Clinical and Imaging-Based Prognostic Models for Recurrence and Local Tumor Progression Following Thermal Ablation of Hepatocellular Carcinoma: A Systematic Review
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Eligibility Criteria
2.3. Methodological and Reporting Quality
3. Results
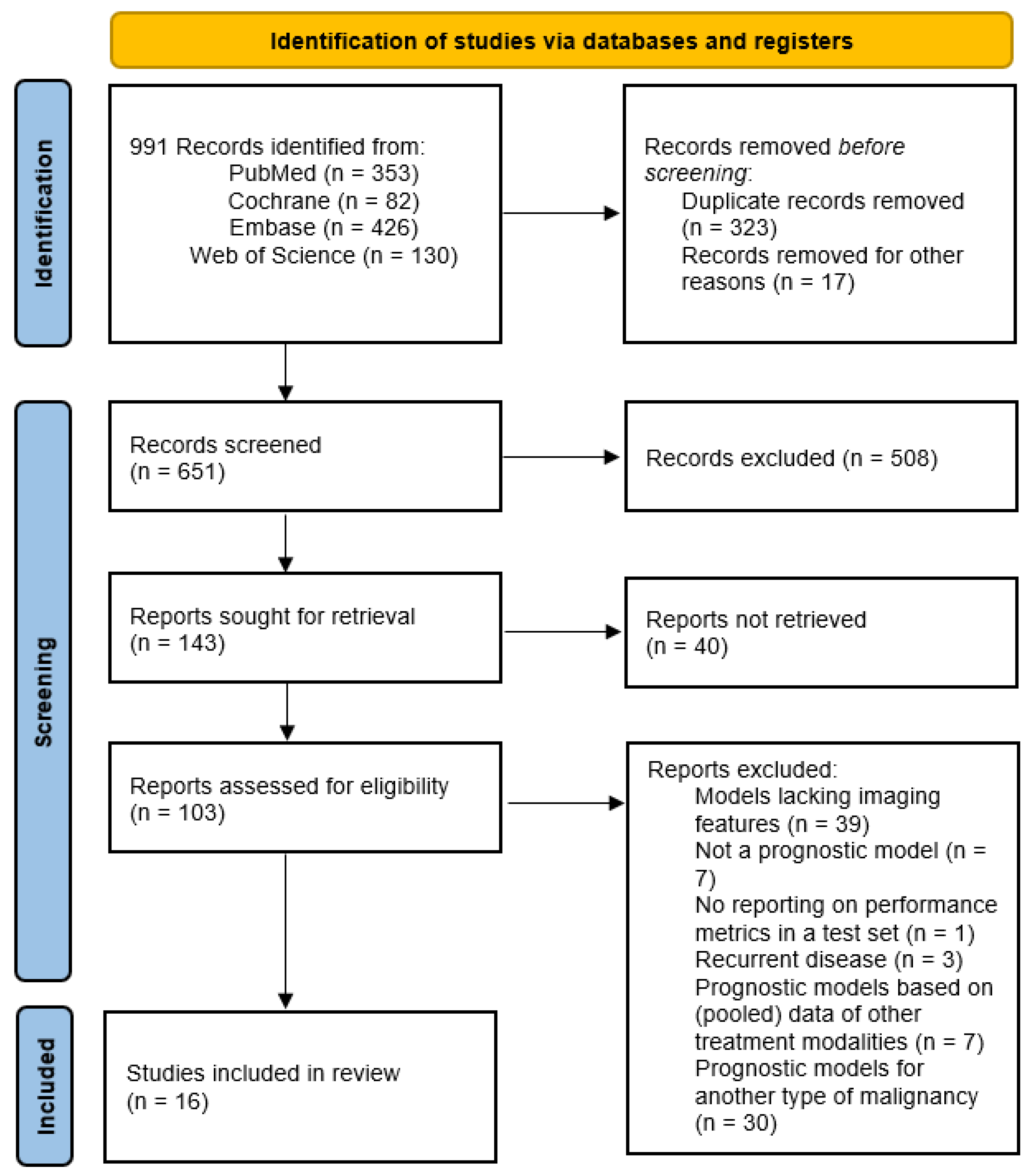
3.1. Study Selection
3.2. Study Characteristics
3.3. Prognostic Model Outcome
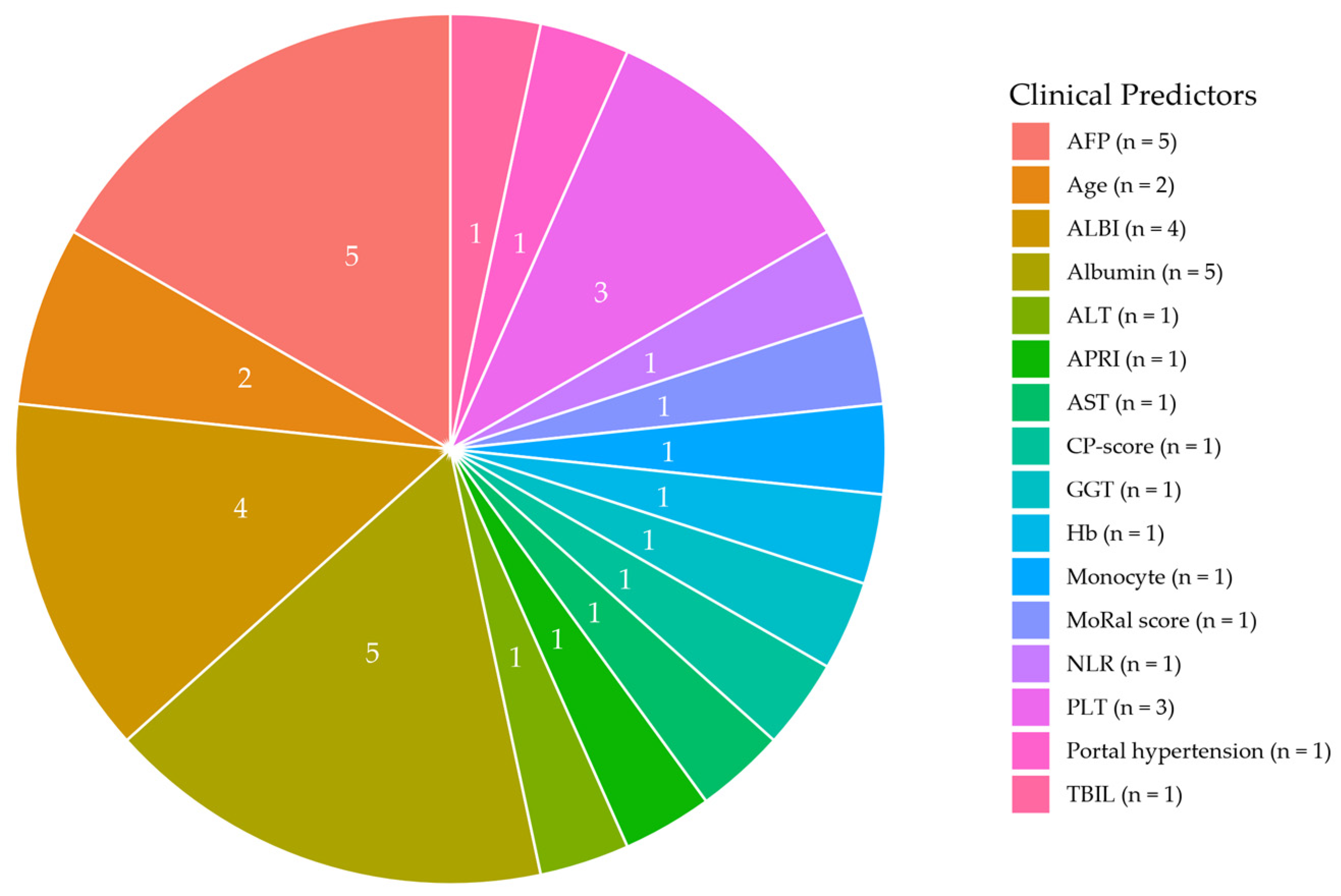
3.4. Prognostic Factors
3.4.1. Clinical Predictors
3.4.2. Imaging-Based Predictors
3.5. Feature Selection Techniques and Model Development
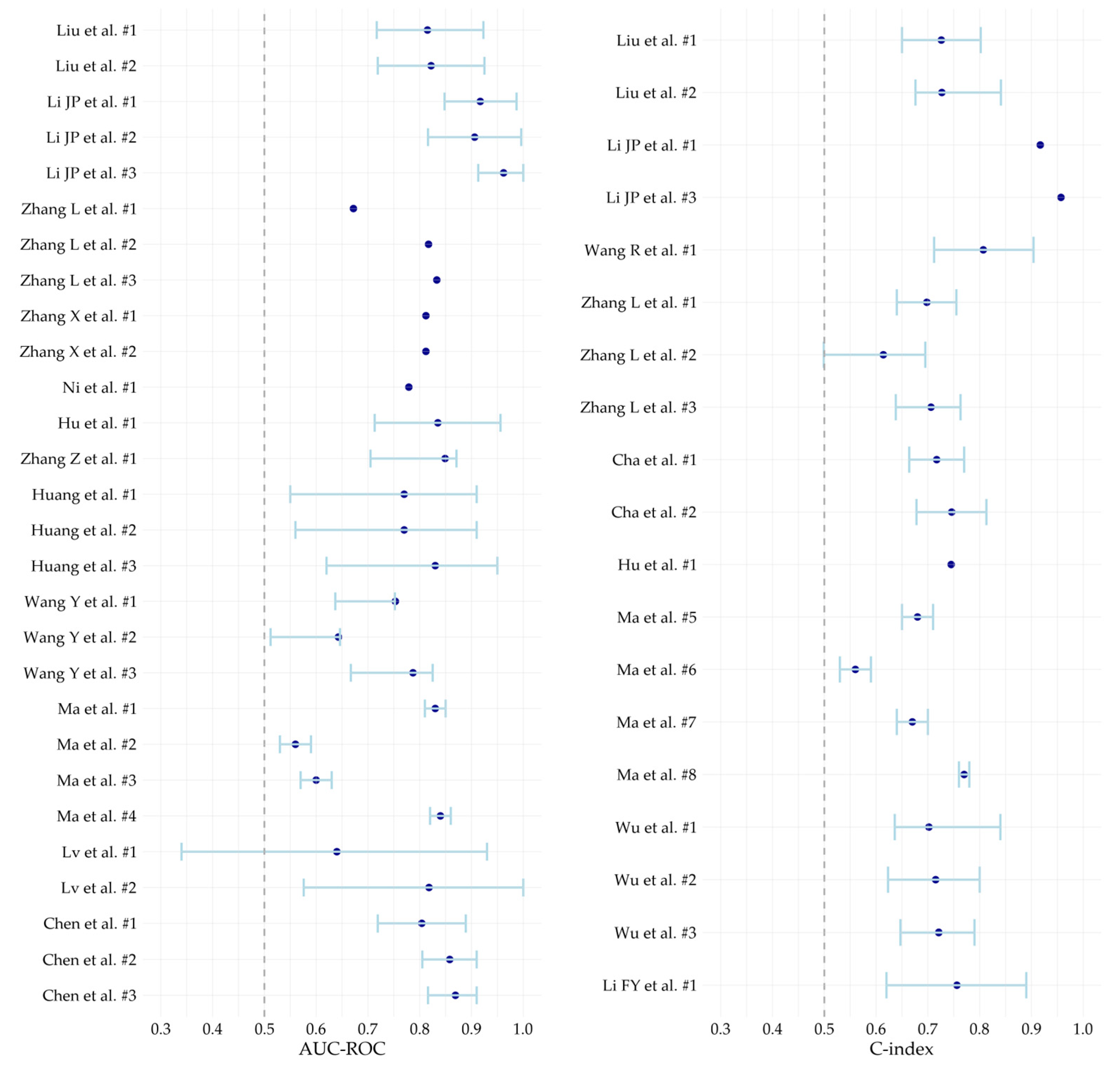
3.6. Model Performance
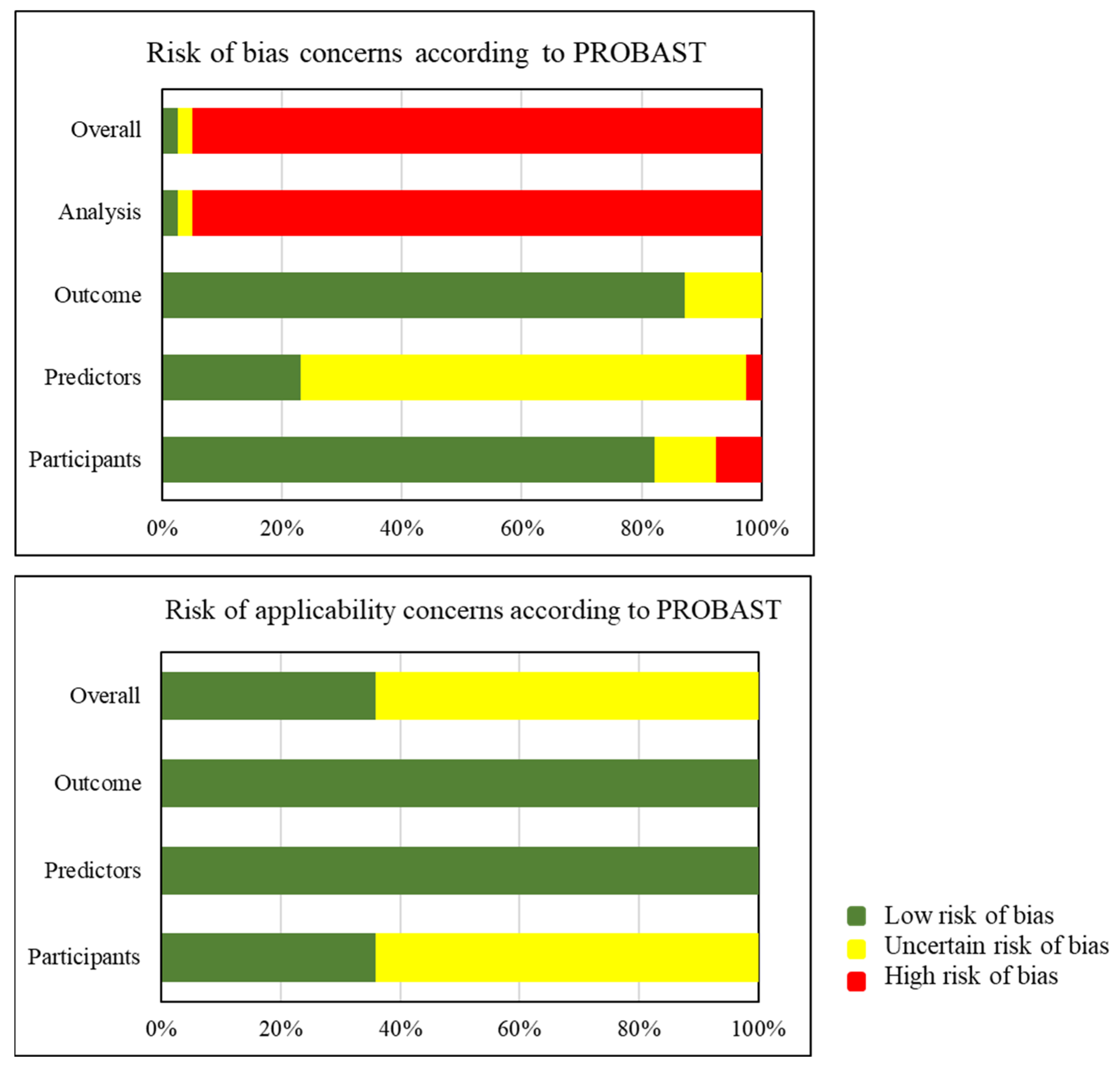
3.7. Risk of Bias Assessment
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| AI | Artificial intelligence |
| AFP | Alpha fetoprotein |
| ALBI | Albumin-bilirubin |
| AUC-ROC | Area under the receiver operating characteristic curve |
| BCLC | Barcelona Clinic Liver Cancer |
| CEUS | Contrast-enhanced ultrasound |
| CNN | Convolutional neural network |
| CI | Confidence interval |
| C-index | Concordance index |
| DL | Deep learning |
| EPV | Event per variable |
| ER | Early recurrence |
| HCC | Hepatocellular carcinoma |
| HL | Hosmer–Lemeshow |
| LI-RADS | Liver Imaging Reporting and Data System |
| LTP | Local tumor progression |
| LTPFS | Local tumor progression free survival |
| LR | Late recurrence |
| MWA | Microwave ablation |
| MV | Multivariate |
| PRISMA | Preferred reporting items for systematic reviews and meta-analysis |
| PROSPERO | International prospective register of systematic review |
| RFA | Radiofrequency ablation |
| RFS | Recurrence free survival |
| ROB | Risk of bias |
| TA | Thermal ablation |
| UV | Univariate |
Appendix A
| Database | Search | Hits |
|---|---|---|
| Pubmed | (((thermoablat*[tiab] OR “RFA”[tiab] OR “MWA”[tiab] OR ((“thermo”[tiab] OR “thermal”[tiab] OR “radiofrequenc*”[tiab] OR “Microwaves”[Majr] OR “Microwave*”[tiab]) AND (“Ablation techniques”[Mesh] OR “Radiofrequency Ablation”[Majr] OR “ablat*”[tiab]))) AND (“Liver Neoplasms”[Mesh] OR HCC[tiab] OR ((“liver”[tiab] OR “livers”[tiab] OR “hepatic”[tiab] OR hepatocellular[tiab]) AND (“neoplasms”[tiab] OR “neoplasm”[tiab] OR “cancer”[tiab] OR “cancers”[tiab] OR “tumor”[tiab] OR “tumors”[tiab] OR “tumour”[tiab] OR “tumours”[tiab] OR “malignan*”[tiab] OR “carcinom*”[tiab]))) AND ((“Nomograms”[Mesh] OR nomogram*[tiab] OR nomograph*[tiab] OR “prognostic model*”[tiab] OR “predictive model*”[tiab] OR “prediction model*”[tiab] OR ((“prognos*”[ti] OR “predict*”[ti]) AND model*[ti])) OR ((“prognos*”[ti] OR “predict*”[ti] OR “prognosis”[majr]) AND (“Artificial Intelligence”[Mesh] OR “AI”[ti] OR “artificial intelligen*”[tiab] OR “AI”[tiab] OR “machine learn*”[tiab] OR “deep learn*”[tiab] OR “neural network*”[tiab] OR “support vector machine*”[tiab] OR “reinforcement learning”[tiab] OR “Markov”[tiab] OR “decision tree*”[tiab] OR “random forest”[tiab] OR “Bayesian network*”[tiab] OR “convolutional network”[tiab] OR “radiomic*”[tiab] OR “gradient boost*” [tiab] OR “feature selection*”[tiab])))) OR ((thermoablat*[tiab] OR “RFA”[tiab] OR “MWA”[tiab] OR ((“thermo”[tiab] OR “thermal”[tiab] OR “radiofrequenc*”[tiab] OR “Microwaves”[Majr] OR “Microwave*”[tiab]) AND (“Ablation techniques”[Mesh] OR “Radiofrequency Ablation”[Majr] OR “ablat*”[tiab]))) AND (“Carcinoma, Hepatocellular”[Majr] OR “HCC”[tiab] OR “hepatocellular carcinoma*”[tiab]) AND (“primary”[tiab]) AND ((“Nomograms”[Mesh] OR nomogram*[tiab] OR nomograph*[tiab] OR “prognostic model*”[tiab] OR “predictive model*”[tiab] OR “prediction model*”[tiab] OR ((“prognos*”[ti] OR “predict*”[ti]) AND model*[ti])) OR (((“prognos*”[ti] OR “predict*”[ti] OR “prognosis”[majr]) AND (“Artificial Intelligence”[Mesh] OR “AI”[ti] OR “artificial intelligen*”[tiab] OR “AI”[tiab] OR “machine learn*”[tiab] OR “deep learn*”[tiab] OR “neural network*”[tiab] OR “support vector machine*”[tiab] OR “reinforcement learning”[tiab] OR “Markov”[tiab] OR “decision tree*”[tiab] OR “random forest”[tiab] OR “Bayesian network*”[tiab] OR “convolutional network”[tiab] OR “radiomic*”[tiab] OR “gradient boost*”[tiab] OR “feature selection*” [tiab])))))) OR ((thermoablat*[tiab] OR “RFA”[tiab] OR “MWA”[tiab] OR ((“thermo”[tiab] OR “thermal”[tiab] OR “radiofrequenc*”[tiab] OR “Microwaves”[Mesh] OR “Microwave*”[tiab]) AND (“Ablation techniques”[Mesh] OR “Radiofrequency Ablation”[Mesh] OR “ablat*”[tiab]))) AND ((“Liver Neoplasms”[Mesh] OR ((“liver”[tiab] OR “livers”[tiab] OR “hepatic”[tiab]) AND (“neoplasms”[tiab] OR “neoplasm”[tiab] OR “cancer”[tiab] OR “cancers”[tiab] OR “tumor”[tiab] OR “tumors”[tiab] OR “tumour”[tiab] OR “tumours”[tiab] OR “malignan*”[tiab] OR “carcinom*”[tiab]))) OR (“primary”[tiab] OR “Carcinoma, Hepatocellular”[Mesh] OR “HCC”[tiab] OR “hepatocellular carcinoma*”[tiab])))) AND (“Artificial Intelligence”[Mesh] OR “AI”[ti] OR “artificial intelligen*”[tiab] OR “AI”[tiab] OR “machine learn*”[tiab] OR “deep learn*”[tiab] OR “neural network*”[tiab] OR “support vector machine*”[tiab] OR “reinforcement learning”[tiab] OR “Markov”[tiab] OR “decision tree*”[tiab] OR “random forest”[tiab] OR “Bayesian network*”[tiab] OR “convolutional network”[tiab] OR “radiomic*”[tiab] OR “gradient boost*”[tiab] OR “feature selection*”[tiab])) | 353 |
| WebScience | ((((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND (HCC OR ((“liver” OR “livers” OR “hepatic” OR hepatocellular) AND (“neoplasms” OR “neoplasm” OR “cancer” OR “cancers” OR “tumor” OR “tumors” OR “tumour” OR “tumours” OR malignan* OR carcinom*))) AND ((nomogram* OR nomograph* OR “prognostic model” OR “prognostic models” OR “predictive model” OR “predictive model” OR “prediction model” OR “prediction models” OR ((prognos* OR predict*) AND model*)) OR ((prognos* OR predict*) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian networks” OR “convolutional network” OR “convolutional networks”)))) OR ((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND (“HCC” OR “hepatocellular carcinoma” OR “hepatocellular carcinomas”) AND (“primary”) AND ((nomogram* OR nomograph* OR “prognostic model” OR “prognostic models” OR “predictive model” OR “predictive models” OR “prediction model” OR “prediction models” OR ((prognos* OR predict*) AND model*)) OR (((prognos* OR predict*) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian network” OR “convolutional network” OR “convolutional networks”)))))) OR ((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND ((((“liver” OR “livers” OR “hepatic”) AND (“neoplasms” OR “neoplasm” OR “cancer” OR “cancers” OR “tumor” OR “tumors” OR “tumour” OR “tumours” OR malignan* OR carcinom*))) OR (“primary” OR “HCC” OR “hepatocellular carcinoma” OR “hepatocellular carcinomas”))) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian networks” OR “convolutional network” OR “convolutional networks”))) | 130 |
| Embase | (((thermoablat*.ti,ab. OR “RFA”.ti,ab. OR “MWA”.ti,ab. OR ((“thermo”.ti,ab. OR “thermal”.ti,ab. OR “radiofrequenc*”.ti,ab. OR exp *microwave radiation/ OR “Microwave*”.ti,ab.) AND (exp *radiofrequency ablation/ OR “ablat*”.ti,ab.))) AND (exp liver tumor/ OR HCC.ti,ab. OR ((“liver”.ti,ab. OR “livers”.ti,ab. OR “hepatic”.ti,ab. OR hepatocellular.ti,ab.) AND (“neoplasms”.ti,ab. OR “neoplasm”.ti,ab. OR “cancer”.ti,ab. OR “cancers”.ti,ab. OR “tumor”.ti,ab. OR “tumors”.ti,ab. OR “tumour”.ti,ab. OR “tumours”.ti,ab. OR “malignan*”.ti,ab. OR “carcinom*”.ti,ab.))) AND ((exp nomogram/ OR nomogram*.ti,ab. OR nomograph*.ti,ab. OR “prognostic model*”.ti,ab. OR “predictive model*”.ti,ab. OR “prediction model*”.ti,ab. OR ((“prognos*”.ti. OR “predict*”.ti.) AND model*.ti.)) OR ((“prognos*”.ti. OR “predict*”.ti. OR exp *prognosis/) AND (exp artificial intelligence/ OR “artificial intelligen*”.ti,ab. OR “AI”.ti,ab. OR “machine learn*”.ti,ab. OR “deep learn*”.ti,ab. OR “neural network*”.ti,ab. OR “support vector machine*”.ti,ab. OR “reinforcement learning”.ti,ab. OR “Markov”.ti,ab. OR “decision tree*”.ti,ab. OR “random forest”.ti,ab. OR “Bayesian network*”.ti,ab. OR “convolutional network”.ti,ab. OR “radiomic*”.ti,ab. OR “gradient boost*”.ti,ab. OR “feature selection*”.ti,ab.)))) OR ((thermoablat*.ti,ab. OR “RFA”.ti,ab. OR “MWA”.ti,ab. OR ((“thermo”.ti,ab. OR “thermal”.ti,ab. OR “radiofrequenc*”.ti,ab. OR exp *microwave radiation/ OR “Microwave*”.ti,ab.) AND (exp *radiofrequency ablation/ OR “ablat*”.ti,ab.))) AND (exp *hepatocellular carcinoma cell line/ OR exp *fibrolamellar hepatocellular carcinoma/ OR “HCC”.ti,ab. OR “hepatocellular carcinoma*”.ti,ab.) AND (primary”.ti,ab.) AND ((exp nomogram/ OR nomogram*.ti,ab. OR nomograph*.ti,ab. OR “prognostic model*”.ti,ab. OR “predictive model*”.ti,ab. OR “prediction model*”.ti,ab. OR ((“prognos*”.ti. OR “predict*”.ti.) AND model*.ti.)) OR (((“prognos*”.ti. OR “predict*”.ti. OR exp *prognosis/) AND (exp artificial intelligence/ OR “AI”.ti. OR “artificial intelligen*”.ti,ab. OR “AI”.ti,ab. OR “machine learn*”.ti,ab. OR “deep learn*”.ti,ab. OR “neural network*”.ti,ab. OR “support vector machine*”.ti,ab. OR “reinforcement learning”.ti,ab. OR “Markov”.ti,ab. OR “decision tree*”.ti,ab. OR “random forest”.ti,ab. OR “Bayesian network*”.ti,ab. OR “convolutional network”.ti,ab. OR “radiomic*”.ti,ab. OR “gradient boost*”.ti,ab. OR “feature selection*”.ti,ab.)))))) OR ((thermoablat*.ti,ab. OR “RFA”.ti,ab. OR “MWA”.ti,ab. OR ((“thermo”.ti,ab. OR “thermal”.ti,ab. OR “radiofrequenc*”.ti,ab. OR exp microwave radiation/ OR “Microwave*”.ti,ab.) AND (exp radiofrequency ablation/ OR “ablat*”.ti,ab.))) AND ((exp liver tumor/ OR ((“liver”.ti,ab. OR “livers”.ti,ab. OR “hepatic”.ti,ab.) AND (“neoplasms”.ti,ab. OR “neoplasm”.ti,ab. OR “cancer”.ti,ab. OR “cancers”.ti,ab. OR “tumor”.ti,ab. OR “tumors”.ti,ab. OR “tumour”.ti,ab. OR “tumours”.ti,ab. OR “malignan*”.ti,ab. OR “carcinom*”.ti,ab.))) OR (“primary”.ti,ab. OR exp hepatocellular carcinoma cell line/ OR exp fibrolamellar hepatocellular carcinoma/OR “HCC”.ti,ab. OR “hepatocellular carcinoma*”.ti,ab. AND (“neoplasms”.ti,ab. OR “neoplasm”.ti,ab. OR “cancer”.ti,ab. OR “cancers”.ti,ab. OR “tumor”.ti,ab. OR “tumors”.ti,ab. OR “tumou]r”.ti,ab. OR “tumours”.ti,ab. OR “malignan*”.ti,ab. OR “carcinom*”.ti,ab.))) AND (exp Artificial Intelligence/ OR “AI”.ti. OR “artificial intelligen*”.ti,ab. OR “AI”.ti,ab. OR “machine learn*”.ti,ab. OR “deep learn*”.ti,ab. OR “neural network*”.ti,ab. OR “support vector machine*”.ti,ab. OR “reinforcement learning”.ti,ab. OR “Markov”.ti,ab. OR “decision tree*”.ti,ab. OR “random forest”.ti,ab. OR “Bayesian network*”.ti,ab. OR “convolutional network”.ti,ab. OR “radiomic*”.ti,ab. OR “gradient boost*”.ti,ab. OR “feature selection*”.ti,ab.)) | 426 |
| Cochrane | (((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND (HCC OR ((“liver” OR “livers” OR “hepatic” OR hepatocellular) AND (“neoplasms” OR “neoplasm” OR “cancer” OR “cancers” OR “tumor” OR “tumors” OR “tumour” OR “tumours” OR malignan* OR carcinom*))) AND ((nomogram* OR nomograph* OR “prognostic model” OR “prognostic models” OR “predictive model” OR “predictive model” OR “prediction model” OR “prediction models” OR ((prognos* OR predict*) AND model*)) OR ((prognos* OR predict*) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian networks” OR “convolutional network” OR “convolutional networks”)))) OR ((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND (“HCC” OR “hepatocellular carcinoma” OR “hepatocellular carcinomas”) AND (“primary”) AND ((nomogram* OR nomograph* OR “prognostic model” OR “prognostic models” OR “predictive model” OR “predictive models” OR “prediction model” OR “prediction models” OR ((prognos* OR predict*) AND model*)) OR (((prognos* OR predict*) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian network” OR “convolutional network” OR “convolutional networks”)))))) OR ((thermoablat* OR “RFA” OR “MWA” OR ((“thermo” OR “thermal” OR radiofrequenc* OR Microwave*) AND (ablat*))) AND ((((“liver” OR “livers” OR “hepatic”) AND (“neoplasms” OR “neoplasm” OR “cancer” OR “cancers” OR “tumor” OR “tumors” OR “tumour” OR “tumours” OR malignan* OR carcinom*))) OR (“primary” OR “HCC” OR “hepatocellular carcinoma” OR “hepatocellular carcinomas”)AND (“neoplasms” OR “neoplasm” OR “cancer” OR “cancers” OR “tumor” OR “tumors” OR “tumour” OR “tumours” OR malignan* OR carcinom*))) AND (“artificial intelligence” OR “AI” OR “machine learning” OR “deep learning” OR “neural network” OR “neural networks” OR “Machine Intelligence” OR “transfer learning” OR “support vector machine” OR “support vector machines” OR “reinforcement learning” OR “Markov” OR “decision tree” OR “decision trees” OR “random forest” OR “Bayesian network” OR “Bayesian networks” OR “convolutional network” OR “convolutional networks”)) | 82 |
| Reference | Radiomic Feature | Category | ||||||
|---|---|---|---|---|---|---|---|---|
| First Order Statistics | GLCM | GLDM | GLSZM | GLRLM | NGTDM | Shape Features | ||
| Chen et al. [17] | 1. original_firstorder_Variance (T1W+C-Post) | |||||||
| 2. wavelet-LLH_glcm_DependenceEntropy (T1W+C-Pre Extended) | ||||||||
| 3. wavelet-LLH_ngtdm_Coarseness (DWI-Pre) | ||||||||
| 4. wavelet-LLH_firstorder_Entropy (T1W+C-Post-Indented) | ||||||||
| 5. log-sigma-2-0-mm-3D_glszm_ZonePercentage (T1W+C-Pre) | ||||||||
| 6. wavelet-HHL_glcm_JointAverage (T1W+C-Pre) | ||||||||
| 7. wavelet-HHL_glcm_Idmn (T1W+C-Post) | ||||||||
| 8. wavelet-LLH_ngtdm_Coarseness (T2W-Pre-Extended) | ||||||||
| 9. original_shape_Elongation (DWI-Pre) | ||||||||
| 10. wavelet-LLH_glrm_LongRunHighGrayLevelEmphasis (T1W+C-Post) | ||||||||
| 11. wavelet-LLH_glrm_SmallDependenceLowGrayLevelEmphasis (T1W+C-Post) | ||||||||
| 12. wavelet-HHL_firstorder_RobustMeanAbsoluteDeviation (DWI-Pre) | ||||||||
| Li JP et al. [18] | 1. Median | |||||||
| 2. correlation | ||||||||
| 3. sum squares | ||||||||
| 4. large dependence emphasis | ||||||||
| 5. large dependence high gray level emphasis | ||||||||
| 6. large dependence low gray level emphasis | ||||||||
| Zhang X et al. [26] | 1. lbp-3D-m2_firstorder_InterquartileRange (AP) | |||||||
| 2. lbp-3D-k_gldm_DependenceVariance (AP) | ||||||||
| 3. lbp-3D-k_gldm_ShortRunLowGrayLevelEmphasis (T1lbp-3D) | ||||||||
| 4. k_gldm_ShortRunLowGrayLevelEmphasis (DP- WI) | ||||||||
| 5. lbp-3D-m2_glcm_ClusterShade (AP) | ||||||||
| 6. lbp-3D-m2_firstorder_10Percentile (DP) | ||||||||
| 7. wavelet-HLL_glcm_RunEntropy (AP) | ||||||||
| 8. lbp-3D-m1_gldm_LargeDependenceLowGrayLevelEmphasis (AP) | ||||||||
| 9. wavelet-HLL_glcm_DifferenceEntropy (T1WI) | ||||||||
| Huang et al. [25] | 1. Dependence Variance (PVP-t1) | |||||||
| 2. Large Dependence Emphasis(PVP-t1) | ||||||||
| 3. Large Area Low Gray Level Emphasis (PVP-t1) | ||||||||
| 4. Dependence Variance (PVP-AP) | ||||||||
| 5. Dependence Variance (DP-t1) | ||||||||
| 6. Large Dependence Emphasis (DP-t1) | ||||||||
| 7. Run Variance (DP-t1) | ||||||||
| 8. Dependence Variance (DP-t1) | ||||||||
| 9. Dependence Non Uniformity Normalized (DP-AP) | ||||||||
| 10. Dependence Variance (DP-AP) | ||||||||
| 11. Large Dependence Emphasis (DP-AP) | ||||||||
| 12. Coarseness (DP-AP) | ||||||||
| Ma et al. [23] | ER: | |||||||
| 1. t_wavelet-HLH_gldm_LowGrayLevelEmphasis | ||||||||
| 2. t_wavelet-HHL_glcm_MaximumProbability | ||||||||
| 3. t_wavelet-LLH_glcm_InverseVariance | ||||||||
| LR: | ||||||||
| 1. t_wavelet-LHH_firstorder_Range | ||||||||
| 2. t_square_gldm_LargeDependenceLowGrayLevelEmphasis | ||||||||
| 3. t_wavelet-HLL_glszm_SizeZoneNonUniformity | ||||||||
| 4. t_squareroot_firstorder_Median | ||||||||
| Zhang L et al. [29] | Peritumoral (5 mm): | |||||||
| 1. V_wavelet.HLH_firstorder_Kurtosis | ||||||||
| 2.T1_original_glcm_InverseVariance | ||||||||
| 3. T2_wavelet.HHL_firstorder_Skewness | ||||||||
| 4. T1_gradient_glrlm_LongRunHighGrayLevelEmphasis | ||||||||
| 5. T1_gradient_glrlm_ShortRunLowGrayLevelEmphasis | ||||||||
| 6. HBP_squareroot_glcm_InverseVariance | ||||||||
| Peritumoral (5 mm + 5 mm) | ||||||||
| 7. A_gradient_gldm_DependenceNonUniformityNormalized | ||||||||
| 8. T1_square_glcm_InverseVariance | ||||||||
| 9. T1_gradient_glrlm_ShortRunLowGrayLevelEmphasis | ||||||||
| 10. A_wavelet.HHL_ngtdm_Contrast | ||||||||
| 11. V_wavelet.HLH_firstorder_Kurtosis | ||||||||
| 12. V_wavelet.HHH_glcm_Imc | ||||||||
| Tumoral: | ||||||||
| 13. T1_original_glcm_InverseVariance | ||||||||
| 14. T2_wavelet.HLH_glcm_SumAverage | ||||||||
| 15. T2_wavelet.HLH_glcm_JointAverage | ||||||||
| 16. T1_wavelet.HHH_glcm_MaximumProbability | ||||||||
| 17. V_wavelet.HLH_firstorder_Skewness | ||||||||
| 18. A_wavelet.LLH_ngtdm_Contrast | ||||||||
| 19. T1_gradient_glrlm_ShortRunLowGrayLevelEmphasis | ||||||||
| 20. V_wavelet.HHH_glcm_MCC | ||||||||
| Wang Y et al. [15] | 1. log.sigma.1.0.mm.3D_glcm_InverseVariancer (AC) | |||||||
| 2. wavelet.HLL_glcm_MCC (AC) | ||||||||
| 3. wavelet.LHL_glrlm_LongRunHighGrayLevelEmphasis (AC) | ||||||||
| 4. log.sigma.4.0.mm.3D_glszm_SmallAreaLowGrayLevelEmphasis (VC) | ||||||||
| 5. wavelet.LHH_glszm_ZoneEntropy (VC) | ||||||||
| 6. log.sigma.3.0.mm.3D_firstorder_Skewness (T2) | ||||||||
| 7. wavelet.LHH_firstorder_RootMeanSquared (T2) | ||||||||
| 8. wavelet.LLH_glszm_SmallAreaLowGrayLevelEmphasis (DC) | ||||||||
| 9. wavelet.HLH_firstorder_Mean (DC) | ||||||||
| 10 wavelet.HHH_glszm_SmallAreaLowGrayLevelEmphasis (DC) | ||||||||
| 11. wavelet.HLL_firstorder_Skewness (DC) | ||||||||
| 12. wavelet.HHH_glcm_Imc1 (FLEX) | ||||||||
| 13.log.sigma.1.0.mm.3D_firstorder_Skewness (FLEX) | ||||||||
| 14. wavelet.HHL_firstorder_Mean (FLEX) | ||||||||
| 15. wavelet.LHH_firstorder_Median (FLEX) | ||||||||
| Lv et al. [27] | 1. LowInsentiy-LareAreaEmphasis | |||||||
| 2. RunLengthNonuniformi ty_AllDirection_offset1_SD | ||||||||
| Author Year | Model Number | Risk of Bias | Applicability | Overall | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1.Participants | 2. Predictors | 3. Outcome | 4. Analysis | 1.Participants | 2. Predictors | 3. Outcome | Risk of Bias | Applicability | ||
| Liu et al. [20] | 1 | + | + | + | + | ? | + | + | + | ? |
| 2 | + | + | + | - | ? | + | + | - | ? | |
| Li JP et al. [18] | 1 | + | ? | + | - | + | + | + | - | + |
| 2 | + | + | + | - | + | + | + | - | + | |
| 3 | + | + | + | - | + | + | + | - | + | |
| Wang R et al. [21] | 1 | + | ? | + | - | + | + | + | - | + |
| Zhang L et al. [29] | 1 | + | ? | + | - | ? | + | + | - | ? |
| 2 | + | ? | + | - | + | + | + | - | + | |
| 3 | + | ? | + | - | ? | + | + | - | ? | |
| Cha et al. [19] | 1 | + | ? | + | - | + | + | + | - | + |
| 2 | + | ? | + | - | + | + | + | - | + | |
| Zhang X et al. [26] | 1 | + | ? | + | - | ? | + | + | - | ? |
| 2 | + | ? | + | - | ? | + | + | - | ? | |
| Ni et al. [24] | 1 | + | ? | + | - | + | + | + | - | + |
| Hu et al. [22] | 1 | + | + | + | - | ? | + | + | - | ? |
| Zhang Z et al. [28] | 1 | ? | + | + | - | + | + | + | - | + |
| Huang et al. [25] | 1 | + | + | + | - | + | + | + | - | + |
| 2 | + | + | + | - | ? | + | + | - | ? | |
| 3 | + | + | + | - | + | + | + | - | + | |
| Wang Y et al. [15] | 1 | + | ? | ? | - | ? | + | + | - | ? |
| 2 | + | ? | ? | ? | ? | + | + | ? | ? | |
| 3 | + | - | ? | - | + | + | + | - | + | |
| Ma et al. [23] | 1 | + | ? | + | - | ? | + | + | - | ? |
| 2 | + | ? | + | - | ? | + | + | - | ? | |
| 3 | + | ? | + | - | ? | + | + | - | ? | |
| 4 | + | ? | + | - | ? | + | + | - | ? | |
| 5 | + | ? | + | - | ? | + | + | - | ? | |
| 6 | + | ? | + | - | ? | + | + | - | ? | |
| 7 | + | ? | + | - | ? | + | + | - | ? | |
| 8 | + | ? | + | - | + | + | + | - | + | |
| Wu et al. [30] | 1 | - | ? | + | - | ? | + | + | - | ? |
| 2 | - | ? | + | - | ? | + | + | - | ? | |
| 3 | - | ? | + | - | ? | + | + | - | ? | |
| Lv et al. [27] | 1 | ? | ? | ? | - | ? | + | + | - | ? |
| 2 | ? | ? | ? | - | ? | + | + | - | ? | |
| Li FY et al. [16] | 1 | ? | ? | + | - | ? | + | + | - | ? |
| Chen et al. [17] | 1 | + | ? | + | - | ? | + | + | - | ? |
| 2 | + | ? | + | - | ? | + | + | - | ? | |
| 3 | + | ? | + | - | + | + | + | - | + | |
References
- Safri, F.; Nguyen, R.; Zerehpooshnesfchi, S.; George, J.; Qiao, L. Heterogeneity of hepatocellular carcinoma: From mechanisms to clinical implications. Cancer Gene Therapy 2024, 31, 1105–1112. [Google Scholar] [CrossRef]
- Sangro, B.; Argemi, J.; Ronot, M.; Paradis, V.; Meyer, T.; Mazzaferro, V.; Jepsen, P.; Golfieri, R.; Galle, P.; Dawson, L.; et al. EASL Clinical Practice Guidelines on the management of hepatocellular carcinoma. J. Hepatol. 2025, 82, 315–374. [Google Scholar] [CrossRef]
- Doyle, A.; Gorgen, A.; Muaddi, H.; Aravinthan, A.D.; Issachar, A.; Mironov, O.; Zhang, W.; Kachura, J.; Beecroft, R.; Cleary, S.P.; et al. Outcomes of radiofrequency ablation as first-line therapy for hepatocellular carcinoma less than 3 cm in potentially transplantable patients. J. Hepatol. 2019, 70, 866–873. [Google Scholar] [CrossRef]
- Laimer, G.; Schullian, P.; Jaschke, N.; Putzer, D.; Eberle, G.; Alzaga, A.; Odisio, B.; Bale, R. Minimal ablative margin (MAM) assessment with image fusion: An independent predictor for local tumor progression in hepatocellular carcinoma after stereotactic radiofrequency ablation. Eur. Radiol. 2020, 30, 2463–2472. [Google Scholar] [CrossRef]
- Haghshomar, M.; Rodrigues, D.; Kalyan, A.; Velichko, Y.; Borhani, A. Leveraging radiomics and AI for precision diagnosis and prognostication of liver malignancies. Front. Oncol. 2024, 14, 1362737. [Google Scholar] [CrossRef]
- Jin, J.; Jiang, Y.; Zhao, Y.L.; Huang, P.T. Radiomics-based Machine Learning to Predict the Recurrence of Hepatocellular Carcinoma: A Systematic Review and Meta-analysis. Acad. Radiol. 2024, 31, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Patrick, E.; Yang, J.Y.H.; Feng, D.D.; Kim, J. Deep multimodal graph-based network for survival prediction from highly multiplexed images and patient variables. Comput. Biol. Med. 2023, 154, 106576. [Google Scholar] [CrossRef]
- Beumer, B.R.; Buettner, S.; Galjart, B.; van Vugt, J.L.A.; de Man, R.A.; Ijzermans, J.N.M.; Koerkamp, B.G. Systematic review and meta-analysis of validated prognostic models for resected hepatocellular carcinoma patients. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2022, 48, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, X.; Li, W.; Ding, X.; Zhang, Y.; Chen, J.; Li, W. Prognostic models for outcome prediction in patients with advanced hepatocellular carcinoma treated by systemic therapy: A systematic review and critical appraisal. BMC Cancer 2022, 22, 750. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Spoletini, G.; Mennini, G.; Laureiro, Z.L.; Tsilimigras, D.I.; Pawlik, T.M.; Rossi, M. Prognostic role of artificial intelligence among patients with hepatocellular cancer: A systematic review. World J. Gastroenterol. 2020, 26, 6679–6688. [Google Scholar] [CrossRef]
- Zou, Z.M.; Chang, D.H.; Liu, H.; Xiao, Y.D. Current updates in machine learning in the prediction of therapeutic outcome of hepatocellular carcinoma: What should we know? Insights Imaging 2021, 12, 31. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.M.; de Groot, J.A.H.; Bouwmeester, W.; Vergouwe, Y.; Mallett, S.; Altman, D.G.; Reitsma, J.B.; Collins, G.S. Critical appraisal and data extraction for systematic reviews of prediction modelling studies: The CHARMS checklist. PLoS Med. 2014, 11, e1001744. [Google Scholar] [CrossRef]
- Wolff, R.F.; Moons, K.G.M.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess the Risk of Bias and Applicability of Prediction Model Studies. Ann. Intern. Med. 2019, 170, 51–58. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Xiao, J.; Geng, X.; Han, L.; Luo, J. Multicenter Integration of MR Radiomics, Deep Learning, and Clinical Indicators for Predicting Hepatocellular Carcinoma Recurrence After Thermal Ablation. J. Hepatocell. Carcinoma 2024, 11, 1861–1874. [Google Scholar] [CrossRef]
- Li, F.-Y.; Li, J.-G.; Wu, S.-S.; Ye, H.-L.; He, X.-Q.; Zeng, Q.-J.; Zheng, R.-Q.; An, C.; Li, K. An Optimal Ablative Margin of Small Single Hepatocellular Carcinoma Treated with Image-Guided Percutaneous Thermal Ablation and Local Recurrence Prediction Base on the Ablative Margin: A Multicenter Study. J. Hepatocell. Carcinoma 2021, 8, 1375–1388. [Google Scholar] [CrossRef]
- Chen, C.; Han, Q.; Ren, H.; Wu, S.; Li, Y.; Guo, J.; Li, X.; Liu, X.; Li, C.; Tian, Y. Multiparametric MRI-based model for prediction of local progression of hepatocellular carcinoma after thermal ablation. Cancer Med. 2023, 12, 17529–17540. [Google Scholar] [CrossRef]
- Li, J.P.; Zhao, S.; Jiang, H.J.; Jiang, H.; Zhang, L.H.; Shi, Z.X.; Fan, T.T.; Wang, S. Quantitative dual-energy computed tomography texture analysis predicts the response of primary small hepatocellular carcinoma to radiofrequency ablation. Hepatobiliary Pancreat. Dis. Int. 2022, 21, 569–576. [Google Scholar] [CrossRef]
- Cha, D.I.; Ahn, S.H.; Lee, M.W.; Jeong, W.K.; Song, K.D.; Kang, T.W.; Rhim, H. Risk Group Stratification for Recurrence-Free Survival and Early Tumor Recurrence after Radiofrequency Ablation for Hepatocellular Carcinoma. Cancers 2023, 15, 687. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, D.; Wang, K.; Xie, X.; Su, L.; Kuang, M.; Huang, G.; Peng, B.; Wang, Y.; Lin, M.; et al. Deep Learning Radiomics Based on Contrast-Enhanced Ultrasound Might Optimize Curative Treatments for Very-Early or Early-Stage Hepatocellular Carcinoma Patients. Liver Cancer 2020, 9, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xu, H.; Chen, W.; Jin, L.; Ma, Z.; Wen, L.; Wang, H.; Cao, K.; Du, X.; Li, M. Gadoxetic acid–enhanced MRI with a focus on LI-RADS v2018 imaging features predicts the prognosis after radiofrequency ablation in small hepatocellular carcinoma. Front. Oncol. 2023, 13, 975216. [Google Scholar] [CrossRef]
- Hu, C.; Song, Y.; Zhang, J.; Dai, L.; Tang, C.; Li, M.; Liao, W.; Zhou, Y.; Xu, Y.; Zhang, Y.Y.; et al. Preoperative Gadoxetic Acid-Enhanced MRI Based Nomogram Improves Prediction of Early HCC Recurrence After Ablation Therapy. Front. Oncol. 2021, 11, 649682. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.P.; He, X.L.; Li, K.; Wang, J.F.; Zeng, Q.J.; Xu, E.J.; He, X.Q.; Li, S.Y.; Kun, W.; Zheng, R.Q.; et al. Dynamic Contrast-Enhanced Ultrasound Radiomics for Hepatocellular Carcinoma Recurrence Prediction After Thermal Ablation. Mol. Imaging Biol. 2021, 23, 572–585. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.H.; Wu, B.L.; Li, M.; Han, X.; Hao, X.W.; Zhang, Y.; Cheng, W.; Guo, C.L. Prediction Model and Nomogram of Early Recurrence of Hepatocellular Carcinoma after Radiofrequency Ablation Based on Logistic Regression Analysis. Ultrasound Med. Biol. 2022, 48, 1733–1744. [Google Scholar] [CrossRef]
- Huang, W.; Pan, Y.; Wang, H.; Jiang, L.; Liu, Y.; Wang, S.; Dai, H.; Ye, R.; Yan, C.; Li, Y. Delta-radiomics Analysis Based on Multi-phase Contrast-enhanced MRI to Predict Early Recurrence in Hepatocellular Carcinoma After Percutaneous Thermal Ablation. Acad. Radiol. 2024, 31, 4934–4945. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, C.; Zheng, D.; Liao, Y.; Wang, X.; Huang, Z.; Zhong, Q. Radiomics nomogram based on multi-parametric magnetic resonance imaging for predicting early recurrence in small hepatocellular carcinoma after radiofrequency ablation. Front. Oncol. 2022, 12, 1013770. [Google Scholar] [CrossRef]
- Lv, X.; Chen, M.; Kong, C.; Shu, G.; Meng, M.; Ye, W.; Cheng, S.; Zheng, L.; Fang, S.; Chen, C.; et al. Construction of a novel radiomics nomogram for the prediction of aggressive intrasegmental recurrence of HCC after radiofrequency ablation. Eur. J. Radiol. 2021, 144, 109955. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, J.; Liu, S.; Dong, L.; Liu, T.; Wang, H.; Han, Z.; Zhang, X.; Liang, P. Multiparametric liver MRI for predicting early recurrence of hepatocellular carcinoma after microwave ablation. Cancer Imaging 2022, 22, 42. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, P.; Hou, J.; Luo, M.; Li, Y.; Jiang, X. Radiomics model based on gadoxetic acid disodium-enhanced mr imaging to predict hepatocellular carcinoma recurrence after curative ablation. Cancer Manag. Res. 2021, 13, 2785–2796. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.P.; Ding, W.Z.; Wang, Y.L.; Liu, S.; Zhang, X.Q.; Yang, Q.; Cai, W.J.; Yu, X.L.; Liu, F.Y.; Kong, D.; et al. Radiomics analysis of ultrasound to predict recurrence of hepatocellular carcinoma after microwave ablation. Int. J. Hyperth. 2022, 39, 595–604. [Google Scholar] [CrossRef]
- Santillan, C.; Chernyak, V.; Sirlin, C. LI-RADS categories: Concepts, definitions, and criteria. Abdom. Radiol. 2018, 43, 101–110. [Google Scholar] [CrossRef]
- Chen, W.; Wang, L.; Hou, Y.; Li, L.; Chang, L.; Li, Y.; Xie, K.; Qiu, L.; Mao, D.; Li, W.; et al. Combined Radiomics-Clinical Model to Predict Radiotherapy Response in Inoperable Stage III and IV Non-Small-Cell Lung Cancer. Technol. Cancer Res. Treat. 2022, 21, 15330338221142400. [Google Scholar] [CrossRef] [PubMed]
- Van Calster, B.; McLernon, D.J.; van Smeden, M.; Wynants, L.; Steyerberg, E.W. Calibration: The Achilles heel of predictive analytics. BMC Med. 2019, 17, 230. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Gleisner, A.L.; Anders, R.A.; Assumpcao, L.; Maley, W.; Choti, M.A. Preoperative assessment of hepatocellular carcinoma tumor grade using needle biopsy: Implications for transplant eligibility. Ann. Surg. 2007, 245, 435–442. [Google Scholar] [CrossRef]
- Renzulli, M.; Brocchi, S.; Cucchetti, A.; Mazzotti, F.; Mosconi, C.; Sportoletti, C.; Brandi, G.; Pinna, A.D.; Golfieri, R. Can Current Preoperative Imaging Be Used to Detect Microvascular Invasion of Hepatocellular Carcinoma? Radiology 2016, 279, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Ünal, E.; İdilman İ, S.; Akata, D.; Özmen, M.N.; Karçaaltıncaba, M. Microvascular invasion in hepatocellular carcinoma. Diagn. Interv. Radiol. 2016, 22, 125–132. [Google Scholar] [CrossRef]
- Puijk, R.S.; Ahmed, M.; Adam, A.; Arai, Y.; Arellano, R.; de Baère, T.; Bale, R.; Bellera, C.; Binkert, C.A.; Brace, C.L.; et al. Consensus Guidelines for the Definition of Time-to-Event End Points in Image-guided Tumor Ablation: Results of the SIO and DATECAN Initiative. Radiology 2021, 301, 533–540. [Google Scholar] [CrossRef]
- Galle, P.R.; Forner, A.; Llovet, J.M.; Mazzaferro, V.; Piscaglia, F.; Raoul, J.L.; Schirmacher, P.; Vilgrain, V. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol. 2018, 69, 182–236. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Llovet, J.M.; Yarchoan, M.; Mehta, N.; Heimbach, J.K.; Dawson, L.A.; Jou, J.H.; Kulik, L.M.; Agopian, V.G.; Marrero, J.A.; et al. AASLD Practice Guidance on prevention, diagnosis, and treatment of hepatocellular carcinoma. Hepatology 2023, 78, 1922–1965. [Google Scholar] [CrossRef]
- Qiu, H.; Wang, M.; Wang, S.; Li, X.; Wang, D.; Qin, Y.; Xu, Y.; Yin, X.; Hacker, M.; Han, S.; et al. Integrating MRI-based radiomics and clinicopathological features for preoperative prognostication of early-stage cervical adenocarcinoma patients: In comparison to deep learning approach. Cancer Imaging 2024, 24, 101. [Google Scholar] [CrossRef]
- Zhang, Y.; Cui, Y.; Liu, H.; Chang, C.; Yin, Y.; Wang, R. Prognostic nomogram combining (18)F-FDG PET/CT radiomics and clinical data for stage III NSCLC survival prediction. Sci. Rep. 2024, 14, 20557. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Overfitting, Model Tuning, and Evaluation of Prediction Performance. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer: Berlin/Heidelberg, Germany, 2022; pp. 109–139. [Google Scholar] [CrossRef]
- Collins, G.S.; Reitsma, J.B.; Altman, D.G.; Moons, K.G.M. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): The TRIPOD Statement. BMC Med. 2015, 13, 1. [Google Scholar] [CrossRef]
| Reference | Tumor Related Inclusion Criteria | Outcome | Time Period | Definition of Outcome |
|---|---|---|---|---|
| Liu et al. (2020) [20] | Single tumor ≤ 5 cm | PFS | TP < 2 years following TA | Time to progression (LTP, new intrahepatic tumor, vascular invasion, or distant organ metastases). LTP: tumor adjacent to ablation margin < 1.0 cm |
| Li JP et al. (2022) [18] | Single tumor ≤ 3 cm, or sum of 2 tumors ≤ 3 cm | TP | <12 months following TA | Tumor progression: LT-TR viable lesion |
| Wang R et al. (2023) [21] | Single tumor < 5 cm or ≤3 tumors each ≤ 3 cm | RFS | The interval between the initial date of TLA and the date of the tumor recurrence or last follow-up visit before 1 October 2021. | Tumor recurrence: LR, IDR, and EM |
| Zhang L et al. (2021) [29] | HCC with longest diameter > 10 mm, without capsular, adjacent organ and/or vascular invasion | RFS | The interval between the initial date of TA and the date of the tumor recurrence | Time to recurrence |
| Cha et al. (2023) [19] | Single tumor ≤ 3 cm | ER RFS | ER: <2 years following TA RFS: at 1- or 2- or 5-years after RFA. | Recurrence: LTP, IDR, and EM RFS: Time to the development of recurrence or death |
| Zhang X et al. (2022) [26] | Single tumor < 5 cm, or <3 tumors each < 3 cm | ER | <2 years following TA | New cancerous focus with typical imaging features of the liver or other organs |
| Ni et al. (2022) [24] | Not specified | ER | <2 years following TA | Recurrence: local and distant IH recurrence. IH local recurrence: active tumors found in adjacent or ablated areas < 1 month of follow-up after ablation. IDR: tumors in the liver parenchyma outside the ablation site on any postprocedural image during the follow-up period |
| Hu et al. (2021) [22] | ≤3 tumors each ≤ 5 cm | ER | <2 years following TA | LTP, IDR and EM |
| Zhang Z et al. (2022) [28] | Single tumor < 5 cm, or ≤3 tumors each < 3 cm | ER | <2 years following TA | The presence of new IH and/or EH lesions |
| Huang et al. (2024) [25] | Not specified | RFS ER | RFS: time from the date of operation to the date of the first recurrence ER: <2 years following TA | Recurrence: IH and/or EM |
| Wang Y et al. (2024) [15] | Single tumor < 5 cm or ≤3 tumors each ≤ 3 cm | IR | <2 years following TA | IH recurrence (local or distant) |
| Ma et al. (2021) [23] | Single tumor < 5 cm | ER LR risk | ER: <2 years following TA LR: >2 years following TA | LTP, IDR and ER |
| Wu et al. (2022) [30] | Single tumor < 5 cm, or <3 tumors each < 3 cm | RFS ER LR | RFS: time between the treatment and disease recurrence or death ER: <2 years following TA LR: <5 years following TA | ER: Time to recurrence (excluding LTP) LR: Time to recurrence RFS: Time to recurrence or death |
| Lv et al. (2021) [27] | Not specified | AIR | >6 months of disease-free status following TA | AIR: Simultaneous development of multiple nodular (>3) or infiltrative recurrence in the treated segment of the liver |
| Li FY et al. (2021) [16] | Single HCC ≤ 3 cm, without major vascular infiltration or extrahepatic metastasis | LTPFS | Within 6-, 12-, and 24- months following TA | LTPFS: Time from ablation to the date of LTP. LTP: enhancements in lesion in the arterial phase with a washout lesion in the delayed phase of a contrast-enhanced imaging examination (CEUS, CT, or MRI) inside or abutting the ablation area during follow-up. |
| Chen et al. (2023) [17] | Single tumor <5 cm, or ≤3 tumors < 3 cm | LTP | No predefined time frame. The median follow-up duration for all patients was 22.5 months (IQR, 11.2–55.3 months). | Abnormal nodular, disseminated, and/or unusual patterns of peripheral enhancement around the ablative site on imaging |
| Reference | Data Source | Ablation Technique | Imaging Modality | Sample Size | Test Cohort | |||
|---|---|---|---|---|---|---|---|---|
| Training | Validation | Test | Internal | External | ||||
| Liu et al. (2020) [20] | Single center | RFA | CEUS | 149 | 0 | 65 | + | - |
| Li JP et al. (2022) [18] | Single center | RFA | DECT | 63 | 0 | Model 1:2000 BSR Model 2: 10-Fold CV Model 3: 2000 BSR | + | - |
| Wang R et al. (2023) [21] | Single center | RFA | GAE-MRI | 153 | 0 | 51 | + | - |
| Zhang L et al. (2021) [29] | Single center | RFA MWA | GAE-MRI | 92 | 0 | 1000 BSR | + | - |
| Cha et al. (2023) [19] | Single center | RFA | GAE-MRI | 152 | 0 | 1000 BSR | + | - |
| Zhang X et al. (2022) [26] | Single center | RFA | CEMRI | 63 | 0 | 27 | + | - |
| Ni et al. (2022) [24] | Single center | RFA | CEUS | 60 | 0 | 48 | + | - |
| Hu et al. (2021) [22] | Single center | RFA MWA | GAE-MRI | 112 | 0 | 48 | + | - |
| Zhang Z et al. (2022) [28] | Single center | MWA | CEMRI | 226 | 0 | 113 | + | - |
| Huang et al. (2024) [25] | Single center | RFA MWA | Gadobenate dimeglumine-MRI | 110 | 0 | From different temporal period: 129 From different scanner: 25 | + | + |
| Wang Y et al. (2024) [15] | Multicenter | RFA MWA | CEMRI | 335 | 84 | From two different centers: 116 | - | + |
| Ma et al. (2021) [23] | Single center | RFA MWA | Model nr. 1 & 5: CEUS Model nr. 2 & 6: US Models 4 & 8: CEUS/US | 255 | 5-Fold CV | 63 | + | - |
| Wu et al. (2022) [30] | Single center | MWA | US | 400 | 0 | 113 | + | - |
| Lv et al. (2021) [27] | Single center | RFA | CEMRI | 40 | 0 | 18 | + | - |
| Li FY et al. (2021) [16] | Multicenter | RFA MWA | MRI/CT/CEUS | 296 | 0 | 148 | + | - |
| Chen et al. (2023) [17] | Multicenter | RFA MWA | CEMRI | 151 | 0 | From center 1: 38 From center 2: 135 From center 3: 93 | + | + |
| Reference | Model nr. | Feature Selection Technique | Model Development Technique | Predictors | ||||
|---|---|---|---|---|---|---|---|---|
| Preprocedural | Postprocedural | EPV | Clinical | Imaging | ||||
| Liu et al. (2020) [20] | 1 | Through CNN framework | Cox-CNN proportional hazard model | + | - | NA | None | 64-dimensional vector as DL-based features |
| 2 | MV Cox Regression on CNN features | Nomogram via MV Cox regression | + | - | NA | 1. Age 2. PLT | 1. Tumor size 2. Survival hazard based on radiomics signatures | |
| Li JP et al. (2022) [18] | 1 | UV and MV logistic regression | MV logistic regression | - | + | 2.9 | 1. ALBI 2. λAP(40–100 keV) | Iodine concentration in the AP within the ROI |
| 2 | LASSO algorithm | Linear regression model | - | + | NI | None | 6 Radiomics features from first order statistics (1), GLCM (2), GLDM (3) | |
| 3 | Integration of clinical and radiomics features from models nr. 1 and 2 | Nomogram via MV logistic regression | - | + | 2.5 | Features from model nr. 1 | Radiomics features from model nr. 2 | |
| Wang R et al. (2023) [21] | 1 | UV and MV logistic regression | Nomogram via Cox proportional hazards regression | + | - | NI | AFP > 100 ng/ml | 1. Rim AP hyperenhancement 2.Targetoid restriction on DWI |
| Zhang L et al. (2021) [29] | 1 | UV and MV Cox regression | MV Cox regression | + | - | 2.4 | 1. Albumin 2. GGT 3. AFP | Tumor size |
| 2 | 1. ICC > 0.75 2. RSF with VIMP-based ranking | Random survival forest | + | - | 0.0 | None | 6 peritumoral (5 mm), 6 peritumoral (5 + 5 mm) and 8 tumoral radiomics features from first order statistics (4), GLCM (9), GLRLM (4), GLDM (1), NGTDM (2). | |
| 3 | Integration of features from models nr. 1 and 2 | Random survival forest | + | - | 1.5 | Features from model nr. 1 | 1. Tumor size 2. Radiomics features from model nr. 2 | |
| Cha et al. (2023) [19] | 1 2 | UV and MV Cox regression | Nomogram via MV Cox regression | + | - | Model 1: 2.7 Model 2: 2.8 | 1. Age 2. ALBI-grade 3. MoRal score > 68 | 1. Non-rim AP hyperenhancement 2. Enhancing capsule 3. Low signal intensity on HBP 4. High risk MVI |
| Zhang X et al. (2022) [26] | 1 | 1. AK native algorithm 2. ICC > 0.75 | Logistic regression | + | - | 0.0 | None | Radiomics features from first order statistics (2), GLCM (3), GLDM (4) |
| 2 | Radiomic: Features from model nr. 1 Clinical and radiological: UV and MV logistic regression | Nomogram via MV logistic regression | + | - | 0.0 | Albumin level | 1. Number of tumors 2. Radiomics features from model nr. 1 | |
| Ni et al. (2022) [24] | 1 | UV and MV logistic regression | Nomogram via MV logistic regression | + | - | 1.1 | 1. Neutrophil-to-lymphocyte ratio 2. AFP | 1. Number of tumors 2. CEUS enhancement pattern |
| Hu et al. (2021) [22] | 1 | UV and MV logistic regression | Nomogram via MV logistic regression | + | - | 7.1 | AFP | 1. Tumor number 2. Arterial peritumoral enhancement 3. satellite nodule 4. Peritumoral hypo intensity on HBP |
| Zhang Z et al. (2022) [28] | 1 | UV and MV logistic regression | Multi variable Cox regression | + | + | 7.2 | None | 1. Tumor size 2. MAM 3. Recurrence score: 3.1. Ill-defined ablation margin 3.2. Capsule enhancement 3.3. ADC 3.4. ∆ADC 3.5. EADC |
| Huang et al. (2024) [25] | 1 | UV and MV logistic regression | MV logistic regression | + | - | 2.7 | Child-Pugh score | 1. High-risk tumor location 2. Incomplete or absent tumor capsule |
| 2 | 1. ICC ≤ 0.75 2. Pearson CC (threshold > 0.99) 3.ANOVA 4.Logistic Regression | Logistic regression | + | - | 0.4 | None | 12 radiomics features from GLDM (9), GLSZM (1), GLRLM (1), NGTDM (1) | |
| 3 | Integration of features from models 1 and 2 | Nomogram via MV logistic regression | + | - | 3.2 | Features from model nr. 1 | Features from models nr. 1 and 2 | |
| Wang Y et al. (2024) [15] | 1 | 1. ICC < 0.7 2. Decision tree ranking 3. UV Cox proportional hazards | MV Cox regression | + | - | NI | None | 15 radiomics features from first order statistics (7), GLCM (3), GLRLM (1), GLSZM (4) |
| 2 | CNN framework | 3D-CNN | + | - | NI | None | 128-dimensional DL-based feature vector | |
| 3 | UV and MV logistic regression | MV logistic regression | + | - | NI | Serum albumin level | 1. Number of tumors 2. Features from radiomics and DL models | |
| Ma et al. (2021) [23] | 1 5 | Through DL framework | DL model | + | - | NA | None | Relevant features selected by DL model |
| 2 | LASSO regression with CV | Logistic regression | + | - | 31 | None | 2 radiomics features from GLDM (1), GLCM (2) | |
| 3 | UV and MV logistic regression | MV logistic regression | + | - | 31 | 1. APRI 2. PLT 3. Monocyte | None | |
| 6 | LASSO regression with CV | Logistic regression | + | - | 13.2 | None | 4 radiomics features from first order statistics (2), GLDM (1), GLSZM (1) | |
| 7 | UV and MV Cox proportional hazards regression | MV Cox proportional hazards | + | - | 17.67 | 1.Portal hypertension 2. ALT 3. Hemoglobin | None | |
| 4 8 | Integration of selected features from CEUS, US, and clinical models using logistic regression | Nomogram via MV logistic regression | + | - | Model 4: 18.6 Model 8: 10.6 | Same as clinical model | 1. DL score 2. Radiomics score | |
| Wu et al. (2022) [30] | 1 | Clinical and radiological: UV and MV Cox regression US semantic: Correlation analysis | MV Cox regression | + | - | NA | 1. AFP 2. ALBI 3. AST 4. TBIL | 1. Tumor size 2. Number of tumors 3. US semantic features: 3.1. Echogenicity 3.2. Morphology 3.3. Hypoechoic halo 3.4. Boundary 3.5. Posterior acoustic enhancement 3.6. Intertumoral vascularity |
| 2 | Radiological: UV and MV Cox regression. DL: ResNet18 framework | MV Cox regression | + | - | NA | None | 1. Tumor size 2. Number of tumors 3. DL-based features | |
| 3 | Clinical and radiological: UV and MV Cox regression. DL: ResNet18 framework | MV Cox regression | + | - | NA | 1. AFP 2. PLT | 1. Tumor size 2. Number of tumors 3. DL-based features | |
| Lv et al. (2021) [27] | 1 | LASSO algorithm | MV logistic regression | + | - | 0.0 | None | 2 radiomics features from GLSZM (1), GLRLM (1) |
| 2 | UV and MV logistic regression | MV logistic regression | + | - | 2.5 | None | 1. Tumor shape 2. ADC value 3. DWI signal intensity 4. ΔSI enhancement rate | |
| Li FY et al. (2021) [16] | 1 | UV and MV Cox regression | Nomogram via MV Cox regression | + | + | 2.6 | None | 1. Tumor size 2. Ablation margin |
| Chen et al. (2023) [17] | 1 | UV and MV logistic regression | Support vector machine | + | + | 1.9 | None | 1. Number of tumors 2. Location of abutting major vessels 3. Ablation margin |
| 2 | 1. Reliability evaluation 2. UV regression 3. Boruta method | Support vector machine | + | + | <1 | None | 1. 8 DL-based features 2. 12 radiomics features from first order statistics (3), GLCM (3), NGTDM (2), GLSZM (1), GLRLM (2), shape features (1) | |
| 3 | Integration of features from models nr. 1 and 2 | Support vector machine | + | + | 0.8 | None | Features of models 1 and 2 | |
| Paper | Nr. | Modeling | Predictors | Out Come | AUC-ROC | C-Index | Kaplan–Meier | Calibration | DCA | Cohort | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AI | C | Cl | R | RM | DL | |||||||||
| Liu et al. [20] | 1. | RFS * | 0.81 (0.72–0.93) †† | 0.73 (0.65–0.80) | p < 0.005 | - | + | IV | ||||||
| 2. | RFS * | 0.82 (0.72–0.93) †† | 0.73 (0.68–0.84) | p < 0.005 | HL p = 0.479 | ThP: >30% | IV | |||||||
| Li JP et al. [18] | 1. | RFS * | 0.92 (0.85–0.99) † | 0.92 | - | HL p = 0.792 | + | IV | ||||||
| 2. | RFS * | 0.90 (0.82–1.00) † | - | - | - | + | IV | |||||||
| 3. | RFS * | 0.96 (0.91–1.00) † | 0.96 | - | HL p = 0.71 | + | IV | |||||||
| Wang R et al. [21] | 1. | RFS | - | 0.81 (0.71–0.90) | - | + | + | IV | ||||||
| Zhang L et al. [29] | 1. | RFS | 0.67 †* | 0.70 (0.64–0.76) | - | - | + | IV | ||||||
| 2. | RFS | 0.82 †* | 0.61 (0.50–0.70) | - | - | + | IV | |||||||
| 3. | RFS | 0.83 †* | 0.71 (0.64–0.76) | p = 0.007 | - | + | IV | |||||||
| Cha et al. [19] | 1. | RFS | - | 0.72 (0.66–0.77) | p < 0.001 | - | - | IV | ||||||
| 2. | ER | - | 0.75 (0.68–0.81) | p < 0.001 | - | - | IV | |||||||
| Zhang X et al. [26] | 1. | ER | 0.81 | - | - | - | - | IV | ||||||
| 2. | ER | 0.81 | - | - | + | + | IV | |||||||
| Ni et al. [24] | 1. | ER | 0.78 | - | - | + | ThP: 4.3–87.3% | IV | ||||||
| Hu et al. [22] | 1. | ER | 0.83 (0.71–0.96) | 0.75 | p < 0.001 | HL p = 0.168 | ThP: 24–99% | IV | ||||||
| Zhang Z et al. [28] | 1. | ER | 0.85 (0.71–0.87) | p < 0.05 | - | - | IV | |||||||
| Huang et al. [25] | 1. | ER | 0.77 (0.55–0.91) | - | - | - | + | EV | ||||||
| 2. | ER | 0.77 (0.56–0.91) | - | p < 0.0001 | - | + | EV | |||||||
| 3. | ER | 0.83 (0.62–0.95) | - | p < 0.0001 | HL p = 0.397 | + | EV | |||||||
| Wang Y et al. [15] | 1. | IHER | 0.75 (0.64–0.75) | - | - | - | + | EV | ||||||
| 2. | IHER | 0.64 (0.51–0.65) | - | - | - | + | EV | |||||||
| 3. | IHER | 0.79 (0.67–0.83) | - | - | - | + | EV | |||||||
| Ma et al. [23] | 1. | ER | 0.83 (0.81–0.85) | - | - | - | - | IV | ||||||
| 2. | ER | 0.56 (0.53–0.59) | - | - | - | - | IV | |||||||
| 3. | ER | 0.60 (0.57–0.63) | - | - | - | - | IV | |||||||
| 4. | ER | 0.84 (0.82–0.86) | - | - | + | + | IV | |||||||
| 5. | LR | - | 0.68 (0.65–0.71) | p < 0.0001 | - | - | IV | |||||||
| 6. | LR | - | 0.56 (0.53–0.59) | p = 0.08 | - | - | IV | |||||||
| 7. | LR | - | 0.67 (0.64–0.70) | p < 0.0001 | - | - | IV | |||||||
| 8. | LR | - | 0.77 (0.76–0.78) | p < 0.0001 | + | + | IV | |||||||
| Wu et al. [30] | 1. | ER | - | 0.70 (0.64–0.84) | p < 0.001 | + | - | IV | ||||||
| 2. | LR | - | 0.72 (0.62–0.80) | p < 0.001 | + | - | IV | |||||||
| 3. | RFS | - | 0.72 (0.65–0.79) | p < 0.001 | + | - | IV | |||||||
| Lv et al. [27] | 1. | AIR | 0.64 (0.34–0.93) | - | - | + | ThP: >8% | IV | ||||||
| 2. | AIR | 0.82 (0.58–1.00) | - | - | + | + | IV | |||||||
| Li FY et al. [16] | 1. | LTPFS | - | 0.76 (0.62–0.89) | p = 0.001 | + | - | IV | ||||||
| Chen et al. [17] | 1. | LTP | 0.80 (0.72–0.89) | - | - | - | - | EV | ||||||
| 2. | LTP | 0.86 (0.80–0.91) | - | - | - | - | EV | |||||||
| 3. | LTP | 0.87 (0.82–0.91) | - | p = 0.0021 | + | - | EV | |||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verhagen, C.A.M.; Gholamiankhah, F.; Buijsman, E.C.M.; Broersen, A.; van Erp, G.C.M.; van der Velden, A.L.; Rahmani, H.; van der Leij, C.; Brecheisen, R.; Lanocita, R.; et al. Clinical and Imaging-Based Prognostic Models for Recurrence and Local Tumor Progression Following Thermal Ablation of Hepatocellular Carcinoma: A Systematic Review. Cancers 2025, 17, 2656. https://doi.org/10.3390/cancers17162656
Verhagen CAM, Gholamiankhah F, Buijsman ECM, Broersen A, van Erp GCM, van der Velden AL, Rahmani H, van der Leij C, Brecheisen R, Lanocita R, et al. Clinical and Imaging-Based Prognostic Models for Recurrence and Local Tumor Progression Following Thermal Ablation of Hepatocellular Carcinoma: A Systematic Review. Cancers. 2025; 17(16):2656. https://doi.org/10.3390/cancers17162656
Chicago/Turabian StyleVerhagen, Coosje A. M., Faeze Gholamiankhah, Emma C. M. Buijsman, Alexander Broersen, Gonnie C. M. van Erp, Ariadne L. van der Velden, Hossein Rahmani, Christiaan van der Leij, Ralph Brecheisen, Rodolfo Lanocita, and et al. 2025. "Clinical and Imaging-Based Prognostic Models for Recurrence and Local Tumor Progression Following Thermal Ablation of Hepatocellular Carcinoma: A Systematic Review" Cancers 17, no. 16: 2656. https://doi.org/10.3390/cancers17162656
APA StyleVerhagen, C. A. M., Gholamiankhah, F., Buijsman, E. C. M., Broersen, A., van Erp, G. C. M., van der Velden, A. L., Rahmani, H., van der Leij, C., Brecheisen, R., Lanocita, R., Dijkstra, J., & Burgmans, M. C. (2025). Clinical and Imaging-Based Prognostic Models for Recurrence and Local Tumor Progression Following Thermal Ablation of Hepatocellular Carcinoma: A Systematic Review. Cancers, 17(16), 2656. https://doi.org/10.3390/cancers17162656






