Radiomics Analysis of QUS Spectral Parametric Images for Predicting the Risk of Breast Cancer Recurrence
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Participant Selection
2.2. Data Acquisition
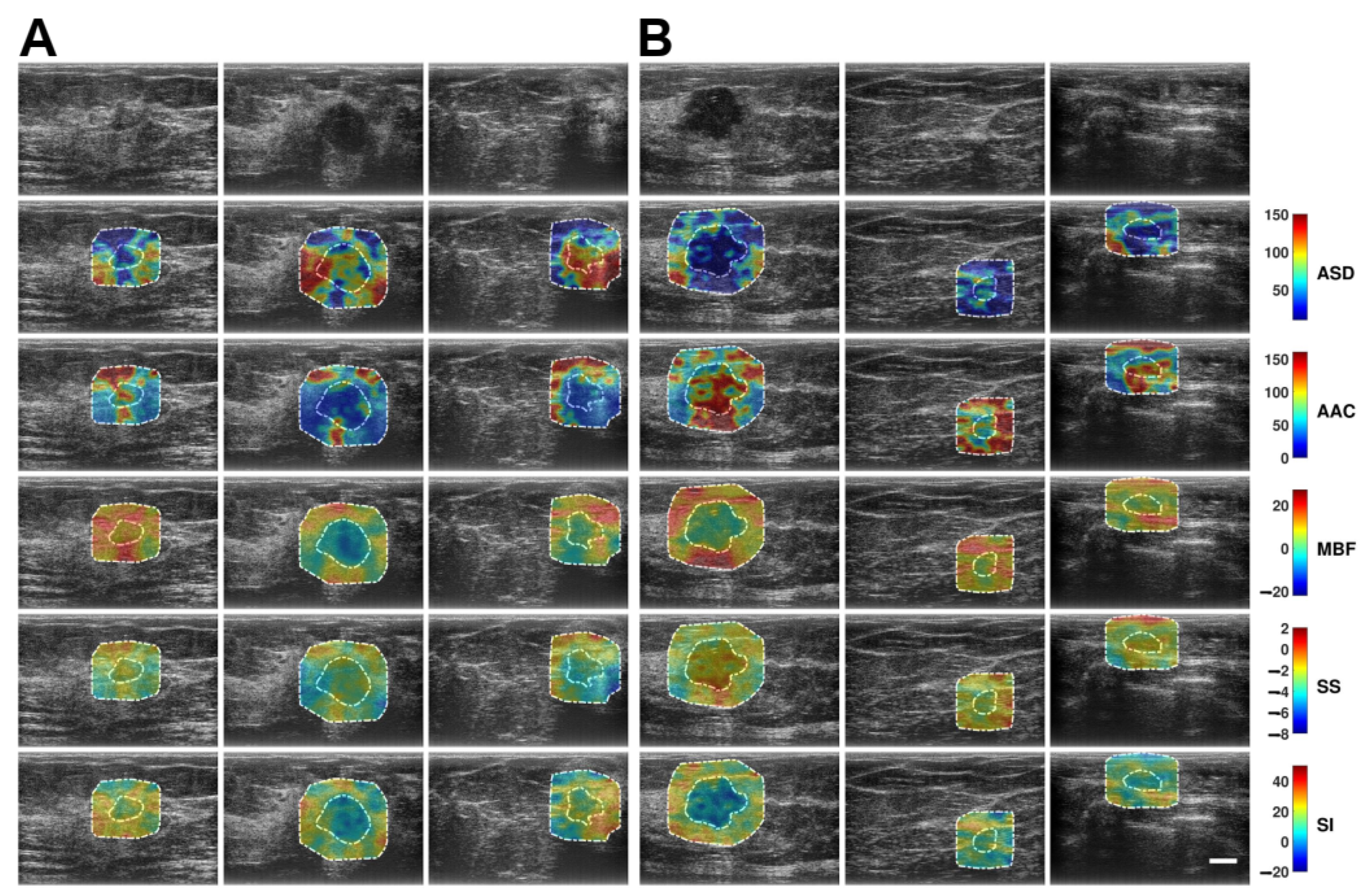
2.3. QUS Spectral Parametric Imaging
2.4. Feature Engineering
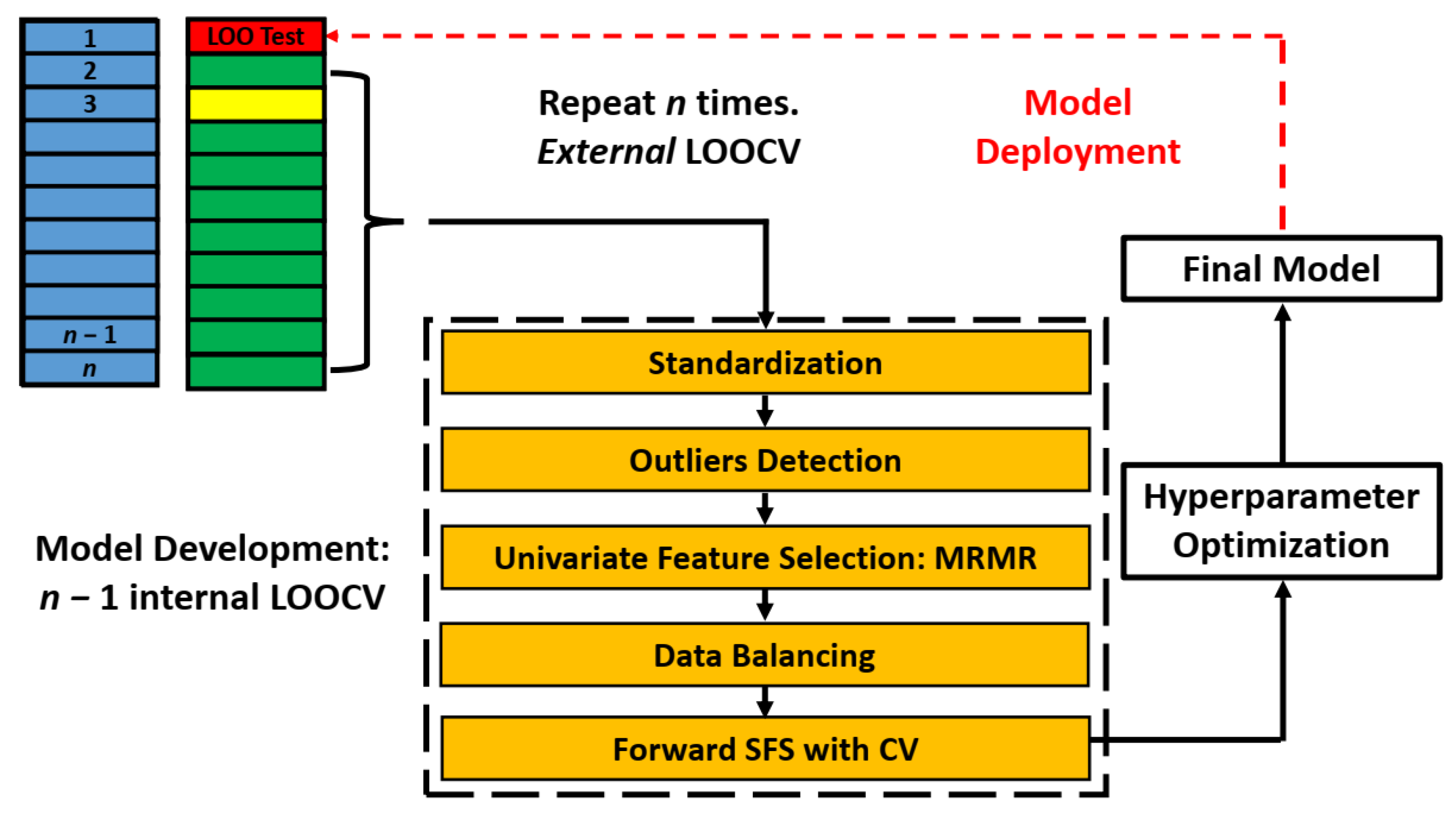
2.5. Data Preprocessing
2.5.1. Data Partitioning
2.5.2. Standardization and Outlier Identification
2.5.3. Feature Selection/Dimension Reduction
2.6. Model Building and Evaluation
2.7. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. QUS Spectral Parametric Images
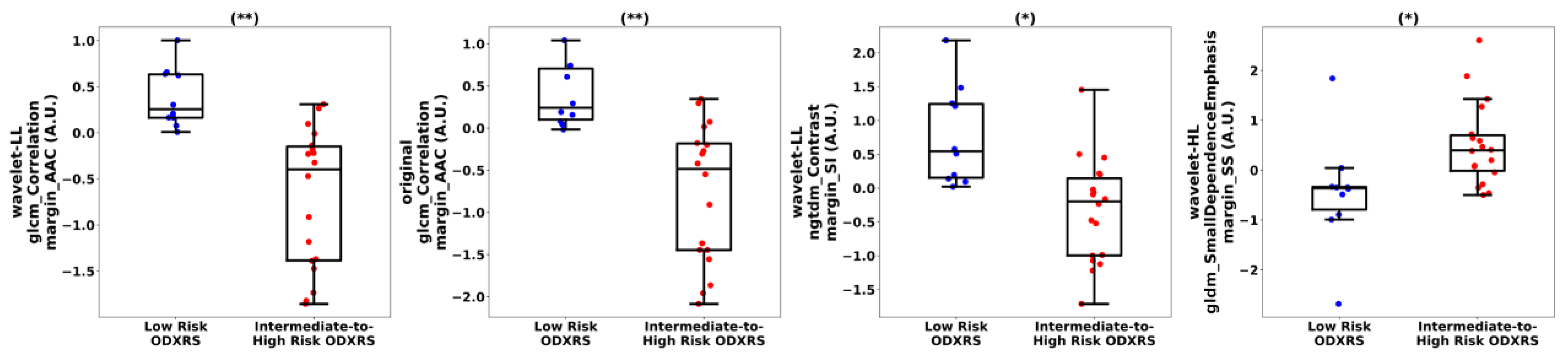
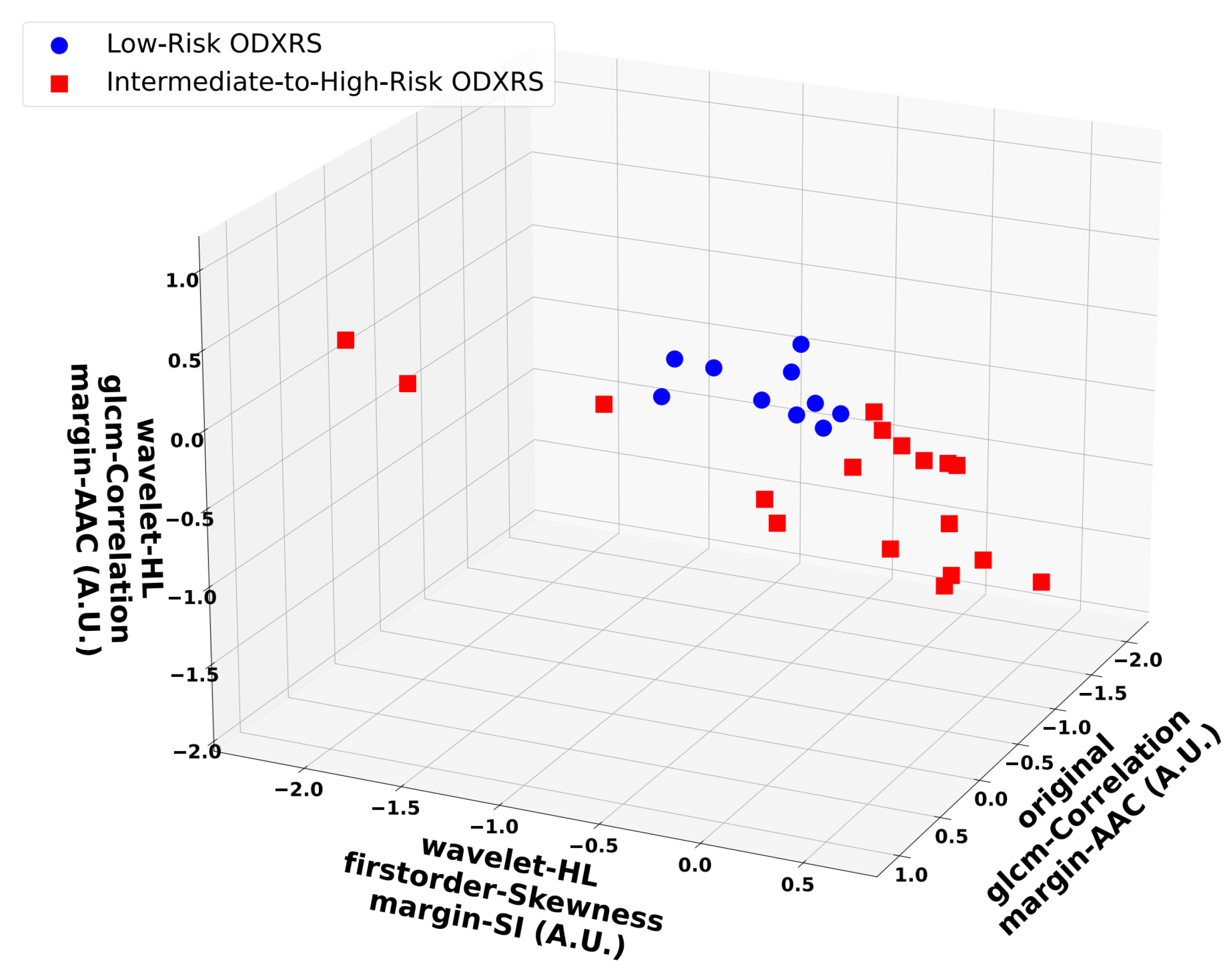
3.3. Feature Analysis
3.4. Classification Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| QUS | Quantitative ultrasound |
| ODXRS | Oncotype DX recurrence score |
| IQR | Inter-quartile range |
| LOOCV | Leave-one-out cross-validation |
| SVM-RBF | Support vector machine–radial basis function |
| AUROC | Area under the receiver operating characteristic curve |
| CI | Confidence interval |
| HR | Hormone receptor |
| HER2 | Human epidermal growth factor receptor 2 |
| LN | Lymph node |
| DRFS | Distant recurrence-free survival |
| RT | Radiation therapy |
| US RF | Ultrasound radio-frequency |
| CT | Computerized tomography |
| MRI | Magnetic resonance imaging |
| PET | Positron emission tomography |
| ER | Estrogen receptor |
| MBF | Mid-band fit |
| SS | Spectral slope |
| SI | Spectral intercept |
| ASD | Average scattering diameter |
| AAC | Average acoustic concentration |
| GLCM | Gray-level co-occurrence matrix |
| GLRM | Gray-level run-length matrix |
| GLSZM | Gray-level size zone matrix |
| NGTDM | Neighboring gray tone difference matrix |
| GLDM | Gray-level dependence matrix |
| MRMR | Maximal relevance minimal redundancy |
| SMOTE | Synthetic minority oversampling technique |
| SFS | Sequential feature selection |
| LDA | Linear discriminant analysis |
| KNN | k-nearest neighbors |
| RF | Random forest |
| PPV | Positive predictive value |
| NPV | Negative predictive value |
| AUPRC | Area under the precision–recall curve |
| IDC | Invasive ductal carcinoma |
| ILC | Invasive lobular carcinoma |
| DCIS | Ductal carcinoma in situ |
| FOV | Field-of-view |
| IDM | Inverse difference moment |
| CNN | Convolutional neural network |
| VIT | Vision transformer |
Appendix A
| Parameters | Values |
|---|---|
| Number of elements | 128 |
| Kerf width [µm] | 25 |
| Element width [µm] | 477 |
| Elevation [mm] | 4 |
| Elevation focus [mm] | 14 |
| Depth of focus [mm] | 16.7 |
| Center frequency [MHz] | 6.3 |
| Bandwidth [MHz] | 3–8 |
| f\# | 1.82 |
| Axial resolution at 15 mm (−6 dB) [µm] | 198 |
| Lateral resolution at 15 mm (−6 dB) [µm] | 483 |
| Feature Class | Feature Name |
|---|---|
| Morphological Features (n = 9) | Mesh Surface |
| Pixel Surface | |
| Perimeter | |
| Perimeter-to-Surface Ratio | |
| Sphericity | |
| Spherical Disproportion | |
| Maximum 2D Diameter | |
| Major Axis Length | |
| Minor Axis Length | |
| Elongation | |
| First-Order Statistical Features (n = 18) | 10th Percentile |
| 90th Percentile | |
| Energy | |
| Entropy | |
| Interquartile Range | |
| Kurtosis | |
| Maximum | |
| Mean Absolute Deviation (MAD) | |
| Mean | |
| Median | |
| Minimum | |
| Range | |
| Robust Mean Absolute Deviation (rMAD) | |
| Root Mean Squared (RMS) | |
| Skewness | |
| Total Energy | |
| Uniformity | |
| Variance | |
| GLCM (n = 24) | Autocorrelation |
| Cluster Prominence | |
| Cluster Shade | |
| Cluster Tendency | |
| Contrast | |
| Correlation | |
| Difference Average | |
| Difference Entropy | |
| Difference Variance | |
| Inverse Difference (ID) | |
| Inverse Difference Moment (IDM) | |
| Inverse Difference Moment Normalized (IDMN) | |
| Inverse Difference Normalized (IDN) | |
| Informational Measure of Correlation (IMC) 1 | |
| Informational Measure of Correlation (IMC) 2 | |
| Inverse Variance | |
| Joint Average | |
| Joint Energy | |
| Joint Entropy | |
| Maximal Correlation Coefficient (MCC) | |
| Maximum Probability | |
| Sum Average | |
| Sum Entropy | |
| Sum Squares | |
| GRLM (n = 16) | Gray-Level Nonuniformity |
| Gray-Level Nonuniformity Normalized | |
| Gray-Level Variance | |
| High Gray-Level Run Emphasis | |
| Long Run Emphasis | |
| Long Run High Gray-Level Emphasis | |
| Long Run Low Gray-Level Emphasis | |
| Low Gray-Level Run Emphasis | |
| Run Entropy | |
| Run Length Nonuniformity | |
| Run Length Nonuniformity Normalized | |
| Run Percentage | |
| Run Variance | |
| Short Run Emphasis | |
| Short Run High Gray-Level Emphasis | |
| Short Run Low Gray-Level Emphasis | |
| GLSZM (n = 16) | Gray-Level Nonuniformity |
| Gray-Level Nonuniformity Normalized | |
| Gray-Level Variance | |
| High Gray-Level Zone Emphasis | |
| Large Area Emphasis | |
| Large Area High Gray-Level Emphasis | |
| Large Area Low Gray-Level Emphasis | |
| Low Gray-Level Zone Emphasis | |
| Size Zone Nonuniformity | |
| Size Zone Nonuniformity Normalized | |
| Small Area Emphasis | |
| Small Area High Gray-Level Emphasis | |
| Small Area Low Gray-Level Emphasis | |
| Zone Entropy | |
| Zone Percentage | |
| Zone Variance | |
| GLDM (n = 14) | Dependence Entropy |
| Dependence Nonuniformity | |
| Dependence Nonuniformity Normalized | |
| Dependence Variance | |
| Gray-Level Nonuniformity | |
| Gray-Level Variance | |
| High Gray-Level Emphasis | |
| Large Dependence Emphasis | |
| Large Dependence High Gray-Level Emphasis | |
| Large Dependence Low Gray-Level Emphasis | |
| Low Gray-Level Emphasis | |
| Small Dependence Emphasis | |
| Small Dependence High Gray-Level Emphasis | |
| Small Dependence Low Gray-Level Emphasis | |
| NGTDM (n = 5) | Busyness |
| Coarseness | |
| Complexity | |
| Contrast | |
| Strength |
| Classifier | Hyperparameters | Values |
|---|---|---|
| SVM-Linear | C | {1 × 10−4, 1 × 10−3, 1 × 10−2, 1 × 10−1, 1, 10} |
| SVM-RBF | C | {1 × 10−4, 1 × 10−3, 1 × 10−2, 1 × 10−1, 1, 10} |
| γ | {1 × 10−3, 1 × 10−2, 1 × 10−1, 1, 10, 100, 1000} | |
| RF | N Estimators | {5, 10, …, 25} |
| Criterion | {‘gini’, ‘entropy’} | |
| Max Tree Depth | {3, 4} | |
| Max Features | {‘sqrt’, ‘log2’} | |
| Max Samples | {0.5, 0.75, 0.9} |
References
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Paik, S.; Tang, G.; Shak, S.; Kim, C.; Baker, J.; Kim, W.; Cronin, M.; Baehner, F.L.; Watson, D.; Bryant, J.; et al. Gene Expression and Benefit of Chemotherapy in Women With Node-Negative, Estrogen Receptor-Positive Breast Cancer. J. Clin. Oncol. 2006, 24, 3726–3727. [Google Scholar] [CrossRef]
- Syed, Y.Y. Oncotype DX breast recurrence score: A review of its use in early-stage breast cancer. Mol. Diagn. Ther. 2020, 24, 621–632. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Sparano, J.A.; Gray, R.J.; Ravdin, P.M.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E., Jr.; Dees, E.C.; Goetz, M.P.; et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N. Engl. J. Med. 2019, 380, 2395–2405. [Google Scholar] [CrossRef] [PubMed]
- Chevli, N.; Haque, W.; Tran, K.T.; Farach, A.M.; Schwartz, M.R.; Hatch, S.S.; Butler, E.B.; Teh, B.S. 21-Gene recurrence score predictive for prognostic benefit of radiotherapy in patients age ≥ 70 with T1N0 ER/PR + HER2- breast cancer treated with breast conserving surgery and endocrine therapy. Radiother. Oncol. 2022, 174, 37–43. [Google Scholar] [CrossRef]
- Jagsi, R.; Griffith, K.A.; Harris, E.E.; Wright, J.L.; Recht, A.; Taghian, A.G.; Lee, L.; Moran, M.S.; Small, W., Jr.; Johnstone, C.; et al. Omission of Radiotherapy After Breast-Conserving Surgery for Women with Breast Cancer with Low Clinical and Genomic Risk: 5-Year Outcomes of IDEA. J. Clin. Oncol. 2024, 42, 390–398. [Google Scholar] [CrossRef]
- Wang, S.Y.; Dang, W.; Richman, I.; Mougalian, S.S.; Evans, S.B.; Gross, C.P. Cost-Effectiveness Analyses of the 21-Gene Assay in Breast Cancer: Systematic Review and Critical Appraisal. J. Clin. Oncol. 2018, 36, 1619–1627. [Google Scholar] [CrossRef]
- Osapoetra, L.O.; Sannachi, L.; DiCenzo, D.; Quiaoit, K.; Fatima, K.; Czarnota, G.J. Breast lesion characterization using quantitative ultrasound (QUS) and derivative texture methods. Transl. Oncol. 2020, 13, 100827. [Google Scholar] [CrossRef]
- Destrempes, F.; Trop, I.; Allard, L.; Chayer, B.; Garcia-Duitama, J.; El Khoury, M.; Lalonde, L.; Cloutier, G. Added value of quantitative ultrasound and machine learning in BI-RADS 4-5 assessment of solid breast lesions. Ultrasound Med. Biol. 2020, 46, 436–444. [Google Scholar] [CrossRef]
- Rohrbach, D.; Wodlinger, B.; Wen, J.; Mamou, J.; Feleppa, E. High-frequency Quantitative Ultrasound for Imaging Prostate Cancer using a Novel Micro-Ultrasound Scanner. Ultrasound Med. Biol. 2018, 44, 1341–1354. [Google Scholar] [CrossRef]
- Sannachi, L.; Gangeh, M.; Sadeghi-Naini, A.; Bhargava, P.; Jain, A.; Tran, W.T.; Czarnota, G.J. Quantitative ultrasound monitoring of breast tumour response to neoadjuvant chemotherapy: Comparison of results among clinical scanners. Ultrasound Med. Biol. 2020, 46, 1142–1157. [Google Scholar] [CrossRef]
- Sannachi, L.; Osapoetra, L.O.; DiCenzo, D.; Halstead, S.; Wright, F.; Look-Hong, N.; Slodkowska, E.; Gandhi, S.; Curpen, B.; Kolios, M.C.; et al. A priori prediction of breast cancer response to neoadjuvant chemotherapy using quantitative ultrasound, texture derivative and molecular subtype. Sci. Rep. 2023, 13, 22687. [Google Scholar] [CrossRef] [PubMed]
- Osapoetra, L.O.; Dasgupta, A.; DiCenzo, D.; Fatima, K.; Quiaoit, K.; Saifuddin, M.; Karam, I.; Poon, I.; Husain, Z.; Tran, W.T.; et al. Quantitative US Delta Radiomics to Predict Radiation Response in Individuals with Head and Neck Squamous Cell Carcinoma. Radiology 2024, 6, e340029. [Google Scholar] [CrossRef]
- Mamou, J.; Coron, A.; Hata, M.; Machi, J.; Yanagihara, E.; Laugier, P.; Feleppa, E.J. Three-dimensional high-frequency characterization of cancerous lymph nodes. Ultrasound Med. Biol. 2010, 36, 361–375. [Google Scholar] [CrossRef]
- Liao, Y.Y.; Tsui, P.H.; Li, C.H.; Chang, K.J.; Kuo, W.H.; Chang, C.C.; Yeh, C.K. Classification of scattering media within benign and malignant breast tumours based on ultrasound texture-feature-based and Nakagami-parameter images. Med. Phys. 2011, 38, 2198–2207. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, A.; Razzaque, R.R.; Muhtadi, S.; Shafiullah, A.; Abir, E.U.; Garra, B.S.; Alam, S.K. Ultrasound classification of breast masses using a comprehensive Nakagami imaging and machine learning framework. Ultrasonics 2022, 124, 106744. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Fan, M.; Cui, Y.; You, C.; Liu, L.; Gu, Y.; Peng, W.; Bai, Q.; Gao, X.; Li, L. Radiogenomic Signatures of Oncotype DX Recurrence Score Enable Prediction of Survival in Estrogen Receptor-Positive Breast Cancer: A Multicohort Study. Radiology 2022, 302, 516–524. [Google Scholar] [CrossRef]
- Sutton, E.J.; Oh, J.H.; Dashevsky, B.Z.; Veeraraghavan, H.; Apte, A.P.; Thakur, S.B.; Deasy, J.O.; Morris, E.A. Breast cancer subtype intertumor heterogeneity: MRI-based features predict results of a genomic assay. J. Magn. Reson. Imaging 2015, 42, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Burnside, E.S.; Drukker, K.; Hoadley, K.A.; Fan, C.; Conzen, S.D.; Whitman, G.J.; Sutton, E.J.; Net, J.M.; et al. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology 2016, 281, 382–391. [Google Scholar] [CrossRef]
- Davey, M.G.; Davey, M.S.; Ryan, É.J.; Boland, M.R.; McAnena, P.F.; Lowery, A.J.; Kerin, M.J. Is radiomic MRI a feasible alternative to OncotypeDX recurrence score testing? A systematic review and meta-analysis. BJS Open 2021, 5, zrab081. [Google Scholar] [CrossRef]
- Ha, R.; Chang, P.; Mutasa, S.; Karcich, J.; Goodman, S.; Blum, E.; Kalinsky, K.; Liu, M.Z.; Jambawalikar, S. Convolutional Neural Network Using a Breast MRI Tumor Dataset Can Predict Oncotype Dx Recurrence Score. J. Magn. Reson. Imaging 2019, 49, 518–524. [Google Scholar] [CrossRef]
- Romeo, V.; Cuocolo, R.; Sanduzzi, L.; Carpentiero, V.; Caruso, M.; Lama, B.; Garifalos, D.; Stanzione, A.; Maurea, S.; Brunetti, A. MRI radiomics and machine learning for the prediction of onctoype Dx recurrence score in invasive breast cancer. Cancers 2023, 15, 1840. [Google Scholar] [CrossRef] [PubMed]
- Mao, N.; Yin, P.; Zhang, H.; Zhang, K.; Song, X.; Xing, D.; Chu, T. Mammography-based radiomics for predicting the risk of breast cancer recurrence: A multicenter study. Br. J. Radiol. 2021, 94, 20210348. [Google Scholar] [CrossRef]
- Nam, K.J.; Park, H.; Sook, E.; Lim, Y.; Cho, H.H.; Lee, J.E. Radiomics signature on 4T dynamic contrast-enhanced magnetic resonance imaing for estregon receptor-positive invasive breast cancer. Medicine 2019, 98, e15871. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Adam, R.; Maldijan, T.; Duong, T.Q. Radiomics Analysis of Breast MRI to Predict Oncotype Dx Recurrence Score: Systematic Review. Diagnostics 2025, 15, 1054. [Google Scholar] [CrossRef]
- Mamou, J.; Oelze, M. Quantitative Ultrasound in Soft Tissues; Mamou, J., Oelze, M., Eds.; Springer Nature: Cham, Switzerland, 2023. [Google Scholar]
- Labyed, Y.; Bigelow, T.A.; McFarlin, B.L. Estimate of the attenuation coefficient using a clinical array transducer for the detection of cervical ripening in human pregnancy. Ultrasonics 2011, 51, 34–39. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. In IEEE Transactions on Systems, Man, and Cybernetics; IEEE: New York City, NY, USA, 1973; pp. 610–621. [Google Scholar] [CrossRef]
- Galloway, M.M. Texture analysis using gray level run lengths. Comput. Graph. Image Process. 1975, 4, 172–179. [Google Scholar] [CrossRef]
- Dasarathy, B.V.; Holder, E.B. Image characterizations based on joint gray level run-length distributions. Pattern Recognit. Lett. 1991, 12, 497–502. [Google Scholar] [CrossRef]
- Chu, A.; Sehgal, C.M.; Greenleaf, J.F. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit. Lett. 1990, 11, 415–419. [Google Scholar] [CrossRef]
- Tang, X. Texture information in run-length matrices. IEEE Trans. Image Process. 1998, 7, 1602–1609. [Google Scholar] [CrossRef]
- Thibault, G.; Angulo, J.; Meyer, F. Advanced statistical matrices for texture characterization: Application to cell classification. IEEE Trans. Biomed. Eng. 2014, 61, 630–637. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Sun, C.; Wee, W.G. Neighboring gray level dependence matrix for texture classification. Comput. Vis. Graph. Image Process. 1983, 23, 341–352. [Google Scholar] [CrossRef]
- Liu, F.T.; Ting, K.M.; Zhou, Z.H. Isolation Forest. In Proceedings of the 2008 Eighth IEEE International Conference on Data Mining, Pisa, Italy, 15–19 December 2008; pp. 413–422. [Google Scholar] [CrossRef]
- Peng, H.; Long, F.; Ding, C. Feature selection based on mutual information criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1226–1238. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Flanagan, M.B.; Dabbs, D.J.; Brufsky, A.M.; Beriwal, S.; Bhargava, R. Histopathologic variables predict Oncotype DXTM recurrence score. Mod. Pathol. 2008, 21, 1255–1261. [Google Scholar] [CrossRef]
- Orucevic, A.; Bell, J.L.; McNabb, A.P.; Heidel, R.E. Oncotype DX breast cancer recurrence score can be predicted with a novel nomogram using clinicopathologic data. Breast Cnacer Res. Treat. 2017, 163, 51–61. [Google Scholar] [CrossRef]
- Orucevic, A.; Bell, J.L.; King, M.; McNabb, A.P.; Heidel, R.E. Nomogram update based on TAILORx clinical trial results-Oncotype DX breast cancer recurrence score can be predicted using clinicopathologic data. Breast 2019, 46, 116–125. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef] [PubMed]
- Chalkidou, A.; O’Doherty, M.J.; Marsden, P.K. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS ONE 2015, 10, e0124165. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Osapoetra, L.O.; Moslemi, A.; Moore-Palhares, D.; Halstead, S.; Alberico, D.; Hwang, A.; Sannachi, L.; Curpen, B.; Czarnota, G.J. End-to-end CNN-based deep learning enhances breast lesion characterization using quantitative ultrasound (QUS) spectral parametric images. Sci. Rep. 2025, 15, 32805. [Google Scholar] [CrossRef]
- Pang, J.; Ding, N.; Liu, X.; He, X.; Zhou, W.; Xie, H.; Feng, J.; Li, Y.; He, Y.; Wang, S.; et al. Prognostic value of the baseline systemic immune-inflammation index in HER2-positive metastatic breast cancer: Exploratory analysis of two prospective trials. Ann. Surg. Oncol. 2025, 32, 750–759. [Google Scholar] [CrossRef]
- Botlagunta, M.; Botlagunta, M.; Venkata, M.D.; Kanakapudi, C.; Khan, Z. Correlation-based Comparative Machine Learning Analysis for the Classification of Metastatic Breast Cancer Using Blood Profile. Eurasian J. Med. Oncol. 2024, 8, 152–164. [Google Scholar] [CrossRef]
- Peng, J.; Zhao, M.; Wang, S. Discrimination Model Construction for Non-Lactational Mastitis and Breast Cancer Based on Imaging Features. Br. J. Hosp. Med. 2024, 85, 1–15. [Google Scholar] [CrossRef]
| Characteristics | Low-Risk ODXRS (n = 10) | Intermediate-to-High-Risk ODXRS (n = 21) | All (n = 31) |
|---|---|---|---|
| Age (y) | |||
| Mean (SD) | 54 (8) | 57 (12) | 56 (11) |
| Median (Q1, Q3) | 52 (48, 58) | 56 (49, 68) | 55 (48, 63) |
| Min, max | 46, 74 | 33, 78 | 33, 78 |
| Tumor size (cm) | |||
| Mean (SD) | 2.4 (2.3) | 2.1 (1.1) | 2.2 (1.6) |
| Median (Q1, Q3) | 1.4 (1.2, 2.1) | 1.9 (1.3, 2.8) | 1.7 (1.2, 2.8) |
| Min, max | 1.1, 8.9 | 0.7, 4.6 | 0.7, 8.9 |
| Invasive tumor type n (%) | |||
| Invasive ductal carcinoma | 7 (70%) | 14 (67%) | 21 (68%) |
| Invasive lobular carcinoma | 1 (10%) | 3 (14%) | 4 (13%) |
| Ductal carcinoma in situ | 1 (10%) | 3 (14%) | 4 (13%) |
| Other | 1 (10%) | 1 (5%) | 2 (6%) |
| Histologic tumor grade, n (%) | |||
| Grade I | 2 (20%) | 5 (24%) | 7 (23%) |
| Grade II | 6 (60%) | 12 (57%) | 18 (58%) |
| Grade III | 2 (20%) | 4 (19%) | 6 (19%) |
| Hormone receptor status, n (%) | |||
| ER+, PR+, HER2− | 10 (100%) | 21 (100%) | 31 (100%) |
| Classifier | Recall (%) (CI) | Specificity (%) (CI) | Accuracy (%) (CI) | Balanced Accuracy (%) (CI) | Precision (%) (CI) | NPV (%) (CI) | F1-Score (%) (CI) | AUROC (CI) | AUPRC (CI) |
|---|---|---|---|---|---|---|---|---|---|
| LDA | 67 | 50 | 61 | 58 | 74 | 42 | 70 | 0.67 | 0.82 |
| (14/21) | (5/10) | (19/31) | (14/19) | (5/12) | |||||
| (50–83) | (32–68) | (44–78) | (41–76) | (58–89) | (24–59) | (54–86) | (0.47–0.87) | (0.67–0.97) | |
| KNN k = 5 | 71 | 70 | 71 | 71 | 83 | 54 | 77 | 0.78 | 0.85 |
| (15/21) | (7/10) | (22/31) | (15/18) | (7/13) | |||||
| (56–87) | (54–86) | (55–87) | (55–87) | (70–96) | (36–71) | (62–92) | (0.62–0.94) | (0.72–0.99) | |
| SVM Linear | 71 | 60 | 68 | 66 | 79 | 50 | 75 | 0.54 | 0.76 |
| (15/21) | (6/10) | (21/31) | (15/19) | (6/12) | |||||
| (56–87) | (43–77) | (51–84) | (49–82) | (65–93) | (32–68) | (60–90) | (0.32–0.76) | (0.59–0.93) | |
| SVM-RBF (*) | 86 | 100 | 90 | 93 | 100 | 77 | 92 | 0.95 | 0.98 |
| (18/21) | (10/10) | (28/31) | (18/18) | (10/13) | |||||
| (73–98) | (100–100) | (80–100) | (84–100) | (100–100) | (62–92) | (83–100) | (0.88–1.00) | (0.94–1.00) | |
| RF | 67 | 40 | 58 | 53 | 70 | 36 | 68 | 0.55 | 0.76 |
| (14/21) | (4/10) | (18/31) | (14/20) | (4/11) | |||||
| (50–83) | (23–57) | (41–75) | (36–71) | (54–86) | (19–53) | (52–85) | (0.34–0.77) | (0.59–0.93) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osapoetra, L.O.; Dinniwell, G.; Anzola Pena, M.L.; Alberico, D.; Sannachi, L.; Czarnota, G.J. Radiomics Analysis of QUS Spectral Parametric Images for Predicting the Risk of Breast Cancer Recurrence. Cancers 2025, 17, 3810. https://doi.org/10.3390/cancers17233810
Osapoetra LO, Dinniwell G, Anzola Pena ML, Alberico D, Sannachi L, Czarnota GJ. Radiomics Analysis of QUS Spectral Parametric Images for Predicting the Risk of Breast Cancer Recurrence. Cancers. 2025; 17(23):3810. https://doi.org/10.3390/cancers17233810
Chicago/Turabian StyleOsapoetra, Laurentius Oscar, Graham Dinniwell, Maria Lourdes Anzola Pena, David Alberico, Lakshmanan Sannachi, and Gregory J. Czarnota. 2025. "Radiomics Analysis of QUS Spectral Parametric Images for Predicting the Risk of Breast Cancer Recurrence" Cancers 17, no. 23: 3810. https://doi.org/10.3390/cancers17233810
APA StyleOsapoetra, L. O., Dinniwell, G., Anzola Pena, M. L., Alberico, D., Sannachi, L., & Czarnota, G. J. (2025). Radiomics Analysis of QUS Spectral Parametric Images for Predicting the Risk of Breast Cancer Recurrence. Cancers, 17(23), 3810. https://doi.org/10.3390/cancers17233810









