MRI-Based Radiomics for Outcome Stratification in Pediatric Osteosarcoma †
Simple Summary
Abstract
1. Introduction
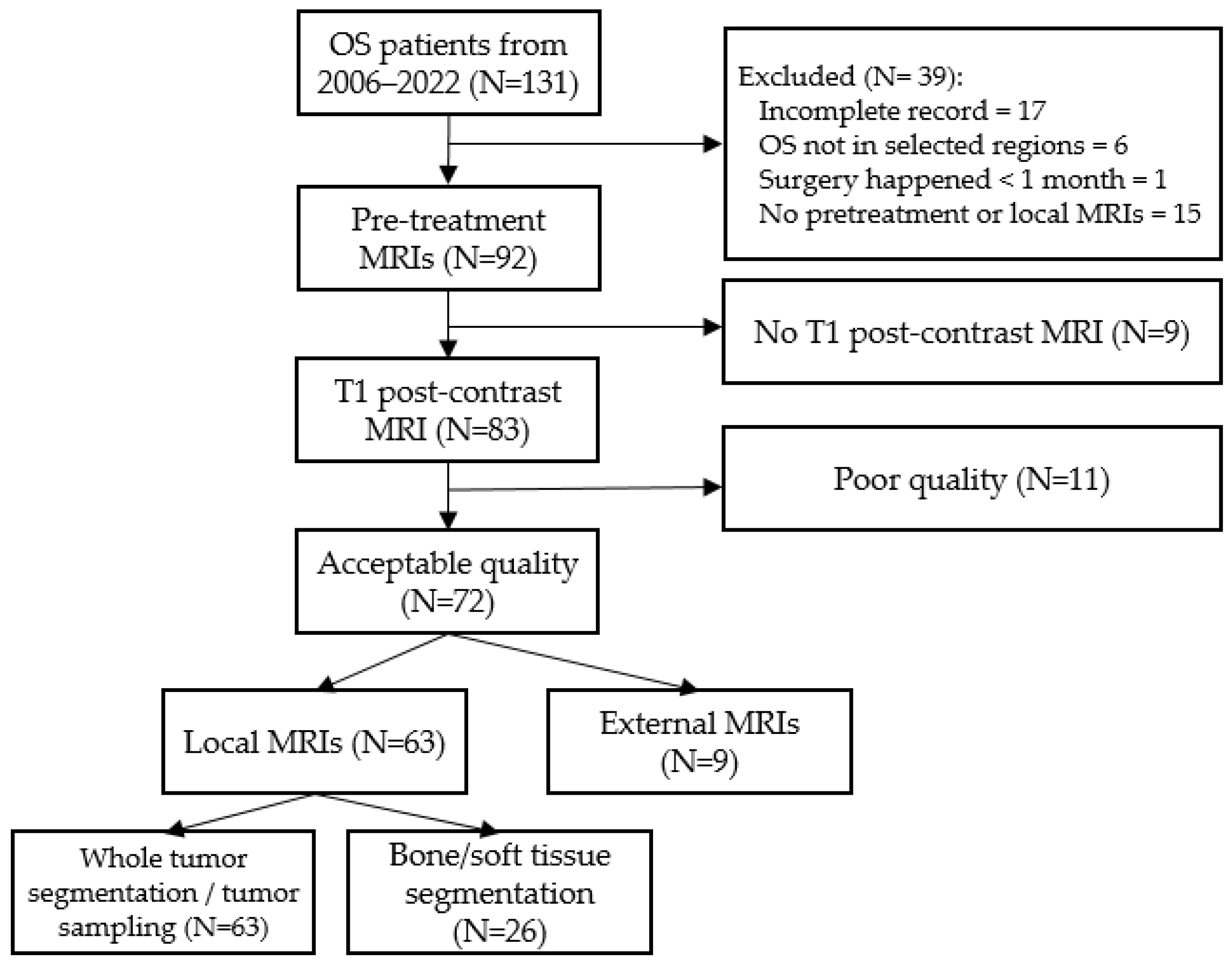
2. Materials and Methods
2.1. Patients’ Cohorts
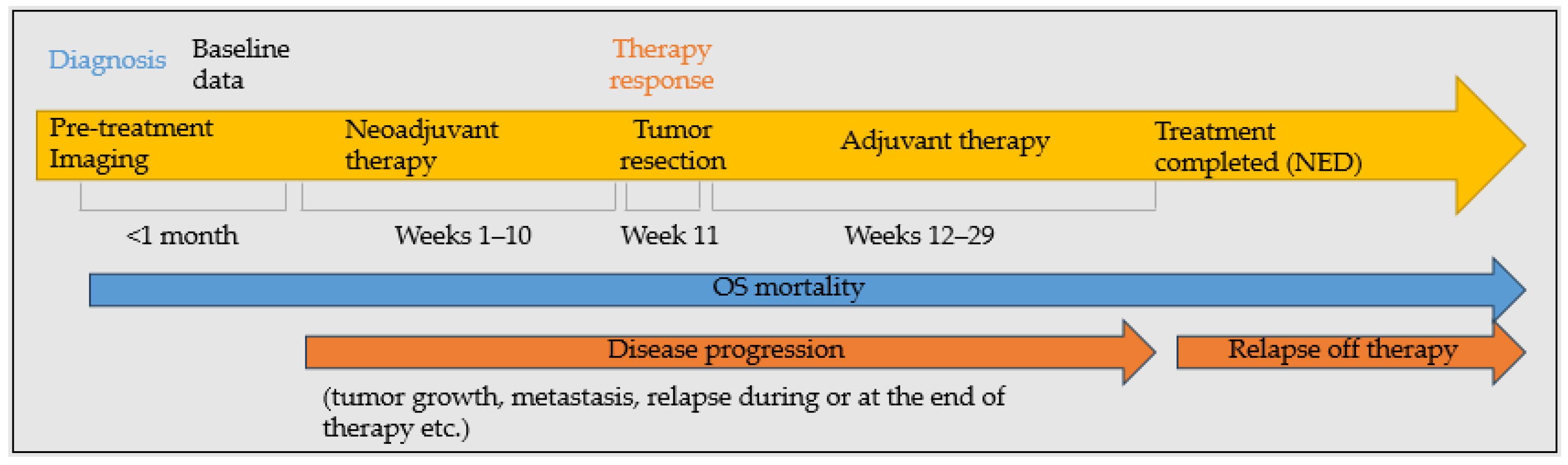
2.2. Evaluated Outcomes
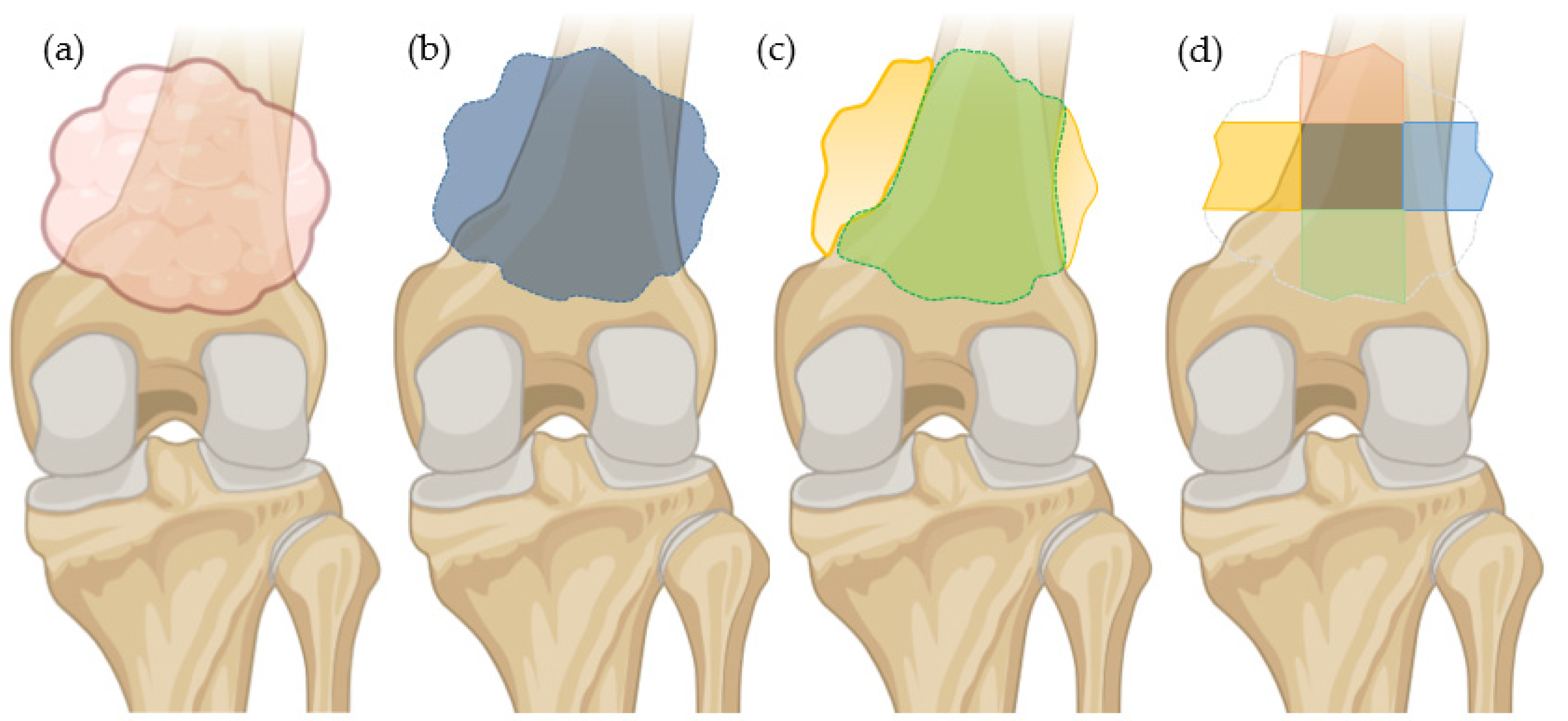
2.3. Segmentation Method
2.4. Data Standardization and Features Sets
2.5. Machine Learning
2.6. Statistical Analysis
3. Results
3.1. Descriptive Statistics
3.2. Outcome Interdependencies
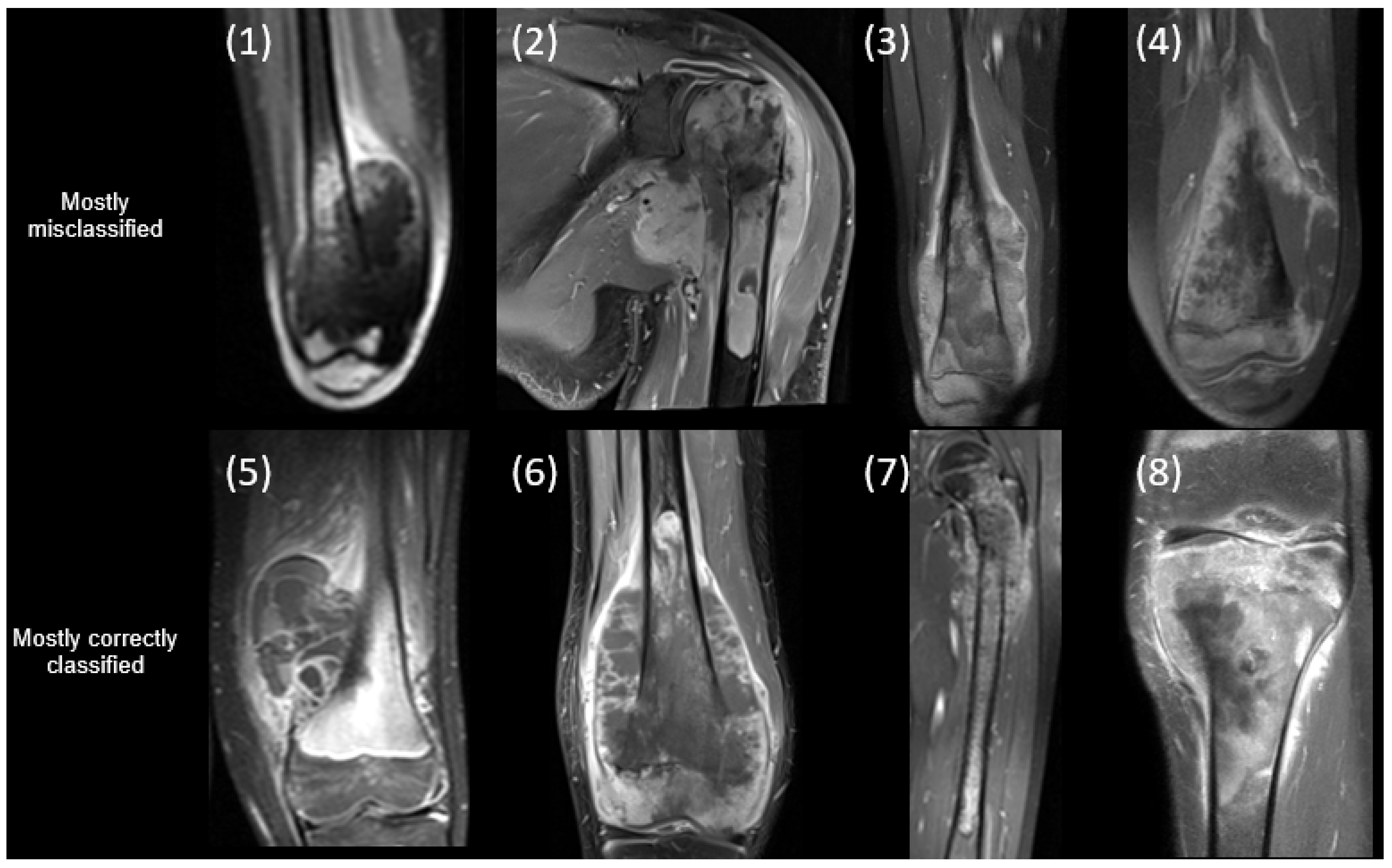
3.3. Classification Results
3.3.1. Progressive Disease
3.3.2. Response to Therapy
3.3.3. Relapse off Therapy
3.3.4. OS-Related Mortality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| OS | Osteosarcoma |
| RF | Radiomic features |
| MRI | Magnetic resonance imaging |
| CT | Computed tomography |
| IBSI | Image Biomarker Standardization Initiative |
| KNN | K-nearest neighbor |
| SVM | Support vector machine |
| LDA | Linear discriminant analysis |
| MLP | Multilayer perceptron classifier |
| CV | Cross-validation |
| ROC AUC | Area under Receiver operating characteristic curve |
| PR AUC | Precision-Recall Area Under the Curve |
References
- Callan, A. OrthoInfo. Osteosarcoma—OrthoInfo—AAOS. Available online: https://www.orthoinfo.org/en/diseases--conditions/osteosarcoma/ (accessed on 21 February 2025).
- American Cancer Society. Survival Rates for Osteosarcoma. Available online: https://www.cancer.org/cancer/types/osteosarcoma/detection-diagnosis-staging/survival-rates.html (accessed on 21 February 2025).
- Aisen, A.; Martel, W.; Braunstein, E.; McMillin, K.; Phillips, W.; Kling, T. MRI and CT evaluation of primary bone and soft-tissue tumors. Am. J. Roentgenol. 1986, 146, 749–756. [Google Scholar] [CrossRef]
- Meyers, P.A.; Gorlick, R. OSTEOSARCOMA. Pediatr. Clin. N. Am. 1997, 44, 973–989. [Google Scholar] [CrossRef]
- Pereira, H.M.; Leite Duarte, M.E.; Ribeiro Damasceno, I.; De Oliveira Moura Santos, L.A.; Nogueira-Barbosa, M.H. Machine learning-based CT radiomics features for the prediction of pulmonary metastasis in osteosarcoma. Br. J. Radiol. 2021, 94, 20201391. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef]
- Coroller, T.P.; Agrawal, V.; Narayan, V.; Hou, Y.; Grossmann, P.; Lee, S.W.; Mak, R.H.; Aerts, H.J.W.L. Radiomic phenotype features predict pathological response in non-small cell lung cancer. Radiother. Oncol. 2016, 119, 480–486. [Google Scholar] [CrossRef]
- Wu, W.; Parmar, C.; Grossmann, P.; Quackenbush, J.; Lambin, P.; Bussink, J.; Mak, R.; Aerts, H.J.W.L. Exploratory Study to Identify Radiomics Classifiers for Lung Cancer Histology. Front. Oncol. 2016, 6, 71. [Google Scholar] [CrossRef]
- Bouhamama, A.; Leporq, B.; Khaled, W.; Nemeth, A.; Brahmi, M.; Dufau, J.; Marec-Bérard, P.; Drapé, J.-L.; Gouin, F.; Bertrand-Vasseur, A.; et al. Prediction of Histologic Neoadjuvant Chemotherapy Response in Osteosarcoma Using Pretherapeutic MRI Radiomics. Radiol. Imaging Cancer 2022, 4, e210107. [Google Scholar] [CrossRef]
- Conti, A.; Duggento, A.; Indovina, I.; Guerrisi, M.; Toschi, N. Radiomics in breast cancer classification and prediction. Semin. Cancer Biol. 2021, 72, 238–250. [Google Scholar] [CrossRef]
- Granzier, R.W.Y.; Verbakel, N.M.H.; Ibrahim, A.; Van Timmeren, J.E.; Van Nijnatten, T.J.A.; Leijenaar, R.T.H.; Lobbes, M.B.I.; Smidt, M.L.; Woodruff, H.C. MRI-based radiomics in breast cancer: Feature robustness with respect to inter-observer segmentation variability. Sci. Rep. 2020, 10, 14163. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Burth, S.; Wick, A.; Götz, M.; Eidel, O.; Schlemmer, H.-P.; Maier-Hein, K.H.; Wick, W.; Bendszus, M.; Radbruch, A.; et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016, 280, 880–889. [Google Scholar] [CrossRef]
- Zhou, M.; Scott, J.; Chaudhury, B.; Hall, L.; Goldgof, D.; Yeom, K.W.; Iv, M.; Ou, Y.; Kalpathy-Cramer, J.; Napel, S.; et al. Radiomics in Brain Tumor: Image Assessment, Quantitative Feature Descriptors, and Machine-Learning Approaches. Am. J. Neuroradiol. 2018, 39, 208–216. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Kotrotsou, A.; Zinn, P.O.; Colen, R.R. Radiomics in Brain Tumors. Magn. Reson. Imaging Clin. N. Am. 2016, 24, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, M.; Tao, Q.; Hu, S.; Hu, C. Non-contrast CT-based radiomics nomogram of pericoronary adipose tissue for predicting haemodynamically significant coronary stenosis in patients with type 2 diabetes. BMC Med. Imaging 2023, 23, 99. [Google Scholar] [CrossRef] [PubMed]
- Hunter, B.; Chen, M.; Ratnakumar, P.; Alemu, E.; Logan, A.; Linton-Reid, K.; Tong, D.; Senthivel, N.; Bhamani, A.; Bloch, S.; et al. A radiomics-based decision support tool improves lung cancer diagnosis in combination with the Herder score in large lung nodules. eBioMedicine 2022, 86, 104344. [Google Scholar] [CrossRef]
- Pan, F.; Feng, L.; Liu, B.; Hu, Y.; Wang, Q. Application of radiomics in diagnosis and treatment of lung cancer. Front. Pharmacol. 2023, 14, 1295511. [Google Scholar] [CrossRef]
- Arthur, A.; Orton, M.R.; Emsley, R.; Vit, S.; Kelly-Morland, C.; Strauss, D.; Lunn, J.; Doran, S.; Lmalem, H.; Nzokirantevye, A.; et al. A CT-based radiomics classification model for the prediction of histological type and tumour grade in retroperitoneal sarcoma (RADSARC-R): A retrospective multicohort analysis. Lancet Oncol. 2023, 24, 1277–1286. [Google Scholar] [CrossRef]
- Peeken, J.C.; Asadpour, R.; Specht, K.; Chen, E.Y.; Klymenko, O.; Akinkuoroye, V.; Hippe, D.S.; Spraker, M.B.; Schaub, S.K.; Dapper, H.; et al. MRI-based delta-radiomics predicts pathologic complete response in high-grade soft-tissue sarcoma patients treated with neoadjuvant therapy. Radiother. Oncol. 2021, 164, 73–82. [Google Scholar] [CrossRef]
- Juntu, J.; Sijbers, J.; De Backer, S.; Rajan, J.; Van Dyck, D. Machine learning study of several classifiers trained with texture analysis features to differentiate benign from malignant soft-tissue tumors in T1-MRI images. Magn. Reson. Imaging 2010, 31, 680–689. [Google Scholar] [CrossRef]
- Fields, B.K.K.; Demirjian, N.L.; Cen, S.Y.; Varghese, B.A.; Hwang, D.H.; Lei, X.; Desai, B.; Duddalwar, V.; Matcuk, G.R. Predicting Soft Tissue Sarcoma Response to Neoadjuvant Chemotherapy Using an MRI-Based Delta-Radiomics Approach. Mol. Imaging Biol. 2023, 25, 776–787. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- White, L.M.; Atinga, A.; Naraghi, A.M.; Lajkosz, K.; Wunder, J.S.; Ferguson, P.; Tsoi, K.; Griffin, A.; Haider, M. T2-weighted MRI radiomics in high-grade intramedullary osteosarcoma: Predictive accuracy in assessing histologic response to chemotherapy, overall survival, and disease-free survival. Skelet. Radiol. 2023, 52, 553–564. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, Q.; Dou, Y.; Cheng, T.; Xia, Y.; Li, H.; Gao, S. Evaluation of the neoadjuvant chemotherapy response in osteosarcoma using the MRI DWI-based machine learning radiomics nomogram. Front. Oncol. 2024, 14, 1345576. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, J.; Cheng, Z.; Lu, X.; Wang, X.; Lu, M.; Li, S.; Xiang, Z.; Zhou, Q.; Liu, Z.; et al. Development and external validation of an MRI-based radiomics nomogram for pretreatment prediction for early relapse in osteosarcoma: A retrospective multicenter study. Eur. J. Radiol. 2020, 129, 109066. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Su, Y.; Duan, J.; Qiu, Q.; Ge, X.; Wang, A.; Yin, Y. Radiomics signature extracted from diffusion-weighted magnetic resonance imaging predicts outcomes in osteosarcoma. J. Bone Oncol. 2019, 19, 100263. [Google Scholar] [CrossRef]
- Huang, B.; Wang, J.; Sun, M.; Chen, X.; Xu, D.; Li, Z.-P.; Ma, J.; Feng, S.-T.; Gao, Z. Feasibility of multi-parametric magnetic resonance imaging combined with machine learning in the assessment of necrosis of osteosarcoma after neoadjuvant chemotherapy: A preliminary study. BMC Cancer 2020, 20, 322. [Google Scholar] [CrossRef]
- Wu, Y.; Xu, L.; Yang, P.; Lin, N.; Huang, X.; Pan, W.; Li, H.; Lin, P.; Li, B.; Bunpetch, V.; et al. Survival Prediction in High-grade Osteosarcoma Using Radiomics of Diagnostic Computed Tomography. EBioMedicine 2018, 34, 27–34. [Google Scholar] [CrossRef]
- Ngan, E.; Mullikin, D.; Theruvath, A.; Annapragada, A.; Ghaghada, K.; Heczey, A.; Starosolski, Z. Classification of osteosarcoma clinical outcomes using contrast-enhanced MRI radiomics and clinical variables. In Proceedings of the SPR 2025 Annual Meeting, Honolulu, HI, USA, 7–11 April 2025. [Google Scholar]
- Chen, H.; Zhang, X.; Wang, X.; Quan, X.; Deng, Y.; Lu, M.; Wei, Q.; Ye, Q.; Zhou, Q.; Xiang, Z.; et al. MRI-based radiomics signature for pretreatment prediction of pathological response to neoadjuvant chemotherapy in osteosarcoma: A multicenter study. Eur. Radiol. 2021, 31, 7913–7924. [Google Scholar] [CrossRef]
| 63 Patients | 26 Patients | 9 Patients | p-Value | |
|---|---|---|---|---|
| Age (Median/Mean ± SD) | 12.29/11.82 ± 3.53 | 12.21/11.83 ± 3.98 | 11.49/12.73 ± 4.20 | 0.9263 |
| Sex (Male) | 43 (68.25%) | 18 (69.23%) | 7 (77.78%) | 0.8451 |
| Race | 0.5845 | |||
| Caucasian | 52 (82.54%) | 22 (84.62%) | 6 (66.66%) | |
| Black/African American | 9 (14.29%) | 3 (11.54%) | 3 (33.33%) | |
| Others | 2 (3.17%) | 1 (3.85%) | 0 (0%) | |
| Hispanics | 31 (49.21%) | 14 (53.85%) | 4 (44.44%) | 0.8690 |
| Laterality (right) | 36 (57.14%) | 16 (61.54%) | 1 (11.11%) | 0.0234 |
| OS location | 0.0532 | |||
| Femur | 41 (65.08%) | 23 (88.46%) | 5 (55.55%) | |
| Tibia | 14 (22.22%) | 3 (11.54%) | 1 (11.11%) | |
| Humus | 6 (9.52%) | 0 (0%) | 1 (11.11%) | |
| Fibula | 2 (3.17%) | 0 (0%) | 2 (22.22%) | |
| Histological subtype | ||||
| Osteoblastic | 50 (79.37%) | 21 (80.77%) | 9 (100%) | 0.4017 |
| Chondroblastic | 21 (33.33%) | 10 (38.46%) | 2 (22.22%) | 0.6798 |
| Telangiectatic | 5 (7.94%) | 1 (3.85%) | 1 (11.11%) | 0.5812 |
| Metastasis (Yes) | 13 (20.63%) | 7 (26.92%) | 5 (55.55%) | 0.0784 |
| Skip lesion (Yes) | 8 (12.70%) | 4 (15.38%) | 1 (11.11%) | 0.8969 |
| Progressive disease during therapy (Yes) | 17 (26.98%) | 6 (23.08%) | 4 (44.44%) | 0.4589 |
| % necrosis (Median/Mean ± SD) | 93/77.32 ± 27.08 | 95/81.21 ± 24.98 | 95/81.11 ± 26.30 | 0.7054 |
| Response on therapy (Adequate) | 37 (58.73%) | 16 (61.54%) | 6 (66.66%) | 0.9509 |
| Relapse off therapy (Yes) | 16 (25.40%) | 7 (26.92%) | 4 (44.44%) | 0.4871 |
| OS related mortality (Yes) | 19 (30.16%) | 6 (23.08%) | 5 (55.55%) | 0.1885 |
| Adequate Response to Therapy (n = 37) | Poor Response to Therapy (n = 26) | p-Value | |
|---|---|---|---|
| Progressive disease during therapy (Yes) | 3 (8.11%) | 14 (53.85%) | <0.0001 |
| Metastasis at diagnosis (Yes) | 11 (29.73%) | 2 (7.69%) | 0.0557 |
| Skip lesion (Yes) | 7 (18.92%) | 1 (3.85%) | 0.1254 |
| Osteoblastic (Yes) | 30 (81.08%) | 20 (76.92%) | 0.6881 |
| Chondroblastic (Yes) | 9 (24.32%) | 12 (46.15%) | 0.0704 |
| Relapse off therapy (n = 16) | No relapse off therapy (n = 47) | p-value | |
| Progressive disease during therapy (Yes) | 4 (25%) | 13 (27.66%) | 0.9999 |
| Response to therapy (Adequate) | 12 (75%) | 25 (53.19%) | 0.1519 |
| % necrosis (mean ± SD) | 81.13 ± 28.78 | 76.03 ± 26.97 | 0.3378 |
| Metastasis at diagnosis (Yes) | 7 (43.75%) | 6 (12.77%) | 0.0082 |
| Skip lesion (Yes) | 4 (25%) | 4 (8.51%) | 0.1857 |
| Osteoblastic (Yes) | 13 (81.25%) | 37 (78.72%) | 0.9999 |
| Chondroblastic (Yes) | 7 (43.75%) | 14 (29.79%) | 0.3062 |
| Deceased (n = 19) | Alive (n = 44) | p-value | |
| Progressive disease during therapy (Yes) | 13 (68.42%) | 4 (9.09%) | <0.0001 |
| Response to therapy (Adequate) | 8 (42.11%) | 29 (65.91%) | 0.0782 |
| Relapse off therapy (Yes) | 9 (47.37%) | 7 (15.91%) | 0.0085 |
| % necrosis (mean ± SD) | 66.58 ± 28.48 | 81.96 ± 25.72 | 0.0762 |
| Metastasis at diagnosis (Yes) | 7 (36.84%) | 6 (13.64%) | 0.0367 |
| Skip lesion (Yes) | 4 (21.05%) | 4 (9.09%) | 0.2286 |
| Osteoblastic (Yes) | 16 (84.21%) | 34 (77.27%) | 0.7375 |
| Chondroblastic (Yes) | 10 (52.63%) | 11 (25%) | 0.0327 |
| Segmentation | RF Type | Validation Set | Testing Set | Best Classifier | No. Features | External Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Segmentation | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ||||
| Whole tumor | IBSI RFs | 0.84 ± 0.14 | 0.70 ± 0.27 | 0.89 ± 0.13 | 0.68 ± 0.29 | 0.66 ± 0.29 | 0.69 | 0.50 | 0.78 | 0.67 | 0.69 | MLP | 4 | 0.33 | 0.25 | 0.40 |
| IBSI RFs + baseline clinical | 0.90 ± 0.11 | 0.93 ± 0.13 | 0.89 ± 0.11 | 0.81 ± 0.17 | 0.87 ± 0.16 | 0.69 | 0.25 | 0.89 | 0.58 | 0.41 | SVM rbf | 8 | 0.44 | 0.25 | 0.60 | |
| All RFs | 0.92 ± 0.08 | 0.93 ± 0.13 | 0.91 ± 0.07 | 0.88 ± 0.13 | 0.91 ± 0.10 | 0.77 | 0.50 | 0.89 | 0.67 | 0.65 | KNN (k = 3) | 13 | 0.44 | 0.25 | 0.60 | |
| All RFs + baseline clinical | 0.96 ± 0.05 | 0.93 ± 0.13 | 0.97 ± 0.06 | 0.92 ± 0.09 | 0.94 ± 0.08 | 0.77 | 0.50 | 0.89 | 0.51 | 0.56 | Random forest | 14 | 0.56 | 0.25 | 0.80 | |
| Whole tumor/tumor sampling | Only baseline clinical | 0.78 ± 0.20 | 0.93 ± 0.13 | 0.71 ± 0.30 | 0.70 ± 0.26 | 0.71 ± 0.19 | 0.77 | 0.75 | 0.78 | 0.88 | 0.78 | Random forest | 12 | 0.22 | 0.50 | 0.00 |
| Tumor sampling | IBSI RFs | 0.82 ± 0.15 | 0.87 ± 0.16 | 0.81 ± 0.18 | 0.75 ± 0.23 | 0.80 ± 0.20 | 0.85 | 0.75 | 0.89 | 0.69 | 0.68 | MLP | 15 | 0.44 | 0.25 | 0.60 |
| IBSI RFs + baseline clinical | 0.90 ± 0.09 | 1.0 ± 0 | 0.87 ± 0.13 | 0.84 ± 0.17 | 0.93 ± 0.09 | 0.69 | 0.50 | 0.78 | 0.63 | 0.56 | Gradient boosting | 6 | 0.67 | 0.50 | 0.80 | |
| All RFs | 0.92 ± 0.12 | 1.0 ± 0 | 0.89 ± 0.17 | 0.91 ± 0.12 | 0.94 ± 0.08 | 0.31 | 1.00 | 0.00 | 0.86 | 0.81 | MLP | 15 | 0.44 | 1.00 | 0.00 | |
| All RFs + baseline clinical | 0.92 ± 0.12 | 0.93 ± 0.13 | 0.93 ± 0.15 | 0.90 ± 0.13 | 0.92 ± 0.09 | 0.31 | 1.00 | 0.00 | 0.89 | 0.83 | MLP | 11 | 0.44 | 1.00 | 0.00 | |
| Bone/soft tissue | IBSI RFs | 0.95 ± 0.07 | 1.0 ± 0 | 0.93 ± 0.09 | 0.94 ± 0.08 | 0.97 ± 0.05 | 0.83 | 0.50 | 1.00 | 0.88 | 0.83 | LDA | 1 | - | - | - |
| IBSI RFs + baseline clinical | 0.95 ± 0.07 | 1.0 ± 0 | 0.93 ± 0.09 | 0.94 ± 0.08 | 0.97 ± 0.05 | 0.83 | 0.50 | 1.00 | 0.88 | 0.83 | LDA | 1 | - | - | - | |
| All RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.67 | 0.50 | 0.75 | 0.75 | 0.75 | SVM rbf | 14 | - | - | - | |
| All RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.67 | 0.50 | 0.75 | 0.81 | 0.58 | Random forest | 15 | - | - | - | |
| Only baseline clinical | 0.79 ± 0.21 | 1.0 ± 0 | 0.73 ± 0.25 | 0.69 ± 0.32 | 0.82 ± 0.23 | 0.67 | 0.00 | 1.00 | 0.75 | 0.75 | MLP | 9 | - | - | - | |
| Segmentation | RF Type | Validation Set | Testing Set | Best Classifier | No. Features | External Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ||||
| Whole tumor | IBSI RFs | 0.72 ± 0.04 | 0.59 ± 0.12 | 0.91 ± 0.11 | 0.83 ± 0.04 | 0.73 ± 0.13 | 0.54 | 0.25 | 1.00 | 0.69 | 0.79 | KNN (k = 10) | 3 | 0.44 | 0.50 | 0.33 |
| IBSI RFs + baseline clinical | 0.78 ± 0.13 | 0.67 ± 0.24 | 0.95 ± 0.10 | 0.86 ± 0.09 | 0.73 ± 0.17 | 0.69 | 0.63 | 0.80 | 0.80 | 0.87 | MLP | 9 | 0.56 | 0.50 | 0.67 | |
| All RFs | 0.88 ± 0.08 | 0.79 ± 0.14 | 1.0 ± 0 | 0.93 ± 0.06 | 0.87 ± 0.12 | 0.62 | 0.63 | 0.60 | 0.78 | 0.88 | SVM rbf | 14 | 0.56 | 0.67 | 0.33 | |
| All RFs + baseline clinical | 0.88 ± 0.08 | 0.79 ± 0.14 | 1.0 ± 0 | 0.93 ± 0.06 | 0.87 ± 0.12 | 0.62 | 0.63 | 0.60 | 0.78 | 0.88 | SVM rbf | 14 | 0.56 | 0.67 | 0.33 | |
| Whole tumor/tumor sampling | Only baseline clinical | 0.76 ± 0.08 | 0.76 ± 0.17 | 0.76 ± 0.16 | 0.84 ± 0.06 | 0.75 ± 0.10 | 0.77 | 0.88 | 0.60 | 0.90 | 0.95 | SVM sigmoid | 15 | 0.67 | 1.00 | 0.00 |
| Tumor sampling | IBSI RFs | 0.80 ± 0.06 | 0.69 ± 0.12 | 0.96 ± 0.08 | 0.89 ± 0.06 | 0.84 ± 0.04 | 0.62 | 0.75 | 0.40 | 0.68 | 0.77 | MLP | 5 | 0.67 | 0.83 | 0.33 |
| IBSI RFs + baseline clinical | 0.88 ± 0.08 | 0.83 ± 0.11 | 0.95 ± 0.10 | 0.94 ± 0.05 | 0.89 ± 0.07 | 0.62 | 0.50 | 0.80 | 0.76 | 0.84 | Random forest | 5 | 0.44 | 0.50 | 0.33 | |
| All RFs | 0.94 ± 0.05 | 0.93 ± 0.09 | 0.95 ± 0.10 | 0.98 ± 0.02 | 0.97 ± 0.03 | 0.46 | 0.25 | 0.80 | 0.60 | 0.72 | SVM Polynomial | 11 | 0.33 | 0.00 | 1.00 | |
| All RFs + baseline clinical | 0.94 ± 0.05 | 0.93 ± 0.09 | 0.95 ± 0.10 | 0.98 ± 0.02 | 0.97 ± 0.03 | 0.46 | 0.25 | 0.80 | 0.60 | 0.72 | SVM Polynomial | 11 | 0.33 | 0.00 | 1.00 | |
| Bone/soft tissue | IBSI RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.50 | 0.25 | 1.00 | 0.75 | 0.92 | Random forest | 10 | - | - | - |
| IBSI RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.33 | 0.25 | 0.50 | 0.63 | 0.80 | Random forest | 12 | - | - | - | |
| All RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | KNN (k = 5) | 4 | - | - | - | |
| All RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.83 | 1.00 | 0.50 | 0.88 | 0.95 | LDA | 3 | - | - | - | |
| Only baseline clinical | 0.90 ± 0.12 | 0.83 ± 0.21 | 1.0 ± 0 | 0.93 ± 0.10 | 0.83 ± 0.21 | 0.50 | 0.25 | 1.00 | 0.75 | 0.92 | Logistic regression | 11 | - | - | - | |
| Segmentation | RF Type | Validation Set | Testing Set | Best Classifier | No. Features | External Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ||||
| Whole tumor | IBSI RFs | 0.84 ± 0.10 | 0.77 ± 0.20 | 0.86 ± 0.16 | 0.70 ± 0.17 | 0.73 ± 0.18 | 0.77 | 0.00 | 1.00 | 0.73 | 0.42 | SVM sigmoid | 4 | 0.67 | 0.25 | 1.00 |
| IBSI RFs + baseline clinical | 0.80 ± 0.09 | 0.80 ± 0.27 | 0.81 ± 0.18 | 0.67 ± 0.11 | 0.80 ± 0.09 | 0.69 | 0.33 | 0.80 | 0.58 | 0.35 | Random forest | 5 | 0.44 | 0.50 | 0.40 | |
| IBSI RFs + baseline clinical + prior outcomes | 0.82 ± 0.08 | 0.70 ± 0.27 | 0.87 ± 0.16 | 0.65 ± 0.14 | 0.75 ± 0.12 | 0.77 | 0.00 | 1.00 | 0.63 | 0.39 | Logistic regression | 3 | 0.56 | 0.00 | 1.00 | |
| All RFs | 0.84 ± 0.10 | 0.93 ± 0.13 | 0.81 ± 0.17 | 0.75 ± 0.18 | 0.84 ± 0.11 | 0.54 | 0.33 | 0.60 | 0.33 | 0.22 | SVM rbf | 11 | 0.78 | 0.75 | 0.80 | |
| All RFs + baseline clinical | 0.88 ± 0.10 | 1.0 ± 0 | 0.83 ± 0.14 | 0.89 ± 0.11 | 0.92 ± 0.08 | 0.46 | 0.33 | 0.50 | 0.57 | 0.36 | SVM rbf | 10 | 0.67 | 1.00 | 0.40 | |
| All RFs + baseline clinical + prior outcomes | 0.90 ± 0.09 | 0.87 ± 0.27 | 0.92 ± 0.10 | 0.78 ± 0.25 | 0.84 ± 0.20 | 0.62 | 1.00 | 0.50 | 0.73 | 0.41 | MLP | 5 | 0.56 | 1.00 | 0.20 | |
| Whole tumor/tumor sampling | Only baseline clinical | 0.74 ± 0.19 | 0.93 ± 0.13 | 0.68 ± 0.24 | 0.68 ± 0.26 | 0.75 ± 0.20 | 0.62 | 0.67 | 0.60 | 0.58 | 0.53 | Random forest | 9 | 0.56 | 0.25 | 0.80 |
| Only baseline clinical + prior outcomes | 0.78 ± 0.22 | 0.93 ± 0.13 | 0.74 ± 0.29 | 0.72 ± 0.29 | 0.76 ± 0.28 | 0.77 | 0.00 | 1.00 | 0.57 | 0.30 | MLP | 8 | 0.56 | 0.00 | 1.00 | |
| Tumor sampling | IBSI RFs | 0.84 ± 0.16 | 0.87 ± 0.16 | 0.83 ± 0.23 | 0.80 ± 0.18 | 0.78 ± 0.18 | 0.77 | 0.33 | 0.90 | 0.67 | 0.43 | SVM sigmoid | 11 | 0.44 | 0.00 | 0.80 |
| IBSI RFs + baseline clinical | 0.94 ± 0.05 | 0.93 ± 0.13 | 0.95 ± 0.07 | 0.86 ± 0.13 | 0.93 ± 0.06 | 0.77 | 0.33 | 0.90 | 0.73 | 0.61 | SVM polynomial | 13 | 0.56 | 0.25 | 0.80 | |
| IBSI RFs + baseline clinical + prior outcomes | 0.94 ± 0.05 | 0.93 ± 0.13 | 0.95 ± 0.07 | 0.86 ± 0.13 | 0.93 ± 0.06 | 0.77 | 0.33 | 0.90 | 0.73 | 0.61 | SVM polynomial | 13 | 0.56 | 0.25 | 0.80 | |
| All RFs | 0.92 ± 0.08 | 0.93 ± 0.13 | 0.92 ± 0.11 | 0.90 ± 0.08 | 0.93 ± 0.07 | 0.23 | 1.00 | 0.00 | 0.73 | 0.41 | MLP | 15 | 0.44 | 1.00 | 0.00 | |
| All RFs + baseline clinical | 0.94 ± 0.05 | 1.0 ± 0 | 0.92 ± 0.07 | 0.93 ± 0.07 | 0.97 ± 0.03 | 0.31 | 1.00 | 0.10 | 0.67 | 0.64 | MLP | 10 | 0.44 | 1.00 | 0.00 | |
| All RFs + baseline clinical + prior outcomes | 0.94 ± 0.05 | 1.0 ± 0 | 0.92 ± 0.07 | 0.93 ± 0.07 | 0.97 ± 0.03 | 0.31 | 1.00 | 0.10 | 0.67 | 0.64 | MLP | 10 | 0.44 | 1.00 | 0.00 | |
| Bone/soft tissue | IBSI RFs | 0.95 ± 0.07 | 1.0 ± 0 | 0.93 ± 0.09 | 0.94 ± 0.08 | 0.97 ± 0.05 | 0.67 | 0.50 | 0.75 | 0.75 | 0.75 | Logistic regression | 3 | - | - | - |
| IBSI RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.67 | 0.50 | 0.75 | 0.75 | 0.75 | SVM rbf | 3 | - | - | - | |
| IBSI RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.67 | 0.50 | 0.75 | 0.75 | 0.75 | SVM rbf | 3 | - | - | - | |
| All RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.50 | 0.50 | 0.50 | 0.63 | 0.50 | MLP | 15 | - | - | - | |
| All RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.50 | 0.50 | 0.50 | 0.75 | 0.75 | SVM rbf | 10 | - | - | - | |
| All RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.50 | 0.50 | 0.50 | 0.75 | 0.75 | SVM rbf | 10 | - | - | - | |
| Only baseline clinical | 0.95 ± 0.07 | 1.0 ± 0 | 0.93 ± 0.09 | 0.94 ± 0.08 | 0.97 ± 0.05 | 0.67 | 0.50 | 0.75 | 0.75 | 0.58 | MLP | 11 | - | - | - | |
| Only baseline clinical + prior outcomes | 0.91 ± 0.14 | 1.0 ± 0 | 0.87 ± 0.19 | 0.92 ± 0.12 | 0.93 ± 0.09 | 0.67 | 0.50 | 0.75 | 0.75 | 0.75 | SVM sigmoid | 2 | - | - | - | |
| Segmentation | RF Type | Validation Set | Testing Set | Best Classifier | No. Features | External Set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ROC AUC | PR AUC | Accuracy | Sensitivity | Specificity | ||||
| Whole tumor | IBSI RFs | 0.80 ± 0.13 | 0.80 ± 0.16 | 0.80 ± 0.25 | 0.69 ± 0.16 | 0.73 ± 0.09 | 0.31 | 0.75 | 0.11 | 0.47 | 0.35 | SVM sigmoid | 10 | 0.67 | 1.00 | 0.25 |
| IBSI RFs + baseline clinical | 0.88 ± 0.08 | 0.73 ± 0.25 | 0.94 ± 0.11 | 0.73 ± 0.18 | 0.77 ± 0.17 | 0.69 | 0.25 | 0.89 | 0.31 | 0.33 | KNN (k = 15) | 8 | 0.56 | 0.20 | 1.00 | |
| IBSI RFs + baseline clinical + prior outcomes | 0.98 ± 0.04 | 1.0 ± 0 | 0.97 ± 0.06 | 0.98 ± 0.03 | 0.99 ± 0.02 | 0.85 | 1.00 | 0.78 | 0.92 | 0.85 | SVM rbf | 3 | 0.56 | 0.40 | 0.75 | |
| All RFs | 0.92 ± 0.12 | 0.93 ± 0.13 | 0.91 ± 0.11 | 0.91 ± 0.15 | 0.92 ± 0.13 | 0.85 | 0.75 | 0.89 | 0.86 | 0.75 | Random forest | 15 | 0.89 | 1.00 | 0.75 | |
| All RFs + baseline clinical | 0.92 ± 0.04 | 0.80 ± 0.16 | 0.97 ± 0.06 | 0.87 ± 0.08 | 0.87 ± 0.11 | 0.77 | 0.75 | 0.78 | 0.81 | 0.57 | Random forest | 6 | 0.56 | 0.20 | 1.00 | |
| All RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.92 | 1.00 | 0.89 | 0.97 | 0.95 | KNN (k = 5) | 6 | 1.00 | 1.00 | 1.00 | |
| whole tumor/tumor sampling | Only baseline clinical | 0.82 ± 0.12 | 0.73 ± 0.13 | 0.86 ± 0.16 | 0.69 ± 0.21 | 0.69 ± 0.19 | 0.62 | 0.50 | 0.67 | 0.67 | 0.46 | SVM polynomial | 15 | 0.44 | 0.20 | 0.75 |
| Only baseline clinical + prior outcomes | 0.98 ± 0.04 | 1.0 ± 0 | 0.97 ± 0.06 | 0.98 ± 0.03 | 0.99 ± 0.02 | 0.85 | 1.00 | 0.78 | 0.92 | 0.85 | SVM rbf | 3 | 0.56 | 0.40 | 0.75 | |
| Tumor sampling | IBSI RFs | 0.90 ± 0.06 | 0.73 ± 0.25 | 0.97 ± 0.06 | 0.80 ± 0.15 | 0.81 ± 0.18 | 0.62 | 0.25 | 0.78 | 0.61 | 0.53 | Random forest | 5 | 0.33 | 0.00 | 0.75 |
| IBSI RFs + baseline clinical | 0.84 ± 0.10 | 0.87 ± 0.16 | 0.83 ± 0.17 | 0.77 ± 0.17 | 0.84 ± 0.14 | 0.69 | 0.50 | 0.78 | 0.72 | 0.47 | Random forest | 7 | 0.33 | 0.00 | 0.75 | |
| IBSI RFs + baseline clinical + prior outcomes | 0.98 ± 0.04 | 1.0 ± 0 | 0.97 ± 0.06 | 0.98 ± 0.03 | 0.99 ± 0.02 | 0.85 | 1.00 | 0.78 | 0.92 | 0.85 | SVM rbf | 3 | 0.56 | 0.40 | 0.75 | |
| All RFs | 0.94 ± 0.08 | 1.0 ± 0 | 0.91 ± 0.11 | 0.91 ± 0.11 | 0.95 ± 0.06 | 0.39 | 1.00 | 0.11 | 0.81 | 0.67 | Random forest | 12 | 0.67 | 1.00 | 0.25 | |
| All RFs + baseline clinical | 0.94 ± 0.08 | 0.93 ± 0.13 | 0.94 ± 0.11 | 0.91 ± 0.11 | 0.93 ± 0.08 | 0.62 | 1.00 | 0.44 | 0.85 | 0.73 | Random forest | 5 | 0.56 | 1.00 | 0.00 | |
| All RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 0.94 ± 0.11 | 0.97 ± 0.05 | 0.98 ± 0.04 | 0.85 | 0.75 | 0.89 | 0.94 | 0.92 | KNN (k = 8) | 8 | 0.78 | 0.60 | 1.00 | |
| Bone/soft tissue | IBSI RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | SVM sigmoid | 7 | - | - | - |
| IBSI RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | KNN (k = 6) | 8 | - | - | - | |
| IBSI RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | Random forest | 8 | - | - | - | |
| All RFs | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.83 | 1.00 | 0.75 | 0.88 | 0.83 | SVM rbf | 6 | - | - | - | |
| All RFs + baseline clinical | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.83 | 1.00 | 0.75 | 0.88 | 0.83 | SVM rbf | 6 | - | - | - | |
| All RFs + baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | Random forest | 4 | - | - | - | |
| Only baseline clinical | 0.73 ± 0.29 | 1.0 ± 0 | 0.68 ± 0.35 | 0.57 ± 0.33 | 0.68 ± 0.35 | 0.67 | 0.50 | 0.75 | 0.63 | 0.50 | MLP | 15 | - | - | - | |
| Only baseline clinical + prior outcomes | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 1.0 ± 0 | 0.83 | 0.50 | 1.00 | 0.88 | 0.83 | SVM rbf | 4 | - | - | - | |
| Patient | Gender | Age (y) | Skip Lesion | Meta-Stasis | OS Type | Progressive Disease | % Necrosis | Relapse off Therapy | Mortality |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Female | 14.0 | No | Yes | Osteobl. | No | 100 | Yes | Yes |
| 2 | Male | 16.3 | No | Yes | Osteobl. | No | 99 | No | No |
| 3 | Female | 14.9 | Yes | Yes | Osteobl. | Yes | >99 | Yes | Yes |
| 4 | Male | 10.9 | No | Yes | Chondrobl. | No | >99 | No | No |
| 5 | Female | 9.8 | No | No | Osteobl. and telangiectatic | No | 100 | No | No |
| 6 | Male | 12.3 | Yes | Yes | Osteobl. and chondrob. | Yes | 95 | Yes | Yes |
| 7 | Male | 9.0 | No | No | Osteobl. | No | 87 | No | No |
| 8 | Male | 14.2 | No | No | Osteobl. | Yes | 40 | No | yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngan, E.; Mullikin, D.; Theruvath, A.J.; Annapragada, A.V.; Ghaghada, K.B.; Heczey, A.A.; Starosolski, Z.A. MRI-Based Radiomics for Outcome Stratification in Pediatric Osteosarcoma. Cancers 2025, 17, 2586. https://doi.org/10.3390/cancers17152586
Ngan E, Mullikin D, Theruvath AJ, Annapragada AV, Ghaghada KB, Heczey AA, Starosolski ZA. MRI-Based Radiomics for Outcome Stratification in Pediatric Osteosarcoma. Cancers. 2025; 17(15):2586. https://doi.org/10.3390/cancers17152586
Chicago/Turabian StyleNgan, Esther, Dolores Mullikin, Ashok J. Theruvath, Ananth V. Annapragada, Ketan B. Ghaghada, Andras A. Heczey, and Zbigniew A. Starosolski. 2025. "MRI-Based Radiomics for Outcome Stratification in Pediatric Osteosarcoma" Cancers 17, no. 15: 2586. https://doi.org/10.3390/cancers17152586
APA StyleNgan, E., Mullikin, D., Theruvath, A. J., Annapragada, A. V., Ghaghada, K. B., Heczey, A. A., & Starosolski, Z. A. (2025). MRI-Based Radiomics for Outcome Stratification in Pediatric Osteosarcoma. Cancers, 17(15), 2586. https://doi.org/10.3390/cancers17152586






