistic information. It is pretrained using self-supervised learning on large-scale image–text pairs. BLIP uses a Vision Transformer [
20] as its image encoder to extract visual features and bidirectional encoder representations from transformers [
21] as its natural language-processing module for text generation and analysis.
Figure 4 illustrates the overall architecture of the BLIP. The image encoder converts the input images into feature vectors, whereas the text encoder converts the input text into vector representations. The image-grounded text encoder integrates the visual features into the textual-encoding process, and the image-grounded text decoder generates the final textual output. This architecture enables the efficient integration of visual and textual modalities, facilitating the model to achieve high recognition performance.
GiT [
22] is a large-scale generative model designed to handle visual–language integration. The architecture is illustrated in
Figure 5. GiT comprises a single-image encoder and a single-text decoder. The image encoder is based on a vision transformer and converts input images into a sequence of feature vectors. The text decoder, which is based on a transformer architecture, significantly relies on multihead self-attention mechanisms within its decoder layers. This structure effectively models the complex interdependencies between image and text tokens, allowing the capture of diverse contextual information that cannot be represented by a single attention mechanism. The processed contextual information is further transformed into a suitable form for text generation through the feedforward layers.
This flexible generation framework supports a wide range of vision–language tasks and contributes to an improved model performance. In our experiments, we employed both the base and large versions of the BLIP and GiT models. Although both models were pretrained, all parameters were fine-tuned using the dataset constructed in this study to adapt the models to medical imaging.
These models were implemented using the Transformers (v4.33.3) and PyTorch (v2.6.0) libraries and trained using the AdamW optimization algorithm with a fixed learning rate of 1 × 10−5, a batch size of 2, and 50 training epochs. Data augmentation was applied to suppress overfitting of the training data and improve the generalization performance. Specifically, the original images were rotated by 90°, 180°, and 270° to introduce directional variation and horizontally flipped to further increase the diversity of the visual features.
Training and inference were performed on a PC equipped with an AMD Ryzen 9 5900X CPU and NVIDIA GeForce RTX 3090 GPU.
2.6. Evaluation
2.6.1. Evaluation Metrics
The similarity between the generated and reference reports was assessed using five standard metrics commonly employed in image-captioning tasks to evaluate the accuracy of the generated findings. These metrics are described as follows.
BLEU was developed to evaluate machine translation. This is one of the most widely used methods for calculating the precision of n-gram matches between candidate and reference sentences. It also includes a brevity penalty for overly short outputs.
Metric for evaluation of translation with explicit ordering (METEOR) [
24]:
METEOR is a metric developed for machine translation. It evaluates sentence similarity based on unigram matches and computes the harmonic mean of precision and recall. A penalty is applied based on the fragmentation of matched segments to reflect word-order differences.
Recall-oriented understudy for gisting evaluation (ROUGE) [
25]:
ROUGE is a set of metrics originally developed for evaluating automatic summarization. It tends to favor longer sentences with high recall. ROUGE-N measures recall based on n-gram overlap, whereas ROUGE-L evaluates the length of the longest common subsequence between the candidate and reference.
Consensus-based image description evaluation (CIDEr) [
26]:
CIDEr was specifically developed to evaluate image captions. It uses term frequency–inverse document frequency (TF-IDF) weighting to reduce the influence of commonly occurring n-grams and calculates a weighted average of cosine similarities between TF-IDF vectors for each n-gram.
Semantic propositional image caption evaluation (SPICE) [
27]:
SPICE evaluates the semantic content of image captions. It uses a pretrained dependency parser to convert sentences into dependency trees, which are transformed into scene graphs. It calculates the F-score based on the matching of logical tuples representing semantic propositions between the candidate and reference graphs.
In addition to the evaluation metrics described above, we conducted a clinical relevance assessment of the generated findings. In the diagnosis of lung nodules, three major morphological characteristics—nodule shape, nodule border, and nodule internal structure—are typically evaluated. Therefore, we scored the degree of agreement between the reference and generated findings for each of these aspects. A score of 1 was assigned when the descriptions were completely consistent, 0 when they were contradictory, and an intermediate value when they were partially consistent. The mean and standard deviation of these scores were then calculated. Cases with missing descriptions for any item were excluded from the analysis.
Furthermore, the malignancy classification was analyzed based on the descriptions generated for each case. Statements related to the benignity or malignancy of the nodules were extracted from each report, and the results were compared with ground truth labels. On the basis of this comparison, a confusion matrix was derived, and the sensitivity, specificity, and accuracy were calculated to evaluate classification performance. Because the VLM produces categorical textual outputs rather than continuous probabilities, threshold-dependent analyses such as ROC or PR curves and calibration metrics were not applicable. Instead, classification results were summarized by sensitivity, specificity, accuracy, and their 95% confidence intervals.
The above classification can also be performed using conventional image classification models. To compare the performance of the proposed method with that of conventional models, VGG16 [
28], InceptionV3 [
29], ResNet50 [
30], DenseNet121 [
31] and the Vision Transformer (base model with a 16 × 16 patch) [
20] were all fine-tuned, and their sensitivity, specificity and accuracy were calculated. All models were pretrained on ImageNet. These classification models were implemented using TensorFlow (v2.5) and vit-keras (v0.1.2) libraries and optimized with the Adam algorithm. The learning rate was fixed at 1 × 10
−5, with a batch size of 8. Class weights were not applied in order to match the training conditions of the VLM, which does not perform explicit class balancing.
2.6.2. Cross-Validation
To evaluate classification accuracy, we employed five-fold cross-validation. In this method, the dataset was divided at the patient level into five subsets; in each iteration, one subset was used as the test data, and the remaining four subsets were used for training. This process was repeated five times; each subset was used as test data exactly once, and the overall performance was assessed by aggregating the results across all folds.
In this study, only the original (nonaugmented) images were used as test data, whereas the training data consisted of augmented images generated from the original images that were not used in the corresponding test fold. A schematic overview of this cross-validation process is illustrated in
Figure 6.
3. Results
In this study, we evaluated four models: base and large versions of both BLIP and GiT. The evaluation metrics for image-finding generation for each model are listed in
Table 2. The confusion matrices for malignancy classification are listed in
Table 3, and the sensitivity, specificity, and accuracy are summarized in
Table 4. In addition,
Table 5 shows the classification results obtained using conventional image classification models. Note that in
Table 3 and
Table 5, the 95% confidence intervals (CIs) are presented in parentheses following each metric.
For all the models, the BLEU scores exceeded 0.4, indicating that the generated reports maintained grammatically appropriate sentence structures. Although no significant differences were observed between the models, the BLIP base model achieved the highest values for both the evaluation metrics and classification accuracy.
In terms of classification performance based on confusion matrices, slight differences were observed between the base and large versions of the GiT model. However, the base version of the BLIP model yielded slightly better results than the other models. Comparisons of sensitivity, specificity, and accuracy revealed that all the models tended to exhibit high sensitivity but relatively low specificity. This suggests that although the models were effective in detecting malignant cases, some benign cases were misclassified. Furthermore, the top classification accuracy of the conventional models was 81.8%, as shown in
Table 5, when using DenseNet121. The statistical difference between the proposed method and the conventional classification model was evaluated using McNemar’s test. The resulting
p-value of 0.80 exceeded the significance level (α = 0.05), indicating that no significant difference was observed between the two methods.
Figure 7 shows an example of an actual input image, along with the corresponding output reports generated by each model. These examples demonstrate that the proposed method successfully generated findings based on visual information and that the generated content somewhat reflects the visual characteristics of the lung nodules.
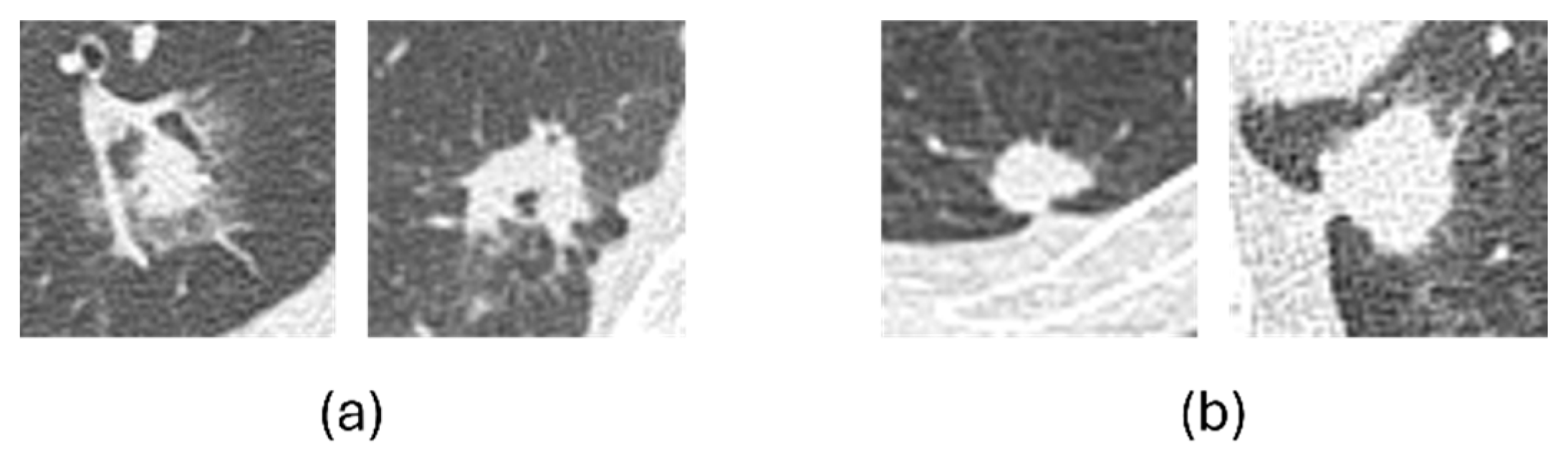
To further examine classification errors,
Figure 8 shows representative examples of misclassified cases. In general, benign nodules that were misclassified as malignant tended to exhibit malignant-like characteristics, such as irregular shapes and indistinct margins, whereas malignant nodules misclassified as benign often appeared relatively round with well-defined borders. These patterns indicate that most errors were associated with overlapping visual characteristics between benign and malignant nodules.
In addition, the clinical relevance scores for nodule shape, border, and internal structure are summarized in
Table 6. Among these, the score for nodule internal structure was the highest, whereas that for nodule shape was relatively lower, which may reflect the large diversity in nodule shapes. Although slight differences were observed between the BLIP and GiT models, the overall trends were consistent, indicating that both models were able to reasonably capture each morphological characteristic.
The parameter counts, training time per fold, and inference time per image for the image classification models and the VLMs are summarized in
Table 7.
4. Discussion
In this study, we used two VLMs—BLIP and GiT—to generate descriptive findings from CT images of lung nodules and perform malignancy classification. In terms of evaluation metrics, all models achieved BLEU scores exceeding 0.4, suggesting that the generated reports maintained grammatical consistency and conformed to the typical structure of the clinical findings. By contrast, metrics that assess informational content and semantic coherence, such as CIDEr and SPICE, yielded relatively lower scores than BLEU. A possible reason for this trend is the use of a predefined clinical vocabulary to prioritize clinical accuracy and consistency in descriptions. Although this vocabulary constraint supports standardization, it may have limited lexical diversity, thereby narrowing the expressive range of the output. This could have negatively impacted the richness of the semantic structures in SPICE and the information density of the n-grams in CIDEr.
For the classification performance in malignancy discrimination, differences were observed, depending on the model type. In the case of BLIP, the base model achieved higher accuracy than the large model. However, for GiT, no significant difference was observed in accuracy between the base and large models. However, in terms of sensitivity, the base models tended to perform slightly better for both architectures than the large models. This suggests that the base models may have been more effective, particularly for detecting malignant cases. This trend may be attributed to the relatively small dataset used in this study, which could have resulted in overfitting or unstable optimization of the larger models. The BLIP large model has more parameters and likely requires more training data to achieve stable convergence. Furthermore, large-scale generative models are known to produce fluent yet potentially inaccurate descriptions, a phenomenon referred to as ‘hallucination’, and these effects may have influenced the observed performance in this study.
These results imply that a larger model capacity does not necessarily lead to better classification performance, particularly when the amount of training data is limited. In such scenarios, achieving an appropriate balance between model size and dataset scale is crucial. In terms of sensitivity and specificity, all models showed high sensitivity but relatively low specificity. This outcome is likely influenced by class imbalance in the dataset, where the number of malignant cases was approximately twice that of the benign cases. Under such an imbalance, models tend to be biased toward the majority class, which can lead to reduced performance in correctly classifying the minority class. Increasing the number of benign cases in future datasets can help achieve more balanced model training and improve the overall classification performance.
The significance of this study is that it provides multifaceted diagnostic support by simultaneously generating both descriptive image findings and classifying lung nodule malignancy. Traditional models for malignancy discrimination have mainly focused on binary classification using architectures, such as TransUNet or CNNs, and they typically provide only classification results. Although these methods often achieve high accuracy, they have low interpretability, which makes it difficult to understand the reasoning behind the decisions of a model. In contrast, the image-captioning approach employed in this study generates textual descriptions based on CT images, enabling visualization of the reasoning of the model and the areas of focus. This suggests its potential utility as an explainable and transparent support tool that can complement physicians’ decisions. In this study, the classification accuracy for malignancy achieved using the BLIP base model was 79.2%. Although this is comparable to or slightly lower than those of conventional classification models, the ability to produce textual diagnostic explanations alongside classification results may offer a different clinical value that traditional approaches cannot provide.
Furthermore, the classification model (DenseNet121) achieved an accuracy rate of 81.8%, while the proposed method (BLIP base) achieved 79.2%. While the classification model made more correct predictions, the McNemar test revealed no significant difference between the two methods (
p = 0.80). Therefore, the proposed method is considered to have equivalent performance to the classification model. In addition, in classification evaluation, the proposed method can present the diagnostic text findings simultaneously. This allows physicians to verify the accuracy of classification assessments based on the generated text, demonstrating the method’s transparency. Thus, the proposed method is a highly transparent and explainable technology that supports physician decision-making and is considered capable of compensating sufficiently for the difference in accuracy rates compared to existing classification methods. As shown in
Table 7, the VLM used in the proposed method requires more computational resources, such as a larger number of parameters and longer processing time, compared to the image classification models. Nevertheless, it provides textual image findings of nodules that enhance explainability, while simultaneously yielding malignancy classification results comparable to conventional models. Therefore, we consider that the performance obtained is well justified by the computational cost.
However, this study has several limitations. First, the dataset used in this study was collected from a single institution, with a limited number of cases and an imbalance between benign and malignant nodules. Therefore, future studies should further validate the generalizability of the proposed method by incorporating multi-institutional data and addressing class imbalance, especially by increasing the number of benign cases to achieve a more balanced dataset. In addition, expanding the training dataset through data augmentation techniques, such as rotation, flipping, and contrast adjustment, or by utilizing publicly available datasets, could help improve model robustness and reduce overfitting. Because the physicians’ findings were originally written in Japanese and translated into English for model input, terminology was normalized during translation to reduce subtle variations, which likely contributed to stable results; however, the potential influence of translation bias should also be considered. Although multilingual VLMs have recently emerged, their current performance remains limited. Therefore, we plan to conduct additional evaluations using multilingual VLMs in future work to assess the potential influence of translation bias. Finally, the generation of image findings in this study was based on pre-extracted nodule regions. Consequently, this method currently does not handle the full clinical workflow, which includes nodule detection from whole-scan CT images. Future studies should address this limitation by building an end-to-end model that integrates existing nodule detection algorithms with image-based report generation and malignancy classification, starting from a full-scan image input.