Homologous Recombination Is Associated with Enhanced Anti-Tumor Innate Immunity and Favorable Prognosis in Head and Neck Cancer
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Clinical Data Analysis
2.2. Survival Analysis
2.3. Correlation Analysis
2.4. Cell Culture and Transfection
2.5. Immunoblotting
3. Results
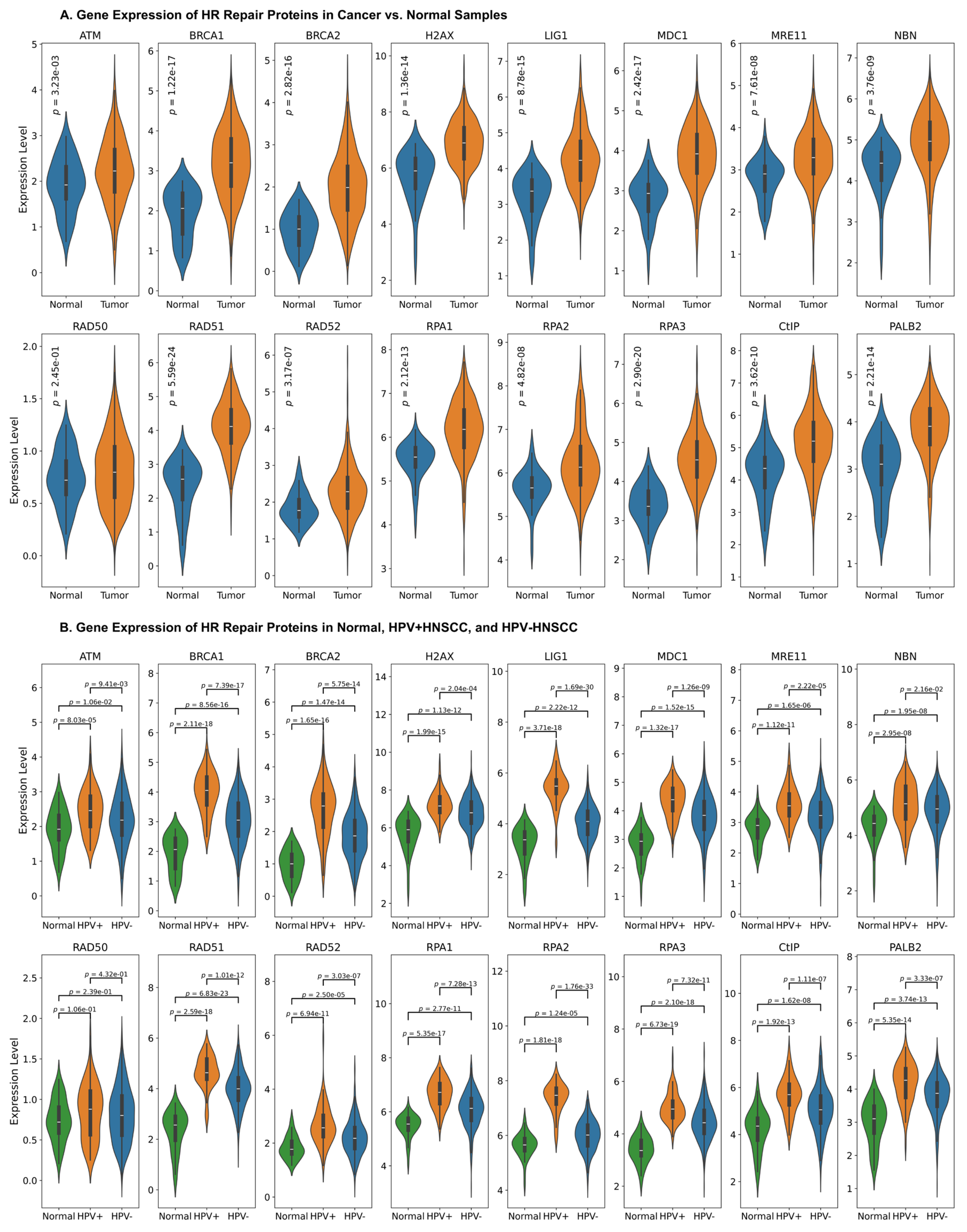
3.1. Upregulation of HR Factors in HNC
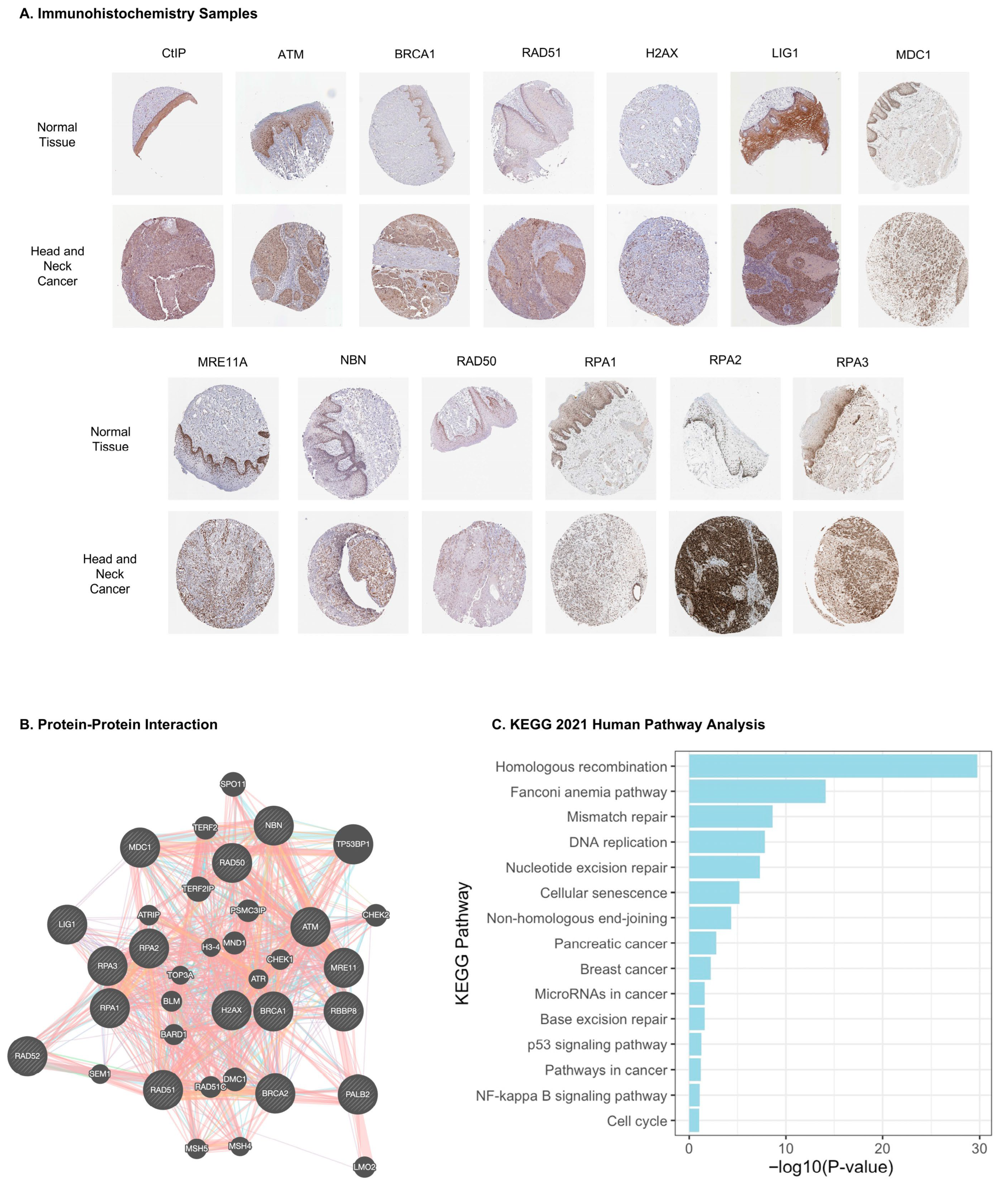
3.2. HR Protein Interaction Network and Pathway Enrichment Analyses
3.3. The Prognostic Value of HR Proteins Expression for HNC Patients
3.4. Correlation Between HR Proteins Expression and DNA Methylation
3.5. Correlation Between HR Proteins and Immune Cell Infiltration in HNC Patients
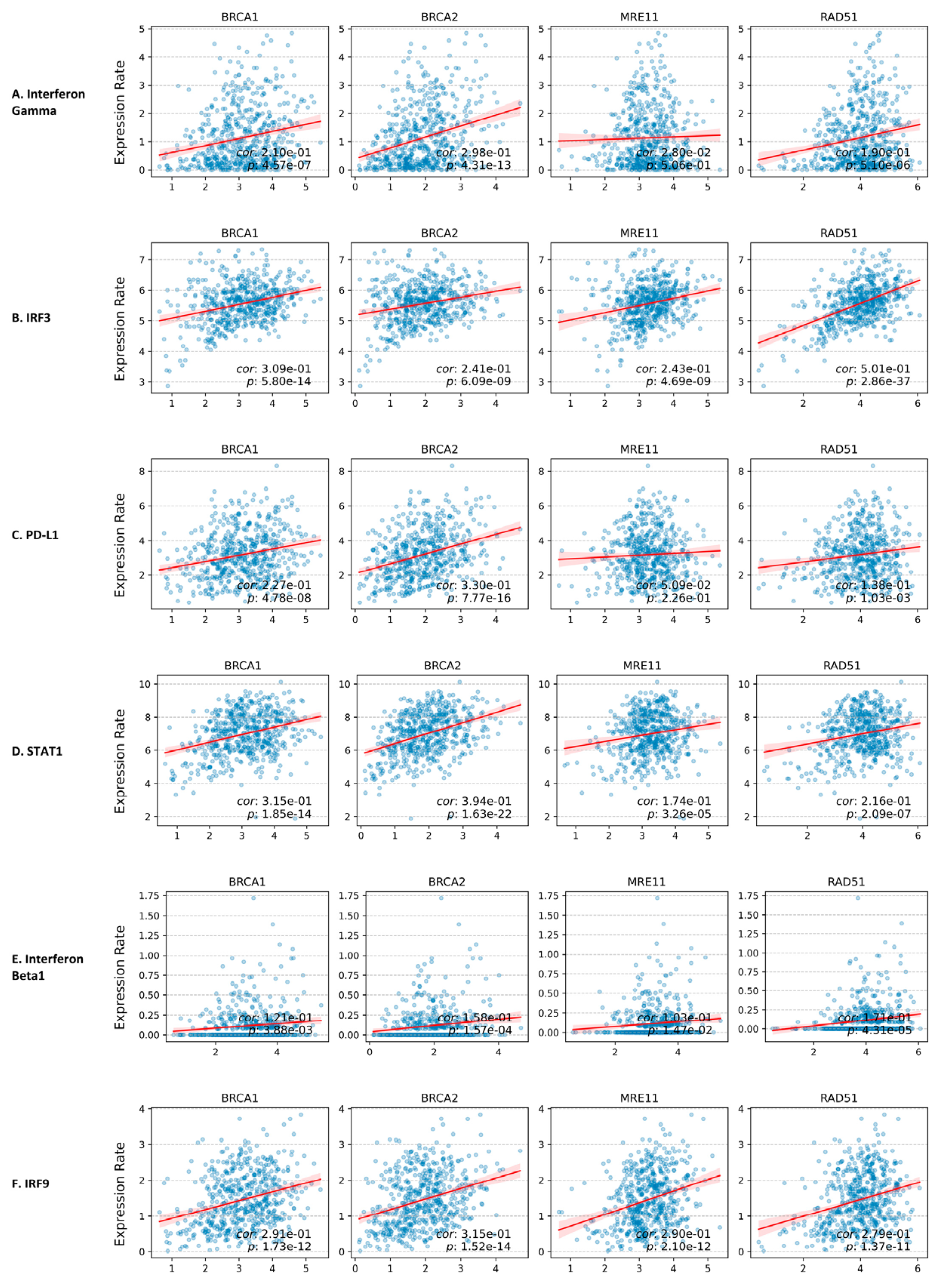
3.6. Correlation Between HR Proteins Gene Expression and cGAS/STING Pathway Downstream Protein Gene Expression
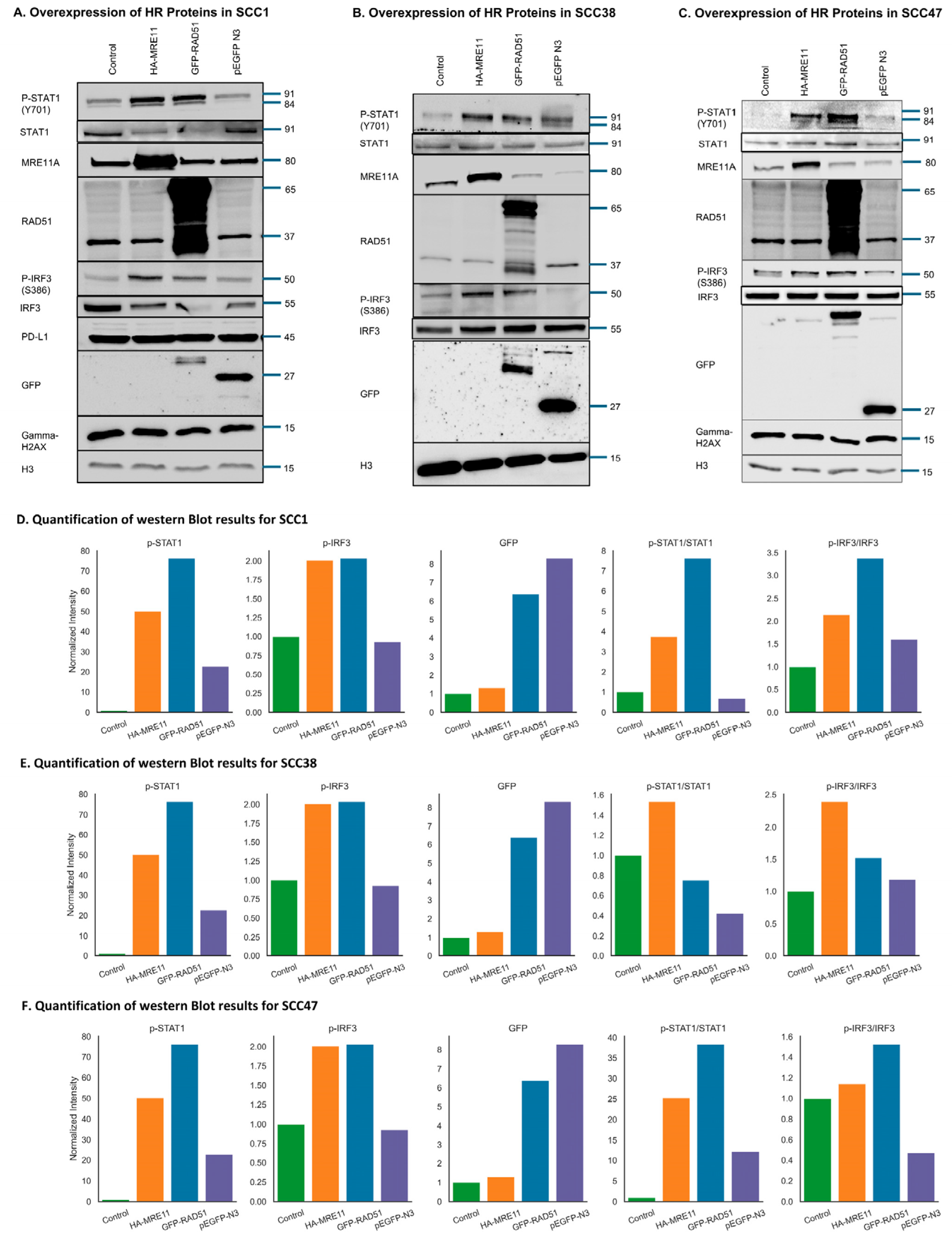
3.7. Activation of the cGAS/STING-Mediated Signaling Pathway by Upregulation of HR Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. Ca 2025, 75, 10. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers 2020, 6, 92. [Google Scholar] [CrossRef] [PubMed]
- Bosch, F.X.; Lorincz, A.; Munoz, N.; Meijer, C.J.L.M.; Shah, K.V. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 2002, 55, 244–265. [Google Scholar] [CrossRef]
- Chaturvedi, A.K.; Zumsteg, Z.S. A snapshot of the evolving epidemiology of oropharynx cancers. Cancer 2018, 124, 2893–2896. [Google Scholar] [CrossRef]
- Wallace, N.A. Catching HPV in the Homologous Recombination Cookie Jar. Trends Microbiol. 2020, 28, 191–201. [Google Scholar] [CrossRef]
- Holcomb, A.J.; Brown, L.; Tawfik, O.; Madan, R.; Shnayder, Y.; Thomas, S.M.; Wallace, N.A. DNA repair gene expression is increased in HPV positive head and neck squamous cell carcinomas. Virology 2020, 548, 174–181. [Google Scholar] [CrossRef]
- Ang, K.K.; Sturgis, E.M. Human Papillomavirus as a Marker of the Natural History and Response to Therapy of Head and Neck Squamous Cell Carcinoma. Semin. Radiat. Oncol. 2012, 22, 128–142. [Google Scholar] [CrossRef]
- Mordzińska-Rak, A.; Telejko, I.; Adamczuk, G.; Trombik, T.; Stepulak, A.; Błaszczak, E. Advancing Head and Neck Cancer Therapies: From Conventional Treatments to Emerging Strategies. Biomedicines 2025, 13, 1046. [Google Scholar] [CrossRef]
- Barsouk, A.; Aluru, J.S.; Rawla, P.; Saginala, K.; Barsouk, A. Epidemiology, risk factors, and prevention of head and neck squamous cell carcinoma. Med. Sci. 2023, 11, 42. [Google Scholar] [CrossRef]
- Dysphagia Section; Oral Care Study Group; Multinational Association of Supportive Care in Cancer/International Society of Oral Oncology; Raber-Durlacher, J.E.; Brennan, M.T.; Verdonck-de Leeuw, I.M.; Gibson, R.J.; Eilers, J.G.; Waltimo, T.; Bots, C.P.; et al. Swallowing dysfunction in cancer patients. Support. Care Cancer 2012, 20, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Vignard, J.; Mirey, G.; Salles, B. Ionizing-radiation induced DNA double-strand breaks: A direct and indirect lighting up. Ra-diother. Oncol. 2013, 108, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Bozzella, M.; Seluanov, A.; Gorbunova, V. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells. Cell Cycle 2008, 7, 2902–2906. [Google Scholar] [CrossRef]
- Mjelle, R.; Hegre, S.A.; Aas, P.A.; Slupphaug, G.; Drabløs, F.; Sætrom, P.; Krokan, H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair 2015, 30, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008, 18, 99–113. [Google Scholar] [CrossRef]
- San Filippo, J.; Sung, P.; Klein, H. Mechanism of Eukaryotic Homologous Recombination. Annu. Rev. Biochem. 2008, 77, 229–257. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Ceccaldi, R.; Shapiro, G.I.; D’ANdrea, A.D. Homologous Recombination Deficiency: Exploiting the Fundamental Vulnerability of Ovarian Cancer. Cancer Discov. 2015, 5, 1137–1154. [Google Scholar] [CrossRef]
- Nguyen, L.W.M.; Martens, J.; Van Hoeck, A.; Cuppen, E. Pan-cancer landscape of homologous recombination deficiency. Nat. Commun. 2020, 11, 5584. [Google Scholar] [CrossRef]
- Toh, M.; Ngeow, J. Homologous Recombination Deficiency: Cancer Predispositions and Treatment Implications. The Oncologist 2021, 26, e1526–e1537. [Google Scholar] [CrossRef]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef]
- Papalouka, C.; Adamaki, M.; Batsaki, P.; Zoumpourlis, P.; Tsintarakis, A.; Goulielmaki, M.; Fortis, S.P.; Baxevanis, C.N.; Zoumpourlis, V. DNA Damage Response Mechanisms in Head and Neck Cancer: Significant Implications for Therapy and Survival. Int. J. Mol. Sci. 2023, 24, 2760. [Google Scholar] [CrossRef] [PubMed]
- Gouttia, O.G.; Zhao, J.; Li, Y.; Zwiener, M.J.; Wang, L.; Oakley, G.G.; Peng, A. The MASTL-ENSA-PP2A/B55 axis modulates cisplatin resistance in oral squamous cell carcinoma. Front. Cell Dev. Biol. 2022, 10, 904719. [Google Scholar] [CrossRef]
- Wang, F.; Gouttia, O.G.; Wang, L.; Peng, A. PARP1 Upregulation in Recurrent Oral Cancer and Treatment Resistance. Front. Cell Dev. Biol. 2022, 9, 804962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chen, T.; Yang, X.; Cheng, H.; Späth, S.S.; Clavijo, P.E.; Chen, J.; Silvin, C.; Issaeva, N.; Su, X.; et al. Attenuated TRAF3 Fosters Activation of Alternative NF-κB and Reduced Expression of Antiviral Interferon, TP53, and RB to Promote HPV-Positive Head and Neck Cancers. Cancer Res. 2018, 78, 4613–4626. [Google Scholar] [CrossRef]
- Brenner, J.C.; Graham, M.P.; Kumar, B.; Saunders, L.M.; Kupfer, R.; Lyons, R.H.; Bradford, C.R.; Carey, T.E. Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 2009, 32, 417–426. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2013, 24, 185–199. [Google Scholar] [CrossRef]
- Li, Y.; Kardell, M.B.; Wang, F.; Wang, L.; Zhu, S.; Bessho, T.; Peng, A. The Sm core components of small nuclear ribonucleo-proteins promote homologous recombination repair. DNA Repair 2021, 108, 103244. [Google Scholar] [CrossRef]
- Driscoll, J.J.; Rixe, O. Overall survival: Still the gold standard: Why overall survival remains the definitive end point in cancer clinical trials. Cancer J. 2009, 15, 401–405. [Google Scholar]
- Eden, A.; Gaudet, F.; Waghmare, A.; Jaenisch, R. Chromosomal Instability and Tumors Promoted by DNA Hypomethylation. Science 2003, 300, 455. [Google Scholar] [CrossRef]
- Luo, X.; Li, Q.; Tang, Y.; Liu, Y.; Zou, Q.; Zheng, J.; Zhang, Y.; Xu, L. Predicting active enhancers with DNA methylation and histone modification. BMC Bioinform. 2023, 24, 414. [Google Scholar] [CrossRef]
- Dvorkin, S.; Cambier, S.; Volkman, H.E.; Stetson, D.B. New frontiers in the cGAS-STING intracellular DNA-sensing pathway. Immunity 2024, 57, 718–730. [Google Scholar] [CrossRef] [PubMed]
- Kastan, M.B.; Bartek, J. Cell-cycle checkpoints and cancer. Nature 2004, 432, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, R.D.; D’Andrea, A.D. DNA Repair Pathways in Clinical Practice: Lessons From Pediatric Cancer Susceptibility Syndromes. J. Clin. Oncol. 2006, 24, 3799–3808. [Google Scholar] [CrossRef]
- Moynahan, M.E.; Jasin, M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 2010, 11, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.-Y.; Yam, C.; Yamaguchi, H.; Hung, M.-C. Biomarkers beyond BRCA: Promising combinatorial treatment strategies in overcoming resistance to PARP inhibitors. J. Biomed. Sci. 2022, 29, 86. [Google Scholar] [CrossRef]
- Pourmasoumi, P.; Moradi, A.; Bayat, M. BRCA1/2 Mutations and Breast/Ovarian Cancer Risk: A New Insights Review. Reprod. Sci. 2024, 31, 3624–3634. [Google Scholar] [CrossRef]
- Arranz-Ledo, M.; Infante, M.; Lastra, E.; Olaverri, A.; Orozco, M.; Mateo, L.C.; Martínez, N.; Hernández, L.; Durán, M. Genetic Features of Tumours Arising in the Context of Suspected Hereditary Cancer Syndromes with RAD50, RAD51C/D, and BRIP1 Germline Mutations, Results of NGS-Reanalysis of BRCA/MMR-Negative Families. Genes 2025, 16, 458. [Google Scholar] [CrossRef]
- Liu, Y.; Lin, Z.; Yan, J.; Zhang, X.; Tong, M.-H. A Rad50-null mutation in mouse germ cells causes reduced DSB formation, abnormal DSB end resection and complete loss of germ cells. Development 2024, 151, dev202312. [Google Scholar] [CrossRef]
- Infante, M.; Arranz-Ledo, M.; Lastra, E.; Olaverri, A.; Ferreira, R.; Orozco, M.; Hernández, L.; Martínez, N.; Durán, M. Profiling of the genetic features of patients with breast, ovarian, colorectal and extracolonic cancers: Association to CHEK2 and PALB2 germline mutations. Clin. Chim. Acta 2023, 552, 117695. [Google Scholar] [CrossRef]
- Lee, J.-H.; Paull, T.T. Cellular functions of the protein kinase ATM and their relevance to human disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 796–814. [Google Scholar] [CrossRef]
- Stucki, M.; Jackson, S.P. γH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair 2006, 5, 534–543. [Google Scholar] [CrossRef]
- Lamarche, B.J.; Orazio, N.I.; Weitzman, M.D. The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett. 2010, 584, 3682–3695. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Nakamura, K.; Taniguchi, Y.; Paull, T.T. Ctp1/CtIP and the MRN Complex Collaborate in the Initial Steps of Ho-mologous Recombination. Mol. Cell 2007, 28, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-C.; Greenhough, L.A.; Masino, L.; Maslen, S.; Bajrami, I.; Tuppi, M.; Skehel, M.; Taylor, I.A.; West, S.C. Mechanism of single-stranded DNA annealing by RAD52–RPA complex. Nature 2024, 629, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Powell, S.N.; Kachnic, L.A. Roles of BRCA1 and BRCA2 in homologous recombination, DNA replication fidelity and the cellular response to ionizing radiation. Oncogene 2003, 22, 5784–5791. [Google Scholar] [CrossRef]
- Lok, B.H.; Powell, S.N. Molecular Pathways: Understanding the Role of Rad52 in Homologous Recombination for Therapeutic Advancement. Clin. Cancer Res. 2012, 18, 6400–6406. [Google Scholar] [CrossRef]
- Sallmyr, A.; Rashid, I.; Bhandari, S.K.; Naila, T.; Tomkinson, A.E. Human DNA ligases in replication and repair. DNA Repair 2020, 93, 102908. [Google Scholar] [CrossRef]
- Psyrri, A.; Gkotzamanidou, M.; Papaxoinis, G.; Krikoni, L.; Economopoulou, P.; Kotsantis, I.; Anastasiou, M.; Souliotis, V. The DNA damage response network in the treatment of head and neck squamous cell carcinoma. ESMO Open 2021, 6, 100075. [Google Scholar] [CrossRef]
- Heitmann, J.; Geeleher, P.; Zuo, Z.; Weichselbaum, R.R.; Vokes, E.E.; Fetscher, S.; Seiwert, T.Y. Poly (ADP-ribose) polymerase inhibitor efficacy in head and neck cancer. Oral Oncol. 2014, 50, 825–831. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Shen, J. Meta-analysis of the association between overexpression of RAD51 family genes and prognosis and clinical features in breast cancer. Sci. Rep. 2025, 15, 4229. [Google Scholar] [CrossRef]
- Saviozzi, S.; Ceppi, P.; Novello, S.; Ghio, P.; Iacono, M.L.; Borasio, P.; Cambieri, A.; Volante, M.; Papotti, M.; Calogero, R.A.; et al. Non–Small Cell Lung Cancer Exhibits Transcript Overexpression of Genes Associated with Homologous Recombination and DNA Replication Pathways. Cancer Res. 2009, 69, 3390–3396. [Google Scholar] [CrossRef] [PubMed]
- Maacke, H.; Jost, K.; Opitz, S.; Miska, S.; Yuan, Y.; Hasselbach, L.; Lüttges, J.; Kalthoff, H.; Stürzbecher, H.-W. DNA repair and recombination factor Rad51 is over-expressed in human pancreatic adenocarcinoma. Oncogene 2000, 19, 2791–2795. [Google Scholar] [CrossRef] [PubMed]
- Walline, H.M.; Goudsmit, C.M.; McHugh, J.B.; Tang, A.L.; Owen, J.H.; Teh, B.T.; McKean, E.; Glover, T.W.; Graham, M.P.; Prince, M.E.; et al. Integration of high-risk human papillomavirus into cellular cancer-related genes in head and neck cancer cell lines. Head Neck 2017, 39, 840–852. [Google Scholar] [CrossRef] [PubMed]
- Pinatti, L.; Walline, H.; Carey, T. Human papillomavirus genome integration and head and neck cancer. J. Dent. Res. 2018, 97, 691–700. [Google Scholar] [CrossRef]
- Wiest, T.; Schwarz, E.; Enders, C.; Flechtenmacher, C.; Bosch, F.X. Involvement of intact HPV16 E6/E7 gene expression in head and neck cancers with unaltered p53 status and perturbed pRb cell cycle control. Oncogene 2002, 21, 1510–1517. [Google Scholar] [CrossRef]
- Zur Hausen, H. Papillomaviruses in the causation of human cancers—A brief historical account. Virology 2009, 384, 260–265. [Google Scholar] [CrossRef]
- Cho, M.-G.; Kumar, R.J.; Lin, C.-C.; Boyer, J.A.; Shahir, J.A.; Fagan-Solis, K.; Simpson, D.A.; Fan, C.; Foster, C.E.; Goddard, A.M.; et al. MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature 2024, 625, 585–592. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soghli, N.; Khormali, A.; Peng, A. Homologous Recombination Is Associated with Enhanced Anti-Tumor Innate Immunity and Favorable Prognosis in Head and Neck Cancer. Cancers 2025, 17, 3999. https://doi.org/10.3390/cancers17243999
Soghli N, Khormali A, Peng A. Homologous Recombination Is Associated with Enhanced Anti-Tumor Innate Immunity and Favorable Prognosis in Head and Neck Cancer. Cancers. 2025; 17(24):3999. https://doi.org/10.3390/cancers17243999
Chicago/Turabian StyleSoghli, Negin, Aminollah Khormali, and Aimin Peng. 2025. "Homologous Recombination Is Associated with Enhanced Anti-Tumor Innate Immunity and Favorable Prognosis in Head and Neck Cancer" Cancers 17, no. 24: 3999. https://doi.org/10.3390/cancers17243999
APA StyleSoghli, N., Khormali, A., & Peng, A. (2025). Homologous Recombination Is Associated with Enhanced Anti-Tumor Innate Immunity and Favorable Prognosis in Head and Neck Cancer. Cancers, 17(24), 3999. https://doi.org/10.3390/cancers17243999





