BRCA Screening and Identification of a Common Haplotype in the Jewish Community of Rome Reveal a Founder Effect for the c.7007G>C, p. (Arg2336Pro) BRCA2 Variant
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection and Statistical Analysis
2.2. BRCA Testing and Haplotype Analysis
2.2.1. BRCA Testing
2.2.2. Haplotype Analysis
3. Results
3.1. Sample Selection
3.2. BRCA Screening
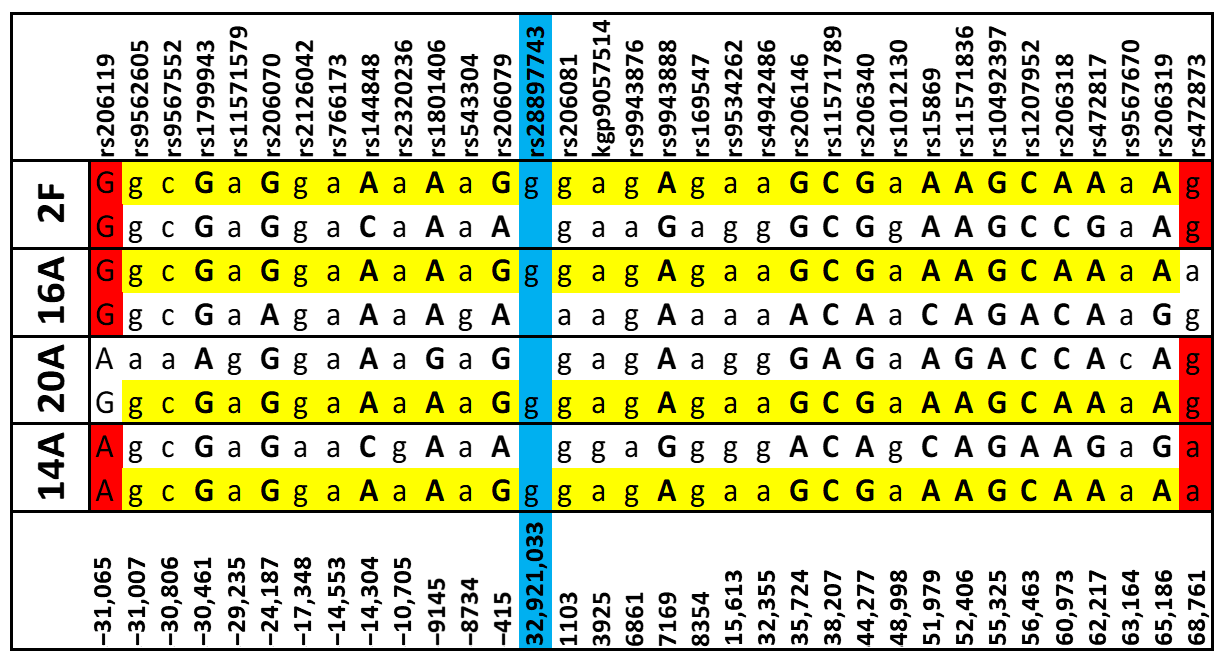
3.3. Haplotype Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACMG | American College of Medical Genetics and Genomics |
| AJ | Ashkenazi Jews |
| BC | Breast cancer |
| HBOC | Hereditary breast/ovarian cancer |
| HGSC | High-grade serous cancer |
| IQR | Interquartile range |
| OC | Ovarian cancer |
| PV | Pathogenic variant |
| VUS | Variant of uncertain significance |
References
- Subbiah, V.; Kurzrock, R. Universal Germline and Tumor Genomic Testing Needed to Win the War Against Cancer: Genomics Is the Diagnosis. J. Clin. Oncol. 2023, 41, 3100–3103. [Google Scholar] [CrossRef] [PubMed]
- Sutton, A.L.; Hurtado-de-Mendoza, A.; Quillin, J. Reducing Disparities in Receipt of Genetic Counseling for Underserved Women at Risk of Hereditary Breast and Ovarian Cancer. J. Womens Health 2020, 29, 1131–1135. [Google Scholar] [CrossRef]
- Gabai-Kapara, E.; Lahad, A.; Kaufman, B.; Friedman, E.; Segev, S.; Renbaum, P.; Beeri, R.; Gal, M.; Grinshpun-Cohen, J.; Djemal, K.; et al. Population-based screening for breast and ovarian cancer risk due to BRCA1 and BRCA2. Proc. Natl. Acad. Sci. USA 2014, 111, 14205–14210. [Google Scholar] [CrossRef]
- Daly, M.B.; Pal, T.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Goggins, M.; Hutton, M.L.; et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, P. BRCA testing launched for people of Jewish ancestry in England. Lancet Oncol. 2024, 25, 284. [Google Scholar] [CrossRef]
- Bedrosian, I.; Somerfield, M.R.; Achatz, M.I.; Boughey, J.C.; Curigliano, G.; Friedman, S.; Kohlmann, W.K.; Kurian, A.W.; Laronga, C.; Lynce, F.; et al. Germline Testing in Patients with Breast Cancer: ASCO-Society of Surgical Oncology Guideline. J. Clin. Oncol. 2024, 42, 584–604. [Google Scholar] [CrossRef] [PubMed]
- Ferla, R.; Calò, V.; Cascio, S.; Rinaldi, G.; Badalamenti, G.; Carreca, I.; Surmacz, E.; Colucci, G.; Bazan, V.; Russo, A. Founder mutations in BRCA1 and BRCA2 genes. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2007, 18 (Suppl. 6), vi93–vi98. [Google Scholar] [CrossRef]
- Barnes-Kedar, I.; Bernstein-Molho, R.; Ginzach, N.; Hartmajer, S.; Shapira, T.; Magal, N.; Lifshitc Kalis, M.; Peretz, T.; Shohat, M.; Basel-Salmon, L.; et al. The yield of full BRCA1/2 genotyping in Israeli high-risk breast/ovarian cancer patients who do not carry the predominant mutations. Breast Cancer Res. Treat. 2018, 172, 151–158. Available online: https://go.gale.com/ps/i.do?p=AONE&sw=w&issn=01676806&v=2.1&it=r&id=GALE%7CA558411577&sid=googleScholar&linkaccess=fulltext (accessed on 20 May 2024). [CrossRef]
- Concolino, P.; Rizza, R.; Mignone, F.; Costella, A.; Guarino, D.; Carboni, I.; Capoluongo, E.; Santonocito, C.; Urbani, A.; Minucci, A. A comprehensive BRCA1/2 NGS pipeline for an immediate Copy Number Variation (CNV) detection in breast and ovarian cancer molecular diagnosis. Clin. Chim. Acta 2018, 480, 173–179. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Fastier-Foster, J.; Grody, W.W.; Hedge, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Parsons, M.T.; de la Hoya, M.; Richardson, M.E.; Tudini, E.; Anderson, M.; Berkofsky-Fessler, W.; Caputo, S.M.; Chan, R.C.; Cline, M.S.; Feng, B.J.; et al. Evidence-based recommendations for gene-specific ACMG/AMP variant classification from the ClinGen ENIGMA BRCA1 and BRCA2 Variant Curation Expert Panel. Am. J. Hum. Genet. 2024, 111, 2044–2058. [Google Scholar] [CrossRef] [PubMed]
- Delaneau, O.; Marchini, J.; Zagury, J.F. A linear complexity phasing method for thousands of genomes. Nat. Methods 2011, 9, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Delaneau, O.; Howie, B.; Cox, A.J.; Zagury, J.F.; Marchini, J. Haplotype estimation using sequencing reads. Am. J. Hum. Genet. 2013, 93, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Papi, L.; Putignano, A.L.; Congregati, C.; Zanna, I.; Sera, F.; Morrone, D.; Falchetti, M.; Rosselli Del Turco, M.; Ottini, L.; Palli, D.; et al. Founder mutations account for the majority of BRCA1-attributable hereditary breast/ovarian cancer cases in a population from Tuscany, Central Italy. Breast Cancer Res. Treat. 2009, 117, 497–504. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Oddoux, C.; Guillen-Navarro, E.; Ditivoli, C.; Dicave, E.; Cilio, M.R.; Clayton, C.M.; Nelson, H.; Sarafoglou, K.; McCain, N.; Peretz, H.; et al. Mendelian diseases among Roman Jews: Implications for the origins of disease alleles. J. Clin. Endocrinol. Metab. 1999, 84, 4405–4409. [Google Scholar] [CrossRef][Green Version]
- Leslie, C.; Dunn, S.P. The Jewish Community of Rome. Sci. Am. 1957, 196, 118–128. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef]
- Laitman, Y.; Friebel, T.M.; Yannoukakos, D.; Fostira, F.; Konstantopoulou, I.; Figlioli, G.; Bonanni, B.; Manoukian, S.; Zuradelli, M.; Tondini, C.; et al. The spectrum of BRCA1 and BRCA2 pathogenic sequence variants in Middle Eastern, North African, and South European countries. Hum. Mutat. 2019, 40, e1–e23. [Google Scholar] [CrossRef]
- Sagi, M.; Eilat, A.; Ben Avi, L.; Goldberg, Y.; Bercovich, D.; Hamburger, T.; Peretz, T.; Lerer, I. Two BRCA1/2 founder mutations in Jews of Sephardic origin. Fam. Cancer 2011, 10, 59–63. [Google Scholar] [CrossRef]
- Laitman, Y.; Simeonov, M.; Herskovitz, L.; Kushnir, A.; Shimon-Paluch, S.; Kaufman, B.; Zidan, J.; Friedman, E. Recurrent germline mutations in BRCA1 and BRCA2 genes in high risk families in Israel. Breast Cancer Res. Treat. 2012, 133, 1153–1157. [Google Scholar] [CrossRef]
- Tsaousis, G.N.; Papadopoulou, E.; Apessos, A.; Agiannitopoulos, K.; Pepe, G.; Kampouri, S.; Diamantopoulos, N.; Floros, T.; Iosifidou, R.; Katopodi, O.; et al. Analysis of hereditary cancer syndromes by using a panel of genes: Novel and multiple pathogenic mutations. BMC Cancer 2019, 19, 535. [Google Scholar] [CrossRef] [PubMed]
- Bernstein-Molho, R.; Laitman, Y.; Schayek, H.; Reish, O.; Lotan, S.; Haim, S.; Zidan, J.; Friedman, E. The yield of targeted genotyping for the recurring mutations in BRCA1/2 in Israel. Breast Cancer Res. Treat. 2018, 167, 697–702. [Google Scholar] [CrossRef]
- Santonocito, C.; Rizza, R.; Paris, I.; De Marchis, L.; Paolillo, C.; Tiberi, G.; Scambia, G.; Capoluongo, E. Spectrum of Germline BRCA1 and BRCA2 Variants Identified in 2351 Ovarian and Breast Cancer Patients Referring to a Reference Cancer Hospital of Rome. Cancers 2020, 12, 1286. [Google Scholar] [CrossRef]
- Patruno, M.; De Summa, S.; Resta, N.; Caputo, M.; Costanzo, S.; Digennaro, M.; Pilato, B.; Bagnulo, R.; Pantaleo, A.; Simone, C.; et al. Spectrum of Germline Pathogenic Variants in BRCA1/2 Genes in the Apulian Southern Italy Population: Geographic Distribution and Evidence for Targeted Genetic Testing. Cancers 2021, 13, 4714. [Google Scholar] [CrossRef] [PubMed]
- Loizzi, V.; Cicinelli, E.; Santamaria, F.; Murgia, F.; Minicucci, V.; Resta, L.; Resta, N.; Natalicchio, M.I.; Ranieri, G.; Cormio, G. BRCAmut and “founder effect”: A prospective study in a single academic institution. Oncotarget 2018, 9, 22353–22358. [Google Scholar] [CrossRef]
- De Matteis, E.; Tumolo, M.R.; Tarantino, P.; Ciccarese, M.; Grassi, T.; Bagordo, F.; De Giorgio, M.R.; Rizzo, E.; Ronzino, G. Prevalence and spectrum of germline BRCA1 and BRCA2 in a cohort of ovarian cancer patients from the Salento peninsula (Southern Italy): A matter of preventive health. Oncotarget 2024, 15, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Milano, A. Storia degli Ebrei italiani nel Levante, 1st ed; Casa Editrice Israel: Firenze, Italy, 1949. [Google Scholar]
- Hamel, N.; Feng, B.J.; Foretova, L.; Stiooa-Lyonnet, D.; Narod, S.A.; Imyanitov, E.; Sinilnikova, O.M.; Tihomirova, L.; Lubinski, J.; Gronwald, J.; et al. On the origin and diffusion of BRCA1 c.5266dupC (5382insC) in European populations. Eur. J. Hum. Genet. 2011, 19, 300–306. [Google Scholar] [CrossRef]
- Houdayer, C.; Caux-Moncoutier, V.; Krieger, S.; Barrois, M.; Bonnet, F.; Bordon, B.; Bronner, M.; Buisson, M.; Coulet, F.; Gaildrat, P.; et al. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum. Mutat. 2012, 33, 1228–1238. [Google Scholar] [CrossRef]
- Serova-Sinilnikova, O.M.; Boutrand, L.; Stoppa-Lyonnet, D.; Bressac-de-Paillerets, B.; Dubois, V.; Lasset, C.; Janin, N.; Bignon, Y.J.; Longy, M.; Maugard, C.; et al. BRCA2 mutations in hereditary breast and ovarian cancer in France. Am. J. Hum. Genet. 1997, 60, 1236. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1712433/ (accessed on 20 May 2024).
- Rebbek, T.R.; Mitra, N.; Wan, F.; Sinilnikova, O.M.; Healey, S.; McGuffog, L.; Mazoyer, S.; Chenevix-Trench, G.; Easton, D.F.; Antoniou, A.C.; et al. Association of Type and Location of BRCA1 and BRCA2 Mutations with Risk of Breast and Ovarian Cancer. JAMA 2015, 313, 1347–1361. [Google Scholar] [CrossRef]
- Sekine, M.; Nishino, K.; Enomoto, T. Differences in Ovarian and Other Cancers Risks by Population and BRCA Mutation Location. Genes 2021, 12, 1050. [Google Scholar] [CrossRef] [PubMed]
- Myers, K.; Davies, S.M.; Harris, R.E.; Spunt, S.L.; Smolarek, T.; Zimmerman, S.; McMasters, R.; Wagner, L.; Mueller, R.; Auerbach, A.D.; et al. The clinical phenotype of children with Fanconi anemia caused by biallelic FANCD1/BRCA2 mutations. Pediatr. Blood Cancer 2012, 58, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Pammi, M.; Saronwala, A.; Magoulas, P.; Ghazi, A.R.; Vetrini, F.; Zhang, J.; He, W.; Dharmadhikari, A.V.; Qu, C.; et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr. 2017, 171, e173438. [Google Scholar] [CrossRef] [PubMed]
- Marano, L.; Verre, L.; Carbone, L.; Poto, G.E.; Fusario, D.; Venezia, D.F.; Calomino, N.; Kaźmierczak-Siedlecka, K.; Polom, K.; Marrelli, D.; et al. Current Trends in Volume and Surgical Outcomes in Gastric Cancer. J. Clin. Med. 2023, 12, 2708. [Google Scholar] [CrossRef]
- Tinterri, C.; Gentile, D.; Caruso, F.; Cortesi, L.; De Laurentiis, M.; Fortunato, L.; Santini, D.; Turchetti, D.; Ferrari, A.; Zambelli, A.; et al. BRCA Testing for Patients Treated in Italy: A National Survey of Breast Centers Associated with Senonetwork. Curr. Oncol. 2024, 31, 3815–3825. [Google Scholar] [CrossRef]
| Total | Roman Jews | Sephardic Jews | |
|---|---|---|---|
| Female n (%) Gender Male n (%) | 44 (100) | 34 (77) | 10 (23) |
| 0 (0) | 0 (0) | 0 (0) | |
| Age at screening Median (IQR) | 64 (55–70) | 65.5 (55–75.25) | 61.5 (53.25–69.25) |
| Breast cancer cases n (%) | 39 (89) | 31 * (79) | 8 (21) |
| Ovarian cancer cases n (%) | 4 (9) | 3 (75) | 1 (25) |
| Breast and ovarian cancer cases n (%) | 1 (2) | 0 (0) | 1 (100) |
| Subjects Enrolled (n = 44) | Roman Jews | Sephardic Jews | |
|---|---|---|---|
| Median Age at diagnosis (IQR) | 56 (47.25–65) | 57.5 (47.25–65.5) | 51.5 (46.5–56) |
| Median Age of BC(IQR) | 56 (45.5–65) | 58 (45–65) | 48 (46–55) |
| Median Age of 2° BC(IQR) * | 76 (62–80) | 76 (62–80) | |
| Median Age of OC(IQR) | 59 (48.5–69) | 49 (NA-NA) | 60.5 (NA-NA) |
| Breast Cancer (n = 40) ^ | 2° Breast Cancer (n = 7) # | Ovarian Cancer (n = 5) ^ | ||||
|---|---|---|---|---|---|---|
| Age at Diagnosis | 56 (45.5–65) | 76 (62–80) | Age at Diagnosis | 59 (48.5–69) | ||
| Histotype N (%) | IDC | 33 (82.5) | 6 (86) | Histotype N (%) | Serous | 5 (100) |
| ILC | 2 (5) | 0 (0) | ||||
| Mucinous | 0 (0) | |||||
| DCIS | 4 (10) | 1 (14) | ||||
| Others | 0 (0) | |||||
| LCIS | 1 (2.5) | 0 (0) | ||||
| Subtype N (%) | LUMINAL | 29 (72.5) | 5 (72) | Grade (Serous OC) N (%) | Low grade | 0 (0) |
| HER2 | 1 (2.5) | 0 (0) | ||||
| TN | 4 (10) | 1 (14) | High grade | 5 (100) | ||
| NA | 6 (15) | 1 (14) | ||||
| Stage N (%) | 0 (pTis) | 5 (12.5) | 1 (14) | Stage N (%) | I | 0 (0) |
| II | 1 (20) | |||||
| I | 19 (47.5) | 1 (14) | ||||
| II | 7 (17.5) | 3 (43) | ||||
| III | 4 (80) | |||||
| III | 2 (5) | 0 (0) | IV | 0 (0) | ||
| IV | 2 (5) | 1 (14) | ||||
| NA | 0 (0) | |||||
| NA | 5 (12.5) | 1 (14) | ||||
| Genotype N (%) | Carriers | 9 * (22.5) | 3 (43) | Genotype N (%) | Carriers | 2 * (40) |
| WT | 28 (70) | 4 (57) | ||||
| WT | 2 (40) | |||||
| VUS° | 3 (7.5) | 0 (0) | VUS° | 1 (20) | ||
| Variants Identified | N° of Families Carriers (%/28) | N° of Subjects Carriers (%/34) | Type of Tumor | ||
|---|---|---|---|---|---|
| BC | OC | ||||
| PV BRCA2 | c.7007G>C | 7 (25) ° | 9 * (26.5) | 7 | 2 |
| c.7962C>T | 1 (3.5) ° | 2 ^ (6) | 2 | 0 | |
| VUS BRCA 1 | c.3691 T>C | 1 (3.5) | 1 (3) | 1 | 0 |
| c.3367 G>T | 1 (3.5) | 1 (3) | 0 | 1 | |
| VUS BRCA 2 | c.1259A>G | 1 (3.5) | 1 (3) | 1 | 0 |
| c.280C>T | 1 (3.5) | 1 (3) | 1 | 0 | |
| Total N° | Median Per Family (IQR) | A/U | Total N ° | Median Per Family (IQR) | ||
|---|---|---|---|---|---|---|
| Subjects | 179 | 25 (15–33) | A | 44 | 5 (3–9) | |
| U | 135 | 20 (13–25) | ||||
| BRCA m subjects ^ | 38 | 4 (4–8) | A | 21 | 2 (2–5) | |
| U | 17 | 2 (1–3) | ||||
| WT subjects # | 29 | 4 (3–5) | A | 4 | 1 (0–1) | |
| U | 25 | 4 (2–4) | ||||
| Tested subjects (carriers) | 12 | 2 (1–2) | A | 12 | 2 (1–2) | |
| U | 0 | 0 (0–0) | ||||
| Tested subjects (non carriers) | 5 | 1 (0–1) | A | 4 | 1 (0–1) | |
| U | 1 | 0 (0–0) | ||||
| Obligate carriers | 8 | 1 (0–2) | A | 7 | 1 (0–2) | |
| U | 1 | 0 (0–0) | ||||
| Segregation analysis (carriers) | 18 | 2 (1–3) | A | 2 | 0 (0–1) | |
| U | 16 | 2 (1–3) | ||||
| Segregation analysis (non carriers) | 24 | 4 (2–4) | A | 0 | 0 (0–0) | |
| U | 24 | 4 (2–4) | ||||
| Unknown Genotype | 112 | 15 (12–23) | A | 19 | 1 (1–5) | |
| U | 93 | 14 (10–19) |
| Total N° of Subjects ° [Median Per Family (IQR)] | Age at Diagnosis Median (IQR) | Total N° of Carriers ° [Median Per Family (IQR)] | Total N° of Non-Carriers ° [Median Per Family (IQR)] | Total N° Untested Subjects ° [Median Per Family (IQR)] | |
|---|---|---|---|---|---|
| Affected subjects * | 44 [5 (3–9)] | 62 (51–69.5) | 21 [2 (2–5)] | 4 [1 (0–1)] | 19 [2 (1–5)] |
| Breast cancer ^ | 29 [5 (0–7)] | 60 (48.75–70) | 13 [1 (0–4)] | 4 [1 (0–1)] | 12 [1.5 (0–3)] |
| Male breast cancer | 2 [0 (0–1)] | 65 (NA) | 2 [0 (0–1)] | 0 | 0 |
| Ovarian cancer | 7 [1 (0–2)] | 55 (49–62) | 5 [0 (0–1)] | 0 | 2 [0 (0–1)] |
| Prostate cancer | 2 [0 (0–1)] | 69.5 (NA) | 1 [0 (0–0)] | 0 | 1 [0 (0–0)] |
| Pancreatic cancer | 1 [0 (0–0)] | 76 (NA) | 1 [0 (0–0)] | 0 | 0 |
| Melanoma | 2 [0 (0–1)] | 62 (NA) | 0 | 0 | 2 [0 (0–1)] |
| Gastric cancer | 3 [0 (0–1)] | 61 (NA) | 2 [0 (0–1)] | 0 | 1 [0 (0–0)] |
| Colorectal cancer | 2 [0 (0–1)] | 49.5 (NA) | 1 [0 (0–0)] | 0 | 1 [0 (0–0)] |
| Malignant glioma | 1 [0 (0–0)] | 70 (NA) | 0 | 0 | 1 [0 (0–0)] |
| Thyroid cancer | 1 [0 (0–0)] | 50 (NA) | 1 [0 (0–0)] | 0 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Marchis, L.; Gelibter, A.J.; Mammone, G.; Madaio, R.A.; Aretini, P.; De Bonis, M.; Zampatti, S.; Peconi, C.; Guadagnolo, D.; Vestri, A.; et al. BRCA Screening and Identification of a Common Haplotype in the Jewish Community of Rome Reveal a Founder Effect for the c.7007G>C, p. (Arg2336Pro) BRCA2 Variant. Cancers 2025, 17, 1906. https://doi.org/10.3390/cancers17121906
De Marchis L, Gelibter AJ, Mammone G, Madaio RA, Aretini P, De Bonis M, Zampatti S, Peconi C, Guadagnolo D, Vestri A, et al. BRCA Screening and Identification of a Common Haplotype in the Jewish Community of Rome Reveal a Founder Effect for the c.7007G>C, p. (Arg2336Pro) BRCA2 Variant. Cancers. 2025; 17(12):1906. https://doi.org/10.3390/cancers17121906
Chicago/Turabian StyleDe Marchis, Laura, Alain Jonathan Gelibter, Giulia Mammone, Raffaele Angelo Madaio, Paolo Aretini, Maria De Bonis, Stefania Zampatti, Cristina Peconi, Daniele Guadagnolo, Annarita Vestri, and et al. 2025. "BRCA Screening and Identification of a Common Haplotype in the Jewish Community of Rome Reveal a Founder Effect for the c.7007G>C, p. (Arg2336Pro) BRCA2 Variant" Cancers 17, no. 12: 1906. https://doi.org/10.3390/cancers17121906
APA StyleDe Marchis, L., Gelibter, A. J., Mammone, G., Madaio, R. A., Aretini, P., De Bonis, M., Zampatti, S., Peconi, C., Guadagnolo, D., Vestri, A., Pizzuti, A., Giardina, E., Capoluongo, E. D., & Minucci, A. (2025). BRCA Screening and Identification of a Common Haplotype in the Jewish Community of Rome Reveal a Founder Effect for the c.7007G>C, p. (Arg2336Pro) BRCA2 Variant. Cancers, 17(12), 1906. https://doi.org/10.3390/cancers17121906







