Simple Summary
This study aimed to pinpoint immune-related genes that show heightened activity in cancerous tissue and explore their correlation with cell infiltration in the tumor microenvironment using bioinformatics analysis. By examining gene expression from both stomach cancer and adjacent healthy tissues, we aimed to uncover their significance in cancer development and their impact on the body’s immune response. We identified several genes, including FN1, COL1A2, THBS2, COL3A1, COL5A1, and BGN, which appear to be associated with poorer outcomes for stomach cancer patients. These genes also demonstrate connections to specific immune cells within cancerous tissue. Understanding the role of these genes in the immune response to cancer could facilitate the development of novel treatments and enhance prognostic capabilities for individuals with stomach cancer.
Abstract
Objective: Gastric carcinoma (GC) is the fifth most commonly diagnosed cancer and the third leading cause of cancer-related deaths globally. The tumor microenvironment plays a significant role in the pathogenesis, prognosis, and response to immunotherapy. However, the immune-related molecular mechanisms underlying GC remain elusive. Bioinformatics analysis of the gene expression of GC and paracancerous healthy tissues from the same patient was performed to identify the key genes and signaling pathways, as well as their correlation to the infiltration of the tumor microenvironment (TME) by various immune cells related to GC development. Methods: We employed GSE19826, a gene expression profile from the Gene Expression Omnibus (GEO), for our analysis. Functional enrichment analysis of Differentially Expressed Genes (DEGs) was conducted using the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes database. Results: Cytoscape software facilitated the identification of nine hub DEGs, namely, FN1, COL1A1, COL1A2, THBS2, COL3A1, COL5A1, APOE, SPP1, and BGN. Various network analysis algorithms were applied to determine their high connectivity. Among these hub genes, FN1, COL1A2, THBS2, COL3A1, COL5A1, and BGN were found to be associated with a poor prognosis for GC patients. Subsequent analysis using the TIMER database revealed the infiltration status of the TME concerning the overexpression of these six genes. Specifically, the abovementioned genes demonstrated direct correlations with cancer-associated fibroblasts, M1 and M2 macrophages, myeloid-derived suppressor cells, and activated dendritic cells. Conclusion: Our findings suggest that the identified hub genes, particularly BGN, FN1, COL1A2, THBS2, COL3A1, and COL5A1, play crucial roles in GC prognosis and TME cell infiltration. This comprehensive analysis enhances our understanding of the molecular mechanisms underlying GC development and may contribute to the identification of potential therapeutic targets and prognostic markers for GC patients.
1. Introduction
Despite marked geographical and regional variations in the frequency of gastric cancer (GC), there has been a consistent decline in its incidence in Western countries in recent decades. Nevertheless, GC continues to hold its position as the fifth most commonly diagnosed cancer and ranks as the third leading cause of cancer-related deaths globally [1].
GC originates from the gastric epithelium and develops through a stepwise progression via precancerous conditions, such as chronic gastritis, advancing towards early carcinomas that infiltrate the mucosa and submucosa. GC has a multifactorial etiology. Endogenous risk factors, such as genetic predisposition, and exogenous factors, like chronic H. pylori-induced gastritis or the increased intake of nitrates, play pivotal roles in its development [2].
The treatment of gastric adenocarcinoma is stage-dependent and involves various multimodal approaches. In recent years, with advancements in endoscopic intervention capabilities, endoscopic submucosal dissection (ESD) and endoscopic mucosal resection (EMR) have emerged as the preferred methods for treating early stage GC, while surgical intervention remains the therapy of choice for more advanced cases [3,4]. Despite progress in understanding the pathogenesis and pathophysiology of GC, perioperative therapy primarily relies on systemic chemotherapy regimens, such as fluorouracil, oxaliplatin, docetaxel, and leucovorin (FLOT) or epirubicin, cisplatin, and capecitabine (ECX), leading to considerable side effects. While neoadjuvant and adjuvant chemotherapy represent essential approaches, immunotherapy plays a limited role in GC treatment compared to many other cancer types [4]. As an example, recent investigations propose that the combination of pembrolizumab and chemotherapy may represent a therapeutic approach for individuals diagnosed with locally advanced or metastatic HER2-negative gastric or gastroesophageal junction adenocarcinoma [5]. Additionally, nivolumab has exhibited superior overall survival when administered alongside chemotherapy compared to sole chemotherapy in previously untreated patients with an advanced gastric, gastroesophageal junction, or esophageal adenocarcinoma [6].
The term tumor microenvironment (TME) refers to the intricate relationships between host cells and malignant cells that develop as a result of tumor cell infiltration [7]. Tumor cells induce substantial molecular, cellular, and physical alterations within the tissues they invade. While the composition of the TME differs among various types of tumors, common characteristics encompass immune cells, stromal cells, blood vessels, and the extracellular matrix (ECM) [8]. The heterogeneity of the TME in GC, particularly with regard to the complexity of the immunological aspects [9], contributes to the lack of widespread acceptance and success of immunotherapy, such as in non-small cell lung cancer or renal cell carcinoma. Therefore, further exploration of the TME and the identification of the key genes related to immune infiltration has garnered significant attention and may serve as a crucial step toward decoding this complexity and the development of new therapies. Through the analysis of the TME, it is hoped that, in the future, it will be possible to predict which GC patients will benefit from immune–oncology treatments and which will not.
Microarray technology, coupled with bioinformatics analysis, has been extensively employed to scrutinize genetic variations across genome sequencing [10]. Despite numerous studies employing these methodologies to explore the clinical relevance of TME infiltrates [11,12], a comprehensive understanding of the diverse array of cells infiltrating the TME remains elusive. Notably, a recent bioinformatics investigation unveiled Stromal-Related Gene Signatures that are intricately linked to macrophage infiltration [13].
The aim of our study was to identify immune-related genes that are overexpressed in cancerous tissue and investigate the relationship of these genes with cell infiltration in the TME through bioinformatics analysis. Subsequently, we determined which of these genes play a significant role in the pathogenesis and progression of GC, potentially serving as targets for pharmaceutical therapies. In our study, we utilized GSE19826 [14], a gene expression profile retrieved from the Gene Expression Omnibus (GEO), for analysis. Subsequently, we conducted a functional enrichment analysis of Differentially Expressed Genes (DEGs), using both the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. We identified hub DEGs demonstrating high connectivity. Following this, we delved into assessing the infiltration status of immune cells in GC using the Tumor Immune Evaluation Resource (TIMER) [15]. Finally, we attempted to establish correlations between these hub DEGs and immune cells, revealing pertinent associations in our research.
2. Materials and Methods
2.1. Microarray Data
The expression profiles of genes associated with gastric adenocarcinoma were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/, accessed on 1 December 2023). One gene expression profile was derived from human gastric adenocarcinoma samples. The GSE19826 series [14] includes 12 samples of GC human tissue and 12 samples of corresponding non-cancerous human tissue from the same patients, along with 3 samples from healthy individuals serving as controls. We utilized the 12 cancerous and 12 non-cancerous samples from the same patients for our analysis. The populations of cancer tissues at each TNM stage were homogenous. Based on the annotation information from the platform, probes were converted into corresponding gene symbols. Additional information regarding the basic characteristics of the patients can be directly acquired from the gene expression profile [14].
2.2. Identification of DEGs between Normal and GC Tissue
The GEO2R tool, available within the GEO series (https://www.ncbi.nlm.nih.gov/geo/geo2r/, accessed on 1 December 2023) [16], was employed to discern DEGs between non-cancerous tissue and GC tissue samples. Genes lacking corresponding gene symbols and those represented by multiple probes were excluded independently. A threshold of |Log2FC| > 1.5 and an adjusted p < 0.05 were applied as criteria for statistical significance.
2.3. Functional Enrichment Analyses of the GC-Related DEGs
In order to explore the biological roles of the identified DEGs within cellular components (CCs), molecular functions (MFs), and biological processes (BPs), the ‘pathfindR’ package [17] was employed to conduct enrichment analyses using GO and the KEGG pathways [18]. The ‘pathfindR’ package was chosen on the basis that it does not solely rely on direct enrichment analysis and thus provides a more comprehensive perspective. Depending solely on a list of significant genes might not provide sufficient information to fully comprehend the underlying disease mechanisms.
2.4. Construction of PPI Network and Identification of Hub DEGs
Furthermore, the GC-associated DEGs identified through the aforementioned methods underwent analysis in the STRING online analysis tool (http://string-db.org, Version: 11.5, accessed on 1 December 2023) [19]. This step was aimed at predicting potential protein interactions among the encoded proteins, utilizing a medium confidence score (>0.4). Utilizing the outcomes obtained from the STRING analysis, a PPI network involving these genes was constructed using the Cytoscape software platform (version: 3.7.10) [20]. To pinpoint the top 10 hub DEGs associated with this network, the ‘cytoHubba’ plugin software in Cytoscape employed a hybrid computation, integrating EPC, Degree, MNC, MCC, and Bottleneck algorithms. This methodology facilitated the further identification of the top 10 hub DEGs.
2.5. Prognostic Value of Hub DEGs as Biomarkers in GC
The prognostic value of the identified top 10 gene hubs was assessed using Kaplan–Meier curves from OncoLnc [21]. OncoLnc encompasses survival information for 8647 individuals across 21 cancer investigations conducted by The Cancer Genome Atlas. These curves were instrumental in evaluating the prognostic efficacy for patients diagnosed with GC, enabling differentiation between those with high and low expression levels of the specified genes.
2.6. Correlation Analyses between Hub DEGs and Infiltrating Immune Cells
The potential associations between the hub DEGs and infiltrating immune cells within the TME were explored using Spearman correlation analysis performed in R [22] using TIMER [15]. TIMER is an open-source server used for a comprehensive analysis of tumor-infiltrating cells in various cancers.
2.7. Expression of Hub Genes in Normal or GC Tissue
The expression analysis of hub genes between normal or GC samples was conducted by Student’s t-test. A p-value < 0.05 was considered significant.
3. Results
3.1. Identification of DEGs between Normal and GC Tissue
In GSE19826, a total of 153 DEGs were confirmed in the cancer tissue, including 91 down-regulated genes and 62 up-regulated genes (Figure 1). The genes are more precisely listed in Supplementary Table S1.
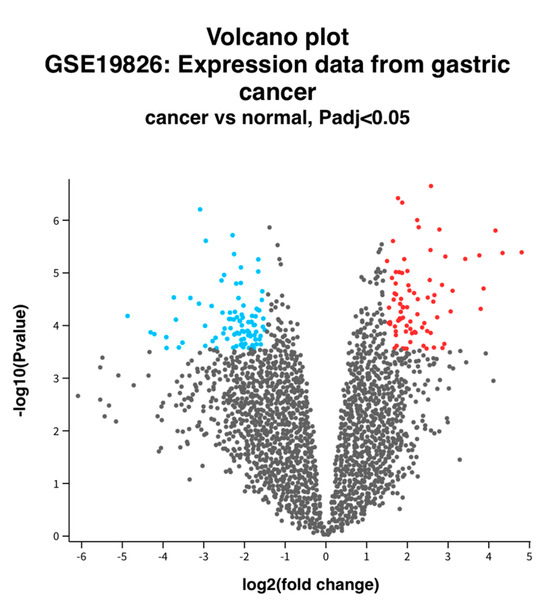
Figure 1.
Volcano plot of DEGs between normal and GC tissue. Volcano plot depicting the up-regulated and down-regulated genes. Red indicates higher expression, while blue indicates lower expression. Log2FC > 1.5 and an adjusted p-value of <0.05 were used.
3.2. Functional Enrichment Analyses
The enriched BPs included collagen fibril organization, skin development, endodermal cell differentiation, neuron projection development, skeletal system development, cardiac right ventricle morphogenesis, protein ubiquitination, negative regulation of the apoptotic signaling pathway, phospholipid efflux, and blood vessel development (Figure 2a, Table 1a). The CCs were endoplasmic reticulum lumen, collagen-containing extracellular matrix, extracellular matrix, endoplasmic reticulum–Golgi intermediate compartment, microfibril, extracellular exosome, very-low-density lipoprotein particle, endocytic vesicle lumen, high-density lipoprotein particle, and Cul3-RING ubiquitin ligase complex (Figure 2b, Table 1b). The enriched MFs included platelet-derived growth factor binding, extracellular matrix structural constituent, protease binding, integrin binding, transcription cis-regulatory region binding, tau protein binding, heparan sulfate proteoglycan binding, proteoglycan binding, extracellular matrix structural constituent conferring tensile strength, and fibronectin binding (Figure 2c, Table 1c). The KEGG pathway analysis revealed enrichment in protein digestion and absorption, ECM–receptor interaction, focal adhesion, platelet activation, proteoglycans in cancer, the AGE-RAGE signaling pathway in diabetic complications, amoebiasis, diabetic cardiomyopathy, the relaxin signaling pathway, and the Wnt signaling pathway (Figure 2d, Table 1d).
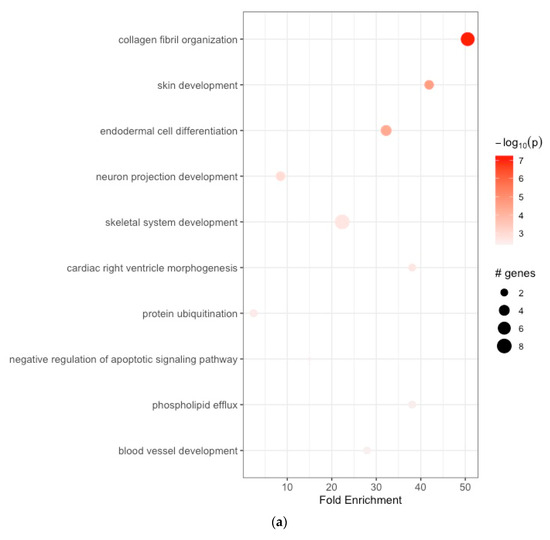
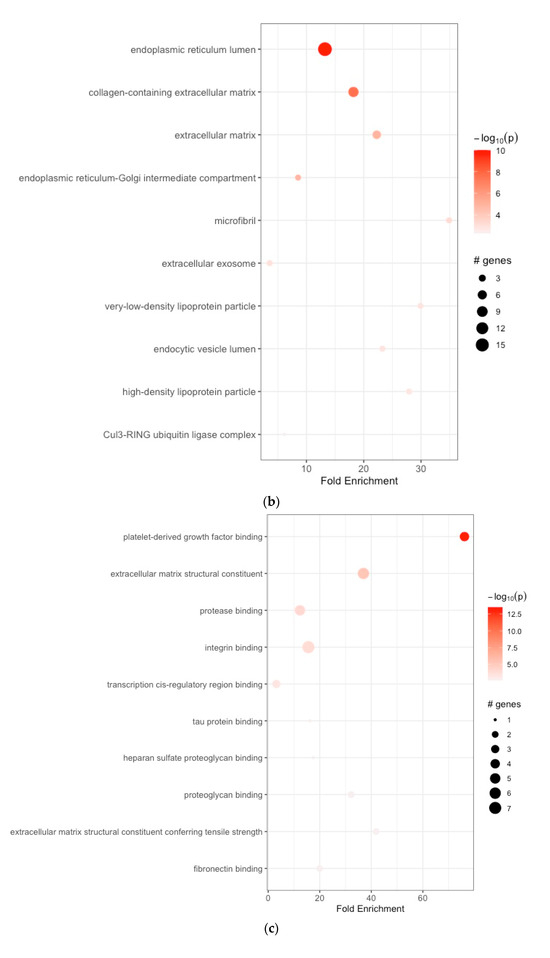
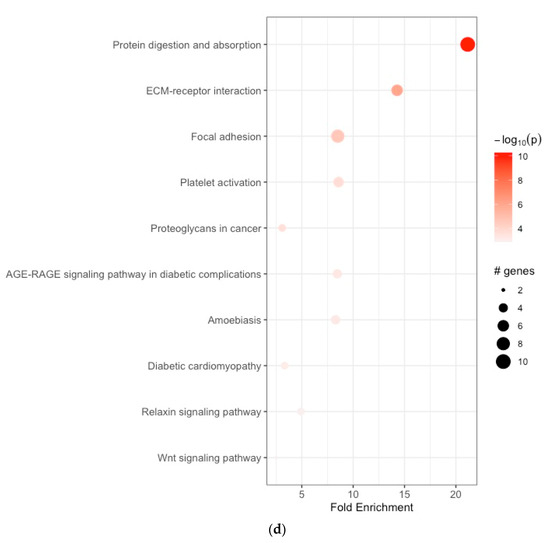
Figure 2.
(a) Enrichment analysis of gastric cancer-related DEGs. GO analysis: enriched biological processes; (b) functional enrichment analyses GO: cellular components; (c) functional enrichment analyses GO: molecular functions; (d) KEGG pathway enrichment analysis.

Table 1.
(a) Functional enrichment analyses GO: enriched biological processes. (b) Functional enrichment analyses GO: cellular components. (c) Functional enrichment analyses GO: molecular functions. (d) KEGG pathway enrichment analysis.
3.3. Construction of PPI Network and Identification of Hub DEGs
A PPI network was created using the DEGs associated with GC (Figure 3). Concurrently, the identification of highly connected DEGs was performed through the utilization of five computational methods within the Cytoscape software platform (Figure 4a–e). This analysis involved the application of EPC, Degree, MNC, MCC, and Bottleneck algorithms (Supplementary Table S2). Following this, nine hub genes (FN1, COL1A1, COL1A2, THBS2, COL3A1, COL5A1, APOE, SPP1, and BGN) were identified from the overlap among the top hub DEGs obtained through the application of the five methods. The differences in expression levels of the previously listed genes between GC and normal tissues were visualized using a heatmap (Figure 5). Additionally, we obtained the variance comparison between GC and normal tissues (Figure 6a–i).
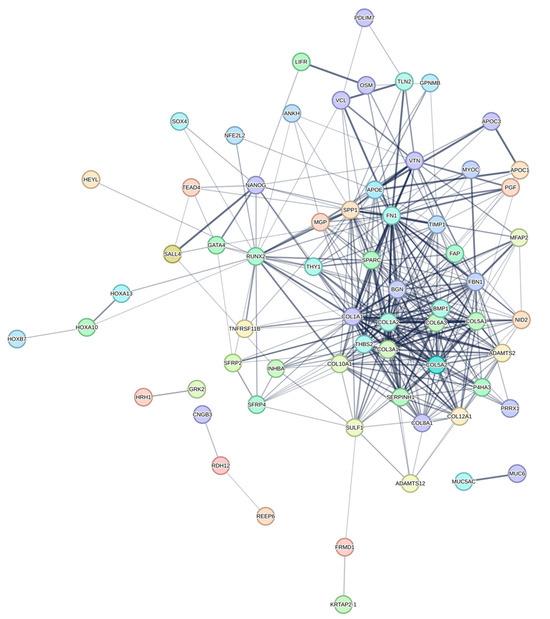
Figure 3.
The PPI network of differentially expressed GC-related genes.
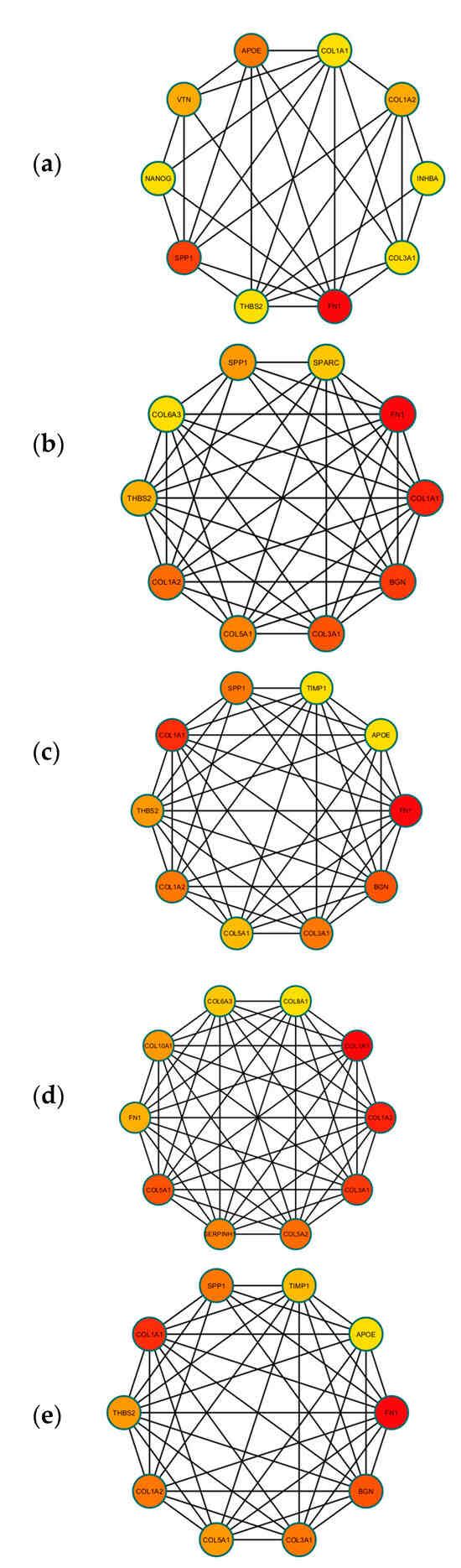
Figure 4.
(a) The top 10 hub DEGS with the highest connectivity extracted by Bottleneck (color depth for ranking of hub DEGs). (b) The top 10 hub DEGS with the highest connectivity extracted by EPC (color depth for ranking of hub DEGs). (c) The top 10 hub DEGS with the highest connectivity extracted by Degree (color depth for ranking of hub DEGs). (d) The top 10 hub DEGS with the highest connectivity extracted by MCC (color depth for ranking of hub DEGs). (e) The top 10 hub DEGS with the highest connectivity extracted by MNC (color depth for ranking of hub DEGs).
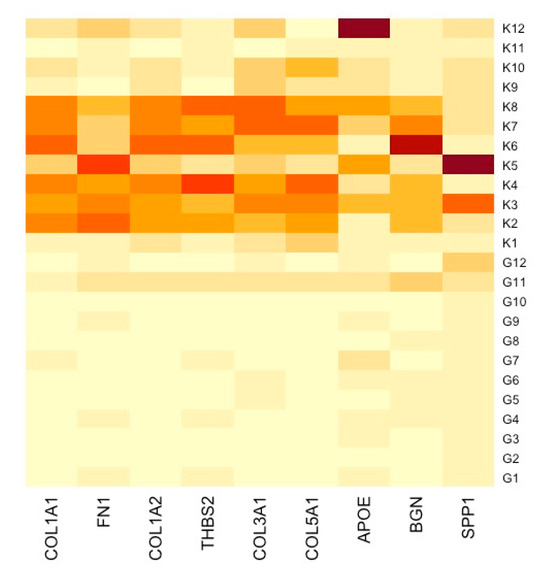
Figure 5.
Heatmap showcasing distinctly the gene expression alterations between GC samples and their respective normal tissue counterparts. K represents normal gastric tissue, and G represents GC tissue.
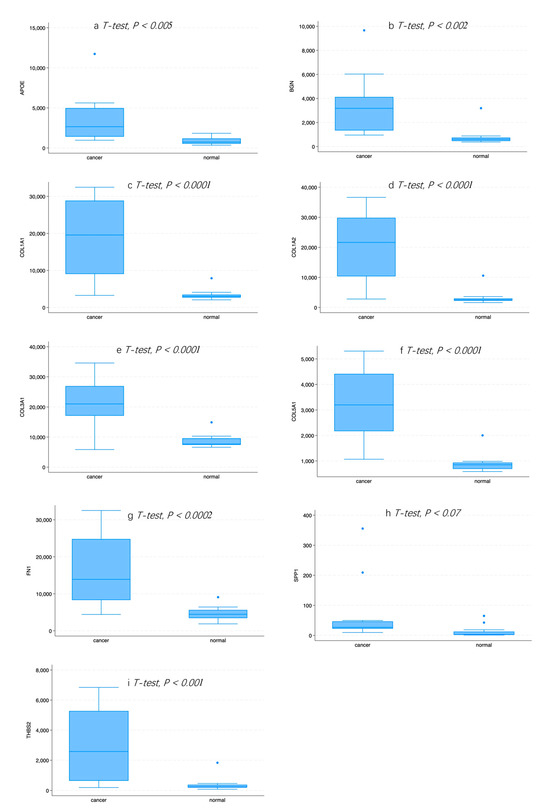
Figure 6.
(a) The expression pattern of APOE in normal and GC tissue. (b) The expression pattern of BGN in normal and GC tissue. (c) The expression pattern of COL1A1 in normal and GC tissue. (d) The expression pattern of COL1A2 in normal and GC tissue. (e) The expression pattern of COL3A1 in normal and GC tissue. (f) The expression pattern of COL5A1 in normal and GC tissue. (g) The expression pattern of FN1 in normal and GC tissue. (h) The expression pattern of SPP1 in normal and GC tissue. (i) The expression pattern of THBS2 in normal and GC tissue.
3.4. Survival Analysis of Hub DEGs in GC
Our results indicate that patients with an elevated expression of six hub DEGs, screened out by the five methods mentioned above, were associated with a poor survival rate. The corresponding Kaplan–Meier curves can be seen in Figure 7a–i. Patients with an increased expression of FN1, COL1A2, THBS2, COL3A1, COL5A1, and BGN showed a poorer prognosis.
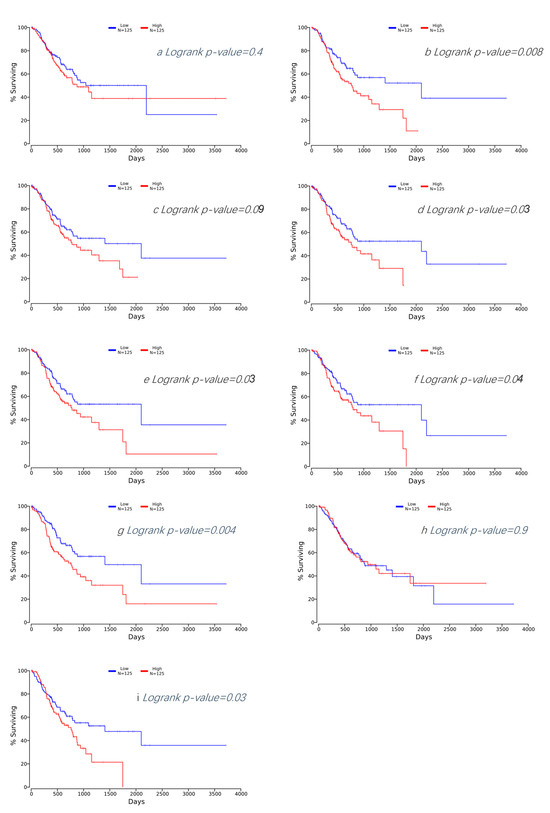
Figure 7.
(a) Kaplan–Meier plot for APOE in GC. (b) Kaplan–Meier plot for BGN in GC. (c) Kaplan–Meier plot for COL1A1 in GC. (d) Kaplan–Meier plot for COL1A2 in GC. (e) Kaplan–Meier plot for COL3A1 in GC. (f) Kaplan–Meier plot for COL5A1 in GC. (g) Kaplan–Meier plot for FN1 in GC. (h) Kaplan–Meier plot for SPP1 in GC. (i) Kaplan–Meier plot for THBS2 in GC.
3.5. TME Evaluation in GC
We investigated the correlation between the expression of the aforementioned selected genes, which were associated with survival, and the infiltration of cancer-associated fibroblasts, CD8+ activated dendritic cells, macrophages, and myeloid-derived suppressor cells (MDSCs). BGN demonstrated a direct correlation with cancer-associated fibroblasts, as well as M1 and M2 macrophages. Conversely, it exhibited an inverse relationship with MDSCs and activated dendritic cells (Figure 8a). FN1 exhibited a positive connection with cancer-associated fibroblasts and M2 macrophages while demonstrating a negative correlation with activated dendritic cells (Figure 8b). THBS2 showcased a positive correlation with M1 and M2 macrophages and cancer-associated fibroblasts. In contrast, it showcased a negative correlation with activated dendritic cells (Figure 8c). COL1A2 displayed a positive association with M1 and M2 macrophages and cancer-associated fibroblasts. Conversely, it exhibited a negative association with activated dendritic cells (Figure 8d). COL3A1 demonstrated a positive correlation with M2 macrophages and cancer-associated fibroblasts. It also showcased a negative correlation with activated dendritic cells and MDSCs (Figure 8e). COL5A1 depicted a positive association with M2 macrophages and cancer-associated fibroblasts. Simultaneously, it portrayed a negative association with activated dendritic cells (Figure 8f). The Pearson correlation curves can be seen in the Supplementary Figures.
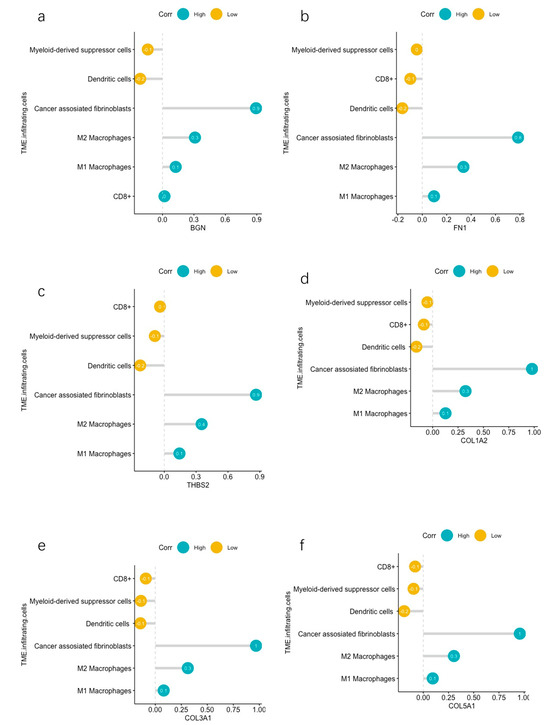
Figure 8.
(a) Correlations between BGN and infiltrating immune cells and CAFs. (b) Correlations between FN1 and infiltrating immune cells and CAFs. (c) Correlations between THBS2 and infiltrating immune cells and CAFs. (d) Correlations between COL1A2 and infiltrating immune cells and CAFs. (e) Correlations between COL3A1 and infiltrating immune cells and CAFs. (f) Correlations between COL5A1 and infiltrating immune cells and CAFs.
4. Discussion
The aim of this study was to identify the key genes involved in the pathogenesis and treatment of gastric adenocarcinoma, particularly linking these genes with components of the TME. This endeavor aimed to ascertain whether these genes and their corresponding proteins could serve as therapeutic targets through immunotherapy. Our study involved analyzing a publicly available GEO dataset to identify DEGs between human GC tissues and non-tumor tissues. A pool of 153 DEGs was identified as potential candidate biomarkers.
The analysis of the enrichment function unveiled the mechanisms driving the DEGs. Gene Ontology profiling highlighted substantial associations of DEGs with CCs, including the endoplasmic reticulum lumen, endoplasmic reticulum–Golgi intermediate compartment, and processes within the extracellular matrix, such as collagen fibril organization and the structural constitution of the collagen-containing extracellular matrix, implying that these genes may play a role in the progression of GC. Furthermore, DEGs were found to be involved in specific biological processes, such as endodermal cell differentiation, neuron projection development, and skeletal system development, as well as functions like platelet-derived growth factor binding, protease binding, integrin binding, and skin development. Additionally, our Kyoto Encyclopedia of Genes and Genomes analysis unveiled their involvement in pathways such as protein digestion and absorption, ECM–receptor interaction, focal adhesion, platelet activation, proteoglycans in cancer, the AGE-RAGE signaling pathway in diabetic complications, amoebiasis, diabetic cardiomyopathy, the relaxin signaling pathway, and the Wnt signaling pathway. Specifically, COL1A1, COL1A2, COL6A3, THBS2, FN1, and SPP1 were found to be associated with both focal adhesion and ECM–receptor interaction pathways. Cell–matrix adhesions are crucial for fundamental biological processes. At these focal adhesions, actin filaments connect to integrin receptors through a complex network of junctional plaque proteins [23,24]. Interactions between cells and the extracellular matrix involve transmembrane molecules like integrins and possibly proteoglycans, CD36, or other cell surface-related components. These interactions directly or indirectly govern cellular activities, such as adhesion, migration, differentiation, proliferation, and apoptosis [23,24]. Signaling pathways involving proteins like FAK initiate downstream effects that trigger the reorganization of the actin cytoskeleton. Notably, the active form of FAK, pFAK, has been linked to a poorer prognosis in patients diagnosed with GC [25]. These findings highlight the emerging significance of the ECM in the pathogenesis and progress of GC.
Another point to emphasize is the significant enrichment of the platelet activation pathway in our KEGG analysis. The genes that were up-regulated were TLN2, COL1A1, COL1A2, COL3A1, and PLA2G4C. Platelet activation and cancer share extensive and intricate interactions [26,27]. These interactions are not unidirectional: the TME can activate platelets, leading to an increased risk of thrombosis and a worse prognosis [28], while conversely, platelets themselves are associated with promoting tumor progression. Specifically, when activated, they release transforming growth factor beta (TGF-β), vascular endothelial growth factor (VEGF), and platelet-derived growth factor (PDGF) [29], which can facilitate angiogenesis and tumoral neovascularization. COL1A1, COL1A2, and COL3A1 belong to the group known as collagen fibers and have the potential to trigger the activation of GPV1. Consequently, this leads to platelet activation and their degranulation. PLA2G4C encodes the enzyme phospholipase A2γ, which hydrolyzes glycerophospholipids to produce free fatty acids and lysophospholipids [30]. Delving into the intricacies of platelet activation within the context of GC progression could potentially yield crucial insights into enhancing patient outcomes and augmenting the effectiveness of immunotherapeutic interventions.
Additionally, we discovered nine major hub genes using the STRING database to establish the PPI network and by employing five computational methods within the Cytoscape software platform. These genes include FN1, COL1A1, COL1A2, THBS2, COL3A1, COL5A1, APOE, SPP1, and BGN. Further survival analysis of these genes indicates that six (FN1, COL3A1, COL5A1, BGN, THBS2, and COL1A2) out of these nine up-regulated genes were significantly associated with an unfavorable prognosis for patients with GC.
The TME is a sophisticated milieu consisting of fibroblasts, endothelial cells, and diverse immune cells [31]. The cells within the TME actively support and stimulate other cell types and extracellular structures. This process fosters an environment that enables tumor cells to evade host immune surveillance and develop increased resistance to cancer therapy. Certain components of the TME deserve special attention, and our study focused on exploring specific aspects. There is mounting evidence showcasing the functional diversity of cancer-associated fibroblasts (CAFs) in studies on gastric adenocarcinoma. Typically, the definition of CAFs serves as an overarching term referring to a heterogeneous group of activated stromal cells. These cells exhibit functions that are distinct and set them apart from those of normal fibroblasts (NFs) [32]. Generally, CAFs play a pivotal role in the tumorigenesis process and cancer progression by releasing various ECM proteins and regulatory molecules [33]. Several studies have demonstrated the capability of CAFs to promote cancer invasion and migration through close interactions with tumor cells [33,34,35]. Additionally, CAFs regulate angiogenesis, immune suppression, and foster chemoresistance in cancer cells [33,36]. It has been demonstrated that CAF infiltration is linked to an immunosuppressive microenvironment and poorer survival outcomes in GC [37]. Cytotoxic CD8+ T cells are pivotal effectors in the anti-cancer immune response. They constitute a critical element of cancer immunotherapy owing to their pleiotropic effects. Solid tumors can be categorized into ‘cold’ and ‘hot’ tumors [38]. This categorization relies, in part, on the quantity of infiltrating T cells, where a ‘hot’ tumor refers to a higher number of CD8+ T cells, whereas a ‘cold’ tumor represents the opposite [39]. They have the ability to migrate into the TME, and once differentiated into cytotoxic cells, they can exert cytotoxic effects against cancer cells [40,41]. Additionally, numerous studies have associated increased CD8+ T cell infiltration in the cancer microenvironment with a better response to immunotherapy [42]. Macrophages are among some of the most prevalent cell types found in the cancer microenvironment [43]. These cells, known as tumor-associated macrophages (TAMs) [44], are broadly classified into two main subtypes: M1-like and M2-like macrophages [45]. This categorization is based on distinct genetic and functional characteristics that align with those observed in normal macrophages. M1 macrophages respond to cytokines and bacterial lipopolysaccharides, which trigger their activation. Consequently, they release various molecules, such as nitric oxide synthase, reactive oxygen species, and the cytokine IL-12, inducing damage to target cells [46,47]. Essentially, these cells exert an anti-cancer effect by eliminating tumor cells [48]. Conversely, the differentiation of macrophages into the M2 phenotype is influenced by factors including CSF-1, IL-4, IL-13, and IL-10 [46,47]. These cells contribute to anti-inflammatory responses and support tumor growth by suppressing the immune system [48]. Under normal conditions, immature myeloid cells migrate from the bone marrow to peripheral organs, where they undergo rapid maturation into macrophages, dendritic cells, or granulocytes [49]. However, within the TME, various factors disrupt the usual maturation process of these immature myeloid cells, leading to the development of an immunosuppressive phenotype [50]. These myeloid-derived suppressor cells (MDSCs) demonstrate a pivotal role in hindering adaptive antitumor immunity by impeding T-cell activation and function while also facilitating the recruitment and stimulation of T regulatory cells [51,52]. This orchestration by MDSCs contributes significantly to immune evasion mechanisms within the tumor environment [51,52]. An increased infiltration of MDSCs in GC has been associated with poorer prognosis [53]. Dendritic cells are among the most potent antigen-presenting cells in the immune system [54]. In a cancer context, dendritic cells participate in a process called cross-priming, wherein they activate CD8+ T cells by presenting cancer antigens [55,56]. Subsequently, these CD8+ T cells undergo training and activation against the presented antigens, thereby initiating a cytotoxic immune response against tumor components [55,56]. This response constitutes a cornerstone of immunity against cancer antigens. Within the TME, various factors, such as IL-6, IL-10, Vascular Endothelial Growth Factor, and Transforming Growth Factor Beta, prevail [57,58]. These factors can negatively regulate the functions of dendritic cells, potentially leading to T-cell tolerance and immune escape rather than immunity [59,60,61].
FN1 is a protein-coding gene responsible for encoding Fibronectin 1 [62]. Fibronectin, a high molecular weight glycoprotein [62], is found in the ECM. It plays crucial roles in various cellular functions, such as cell adhesion, growth, migration, and differentiation, contributing significantly to maintaining cell morphology [63]. Moreover, FN1 displays chemotactic properties, attracting monocytes, and is involved in blood coagulation and wound healing processes [64]. Other bioinformatic in silico analyses have suggested a correlation between the overexpression of FN1 and a poorer prognosis of patients with GC [65,66]. Our investigation substantiated these prior findings and demonstrated a link between FN1 overexpression and heightened infiltration of the TME and M2 macrophages. Concurrently, this gene was found to be associated with the reduced activity of dendritic cells.
COL1A2 is responsible for encoding a chain integral to type I collagen, the predominant fibrillar collagen present in the majority of connective tissues [67]. COL3A1 and COL5A1 encode the alpha-1 chains for type III and V collagen, respectively [67]. COL1A2, COL3A1, and COL5A1 are highly prevalent structural proteins within the ECM. An earlier bioinformatic analysis has also revealed the correlation between COL1A2, COL3A1, COL5A1, FN1, and CAF infiltration in GC [66]. Furthermore, our study demonstrates that the expression of COL1A2, COL3A1, and COL5A1 was positively correlated with M2 macrophages and negatively correlated with activated dendritic cells and CD8+ T-cell infiltration. Hence, we suggest that these genes may play an important role in maintaining and creating an immunosuppressive TME and subsequently promoting tumor progression.
THBS2 encodes the protein Thrombospondin-2, a member of the matricellular calcium-binding glycoprotein family that interacts with growth factors, cell receptors, and the ECM [68]. Its functions include regulating cell proliferation, adhesion, and apoptosis. This gene has been extensively studied in relation to the prognosis and potential influence on the TME of gastric adenocarcinoma [69,70]. Zhang et al. have also constructed a nomogram centered around the expression of THBS2 [70]. Our research validates that the increased expression of these genes correlates with a poorer survival rate linked to elevated levels of CAFs and M2 macrophage infiltration. A negative correlation with activated dendritic cell infiltration was also observed.
BGN is part of the small leucine-rich proteoglycans family and is responsible for encoding Biglycan, a protein that can be modified to form a glycoprotein [71]. BGN plays a role in cell proliferation, the migration of malignant cells, and reducing cell adhesion by interacting with proteins in both the intracellular and extracellular matrix [72,73]. Two in silico analyses have already demonstrated that high BGN expression is significantly associated with poor overall survival in GC patients [74,75]. Additionally, Chen et al. illustrated that BGN exhibited positive correlations with CD8+ T cells, macrophages, and dendritic cell infiltration in GC samples [74]. Zhang et al. also found, among others, a positive correlation between BGN expression and the infiltration of macrophages [75]. Our study partially confirms the former findings. In contrast to Chen et al.’s results [74], we observed a negative correlation with dendritic cells, potentially attributed to our focus on activated dendritic cells. Furthermore, akin to other genes, BGN displays a distinct positive correlation with CAFs. The aforementioned findings support the role of BGN in the occurrence and progression of GC, particularly through its influence on the TME.
5. Conclusions and Limitations
In this study, we identified immune-related genes and investigated the relation of the overexpression of these genes with cell infiltration in GC through bioinformatics analysis. The pathways involving focal adhesion, ECM–receptor interaction, and platelet activation appear to play a significant role in the pathogenesis and progression of GC, potentially serving as targets for pharmaceutical therapies. FN1, COL3A1, COL5A1, BGN, THBS2, and COL1A2 offer promising biomarkers for the prognosis assessment of GC patients. Additionally, we observed a strong correlation between these genes and the composition of the TME. However, our study has several limitations that require careful consideration. Primarily, there is an urgent need for molecular experiments to validate our findings. Furthermore, our comparison was focused on paired GC tissues and their adjacent counterparts without accounting for crucial details such as histological type, GC grade, and the spatial relationship between adjacent and cancerous tissues. These factors could potentially influence the expression patterns of DEGs. Moreover, our identification of DEGs was confined to a single dataset, aiming to retain essential genes but potentially increasing the risk of false positives. Therefore, a larger sample size is essential to authenticate and substantiate the obtained results.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers16071280/s1, Figure S1: Pearson correlation between the infiltration level of the tumor-microenvironment component and the expression level of the genes; Table S1: Down-regulated genes and up-regulated genes in GSE19826; Table S2: Intersection of the top ten Hub DEGs.
Author Contributions
Conceptualization, J.R.-A. and G.K.; methodology, J.R.-A. and G.K.; software, G.K.; validation, K.W., H.H.S., J.R.-A., S.K.-V., G.Z. and G.K.; formal analysis, V.S.T. and G.K.; investigation, M.P., V.S.T., E.K., C.P., G.T. and G.K.; data curation M.P., E.K. and G.K.; writing—original draft preparation, S.K.-V., G.Z., J.R.-A., G.T., E.K. and G.K.; writing—review and editing, C.P., J.R.-A., G.T., S.K.-V., G.Z., H.H.S. and G.K.; visualization, G.K. and J.R.-A.; supervision, J.R.-A.; project administration, J.R.-A. and G.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets used in this study are all available in online public databases.
Acknowledgments
We acknowledge the support of the Open Access Publication Fund of the University of Duisburg-Essen.
Conflicts of Interest
J.R.-A.: Speaker fees from AbbVie and travel support from Gilead. All unrelated to the submitted work.
Abbreviations
| BPs | Biological Processes |
| CAFs | Cancer-associated Fibroblasts |
| CCs | Cellular Components |
| DEGs | Differentially Expressed Genes |
| ECM | Extracellular Matrix |
| ECX | Epirubicin, Cisplatin, and Capecitabine |
| EMR | Endoscopic Mucosal Resection |
| ESD | Endoscopic Submucosal Dissection |
| FDA | Food and Drug Administration |
| FLOT | Fluorouracil, Oxaliplatin, Docetaxel, and Leucovorin |
| GC | Gastric Cancer/Gastric Carcinoma |
| GEO | Gene Expression Omnibus |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MDSCs | Myeloid-derived Suppressor Cells |
| MFs | Molecular Functions |
| NFs | Normal Fibroblasts |
| PD-L1 | Programmed Death-ligand 1 |
| PPI | Protein–Protein Interaction |
| TAMs | Tumor-associated Macrophages |
| TIMER | Tumor Immune Evaluation Resource |
| TME | Tumor Microenvironment |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Carcas, L.P. Gastric cancer review. J. Carcinog. 2014, 13, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Joshi, S.S.; Badgwell, B.D. Current treatment and recent progress in gastric cancer. CA Cancer J. Clin. 2021, 71, 264–279. [Google Scholar] [CrossRef] [PubMed]
- Rha, S.Y.; Oh, D.-Y.; Yañez, P.; Bai, Y.; Ryu, M.-H.; Lee, J.; Rivera, F.; Alves, G.V.; Garrido, M.; Shiu, K.-K. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for HER2-negative advanced gastric cancer (KEYNOTE-859): A multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 2023, 24, 1181–1195. [Google Scholar] [CrossRef] [PubMed]
- Janjigian, Y.Y.; Shitara, K.; Moehler, M.; Garrido, M.; Salman, P.; Shen, L.; Wyrwicz, L.; Yamaguchi, K.; Skoczylas, T.; Bragagnoli, A.C. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet 2021, 398, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hu, X.; Lin, R.; Zhou, G.; Zhao, L.; Zhao, D.; Zhang, Y.; Li, W.; Zhang, Y.; Ma, P. Single-cell landscape reveals active cell subtypes and their interaction in the tumor microenvironment of gastric cancer. Theranostics 2022, 12, 3818. [Google Scholar] [CrossRef] [PubMed]
- Oliver, G.R.; Hart, S.N.; Klee, E.W. Bioinformatics for clinical next generation sequencing. Clin. Chem. 2015, 61, 124–135. [Google Scholar] [CrossRef]
- Zeng, D.; Li, M.; Zhou, R.; Zhang, J.; Sun, H.; Shi, M.; Bin, J.; Liao, Y.; Rao, J.; Liao, W. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 2019, 7, 737–750. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Zhou, R.; Yu, Y.; Luo, Y.; Zhang, J.; Sun, H.; Bin, J.; Liao, Y.; Rao, J.; Zhang, Y. Gene expression profiles for a prognostic immunoscore in gastric cancer. J. Br. Surg. 2018, 105, 1338–1348. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Lu, J.; Lou, J.; Shi, C.; Mo, S.; Shao, Y.; Ni, J.; Zhang, W.; Cheng, X. Gastric cancer tumor microenvironment characterization reveals stromal-related gene signatures associated with macrophage infiltration. Front. Genet. 2020, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wen, Y.-G.; Li, D.-P.; Xia, J.; Zhou, C.-Z.; Yan, D.-W.; Tang, H.-M.; Peng, Z.-H. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med. Oncol. 2012, 29, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef] [PubMed]
- Ulgen, E.; Ozisik, O.; Sezerman, O.U. pathfindR: An R package for comprehensive identification of enriched pathways in omics data through active subnetworks. Front. Genet. 2019, 10, 858. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network analysis and visualization of proteomics data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef]
- Anaya, J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput. Sci. 2016, 2, e67. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2. 0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Giancotti, F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004, 5, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Petit, V.; Thiery, J.-P. Focal adhesions: Structure and dynamics. Biol. Cell 2000, 92, 477–494. [Google Scholar] [CrossRef] [PubMed]
- Peng, K.; Li, S.; Li, Q.; Zhang, C.; Yuan, Y.; Liu, M.; Zhang, L.; Wang, Y.; Yu, S.; Zhang, H. Positive Phospho-Focal Adhesion Kinase in Gastric Cancer Associates With Poor Prognosis After Curative Resection. Front. Oncol. 2022, 12, 953938. [Google Scholar] [CrossRef] [PubMed]
- Gay, L.J.; Felding-Habermann, B. Contribution of platelets to tumour metastasis. Nat. Rev. Cancer 2011, 11, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Acedo, A.L.; Mège, D.; Crescence, L.; Dignat-George, F.; Dubois, C.; Panicot-Dubois, L. Platelets, thrombo-inflammation, and cancer: Collaborating with the enemy. Front. Immunol. 2019, 10, 1805. [Google Scholar] [CrossRef] [PubMed]
- Ferroni, P.; Martini, F.; Portarena, I.; Grenga, I.; Riondino, S.; La Farina, F.; Laudisi, A.; Roselli, M.; Guadagni, F. An activated protein C-dependent thrombin generation assay predicts chemotherapy-associated venous thromboembolism in cancer patients. Thromb. Haemost. 2011, 105, 931–932. [Google Scholar] [PubMed]
- Meikle, C.K.; Kelly, C.A.; Garg, P.; Wuescher, L.M.; Ali, R.A.; Worth, R.G. Cancer and thrombosis: The platelet perspective. Front. Cell Dev. Biol. 2017, 4, 147. [Google Scholar] [CrossRef]
- Asai, K.; Hirabayashi, T.; Houjou, T.; Uozumi, N.; Taguchi, R.; Shimizu, T. Human group IVC phospholipase A2 (cPLA2γ): Roles in the membrane remodeling and activation induced by oxidative stress. J. Biol. Chem. 2003, 278, 8809–8814. [Google Scholar] [CrossRef]
- Albini, A.; Magnani, E.; Noonan, D. The tumor microenvironment: Biology of a complex cellular and tissue society. Q. J. Nucl. Med. Mol. Imaging 2010, 54, 244–248. [Google Scholar] [PubMed]
- Sun, H.; Wang, X.; Wang, X.; Xu, M.; Sheng, W. The role of cancer-associated fibroblasts in tumorigenesis of gastric cancer. Cell Death Dis. 2022, 13, 874. [Google Scholar] [CrossRef] [PubMed]
- Orimo, A.; Gupta, P.B.; Sgroi, D.C.; Arenzana-Seisdedos, F.; Delaunay, T.; Naeem, R.; Carey, V.J.; Richardson, A.L.; Weinberg, R.A. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 2005, 121, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Shan, T.; Qiao, L. Collagen type IV alpha 1 (COL4A1) silence hampers the invasion, migration and epithelial–mesenchymal transition (EMT) of gastric cancer cells through blocking Hedgehog signaling pathway. Bioengineered 2022, 13, 8972–8981. [Google Scholar] [CrossRef] [PubMed]
- Izumi, D.; Ishimoto, T.; Miyake, K.; Sugihara, H.; Eto, K.; Sawayama, H.; Yasuda, T.; Kiyozumi, Y.; Kaida, T.; Kurashige, J. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin β1 clustering and invasiveness in gastric cancer. Int. J. Cancer 2016, 138, 1207–1219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Deng, T.; Liu, R.; Ning, T.; Yang, H.; Liu, D.; Zhang, Q.; Lin, D.; Ge, S.; Bai, M. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer 2020, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yao, L.; Qu, J.; Liu, L.; Lu, N.; Wang, J.; Zhang, J. Cancer-associated fibroblast infiltration in gastric cancer: The discrepancy in subtypes pathways and immunosuppression. J. Transl. Med. 2021, 19, 325. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, J.A.; Sweis, R.F.; Bao, R.; Luke, J.J. T cell–inflamed versus non-T cell–inflamed tumors: A conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol. Res. 2018, 6, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Bruni, D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef]
- Nolz, J.C. Molecular mechanisms of CD8+ T cell trafficking and localization. Cell. Mol. Life Sci. 2015, 72, 2461–2473. [Google Scholar] [CrossRef]
- Slaney, C.Y.; Kershaw, M.H.; Darcy, P.K. Trafficking of T cells into tumors. Cancer Res. 2014, 74, 7168–7174. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, C.; Cai, X.; Xie, Z.; Zhou, L.; Cheng, B.; Zhong, R.; Xiong, S.; Li, J.; Chen, Z. The association between CD8+ tumor-infiltrating lymphocytes and the clinical outcome of cancer immunotherapy: A systematic review and meta-analysis. EClinicalMedicine 2021, 41, 101134. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Zhang, D.; Sun, K.; Zhu, Y.; Xu, J.; Kang, Y.; Zhang, G.; Cai, Y.; Mao, S.; Long, R.; et al. Propofol maintains Th17/Treg cell balance in elderly patients undergoing lung cancer surgery through GABAA receptor. BMC Immunol. 2022, 23, 58. [Google Scholar] [CrossRef] [PubMed]
- Ginhoux, F.; Jung, S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014, 14, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Chanmee, T.; Ontong, P.; Konno, K.; Itano, N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014, 6, 1670–1690. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.Y. M1 and M2 polarization of macrophages: A mini-review. Med. Biol. Sci. Eng. 2019, 2, 1–5. [Google Scholar] [CrossRef]
- Wang, N.; Liang, H.; Zen, K. Molecular mechanisms that influence the macrophage M1–M2 polarization balance. Front. Immunol. 2014, 5, 614. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liang, Y.; Wang, L. Shaping polarization of tumor-associated macrophages in cancer immunotherapy. Front. Immunol. 2022, 13, 888713. [Google Scholar] [CrossRef] [PubMed]
- Schulz, C.; Perdiguero, E.G.; Chorro, L.; Szabo-Rogers, H.; Cagnard, N.; Kierdorf, K.; Prinz, M.; Wu, B.; Jacobsen, S.E.W.; Pollard, J.W. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012, 336, 86–90. [Google Scholar] [CrossRef]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef]
- Solito, S.; Bronte, V.; Mandruzzato, S. Antigen specificity of immune suppression by myeloid-derived suppressor cells. J. Leukoc. Biol. 2011, 90, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Guo, J.; Weng, L.; Tang, W.; Jin, S.; Ma, W. Myeloid-derived suppressor cells—New and exciting players in lung cancer. J. Hematol. Oncol. 2020, 13, 10. [Google Scholar] [CrossRef] [PubMed]
- Gabitass, R.F.; Annels, N.E.; Stocken, D.D.; Pandha, H.A.; Middleton, G.W. Elevated myeloid-derived suppressor cells in pancreatic, esophageal and gastric cancer are an independent prognostic factor and are associated with significant elevation of the Th2 cytokine interleukin-13. Cancer Immunol. Immunother. 2011, 60, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Banchereau, J.; Steinman, R.M. Dendritic cells and the control of immunity. Nature 1998, 392, 245–252. [Google Scholar] [CrossRef]
- Kurts, C.; Robinson, B.W.; Knolle, P.A. Cross-priming in health and disease. Nat. Rev. Immunol. 2010, 10, 403–414. [Google Scholar] [CrossRef] [PubMed]
- Melief, C.J. Cancer immunotherapy by dendritic cells. Immunity 2008, 29, 372–383. [Google Scholar] [CrossRef] [PubMed]
- Gottfried, E.; Kreutz, M.; Mackensen, A. Tumor-induced modulation of dendritic cell function. Cytokine Growth Factor Rev. 2008, 19, 65–77. [Google Scholar] [CrossRef] [PubMed]
- Zong, J.; Keskinov, A.A.; Shurin, G.V.; Shurin, M.R. Tumor-derived factors modulating dendritic cell function. Cancer Immunol. Immunother. 2016, 65, 821–833. [Google Scholar] [CrossRef] [PubMed]
- Bandola-Simon, J.; Roche, P.A. Dysfunction of antigen processing and presentation by dendritic cells in cancer. Mol. Immunol. 2019, 113, 31–37. [Google Scholar] [CrossRef]
- Chrisikos, T.T.; Zhou, Y.; Slone, N.; Babcock, R.; Watowich, S.S.; Li, H.S. Molecular regulation of dendritic cell development and function in homeostasis, inflammation, and cancer. Mol. Immunol. 2019, 110, 24–39. [Google Scholar] [CrossRef]
- Gupta, Y.H.; Khanom, A.; Acton, S.E. Control of dendritic cell function within the tumour microenvironment. Front. Immunol. 2022, 13, 733800. [Google Scholar] [CrossRef] [PubMed]
- Mitrović, S.; Mitrović, D.; Todorović, V. Fibronectin—A multifunctional glycoprotein. Srp. Arh. Za Celok. Lek. 1995, 123, 198–201. [Google Scholar]
- Pankov, R.; Yamada, K.M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. [Google Scholar] [CrossRef] [PubMed]
- Grinnell, F.; Billingham, R.E.; Burgess, L. Distribution of fibronectin during wound healing in vivo. J. Investig. Dermatol. 1981, 76, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, L.; Chen, Z.; Song, Y.; Gu, T.; Liu, X.; Zhao, H.; Yao, N. Identification of critical genes in gastric cancer to predict prognosis using bioinformatics analysis methods. Ann. Transl. Med. 2020, 8, 14. [Google Scholar] [CrossRef]
- Ucaryilmaz Metin, C.; Ozcan, G. Comprehensive bioinformatic analysis reveals a cancer-associated fibroblast gene signature as a poor prognostic factor and potential therapeutic target in gastric cancer. BMC Cancer 2022, 22, 692. [Google Scholar] [CrossRef] [PubMed]
- Gelse, K.; Pöschl, E.; Aigner, T. Collagens—Structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 2003, 55, 1531–1546. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.C.; Lawler, J. The thrombospondins. Int. J. Biochem. Cell Biol. 2004, 36, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Wang, W.; Yu, B.; Tan, S. Thrombospondin-2 acts as a bridge between tumor extracellular matrix and immune infiltration in pancreatic and stomach adenocarcinomas: An integrative pan-cancer analysis. Cancer Cell Int. 2022, 22, 213. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, H.; Xiang, X.; Liu, L.; Huang, H.; Tang, G. THBS2 is closely related to the poor prognosis and immune cell infiltration of gastric cancer. Front. Genet. 2022, 13, 803460. [Google Scholar] [CrossRef]
- Chen, D.; Qin, Y.; Dai, M.; Li, L.; Liu, H.; Zhou, Y.; Qiu, C.; Chen, Y.; Jiang, Y. BGN and COL11A1 regulatory network analysis in colorectal cancer (CRC) reveals that BGN influences CRC cell biological functions and interacts with miR-6828-5p. Cancer Manag. Res. 2020, 12, 13051–13069. [Google Scholar] [CrossRef] [PubMed]
- Cooper, J.; Giancotti, F.G. Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 2019, 35, 347–367. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Layseca, P.; Icha, J.; Hamidi, H.; Ivaska, J. Integrin trafficking in cells and tissues. Nat. Cell Biol. 2019, 21, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Yang, Z. Identification of differentially expressed genes reveals BGN predicting overall survival and tumor immune infiltration of gastric cancer. Comput. Math. Methods Med. 2021, 2021, 5494840. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, H.; Xiang, X.; Liu, L.; Huang, H.; Tang, G. BGN may be a potential prognostic biomarker and associated with immune cell enrichment of gastric cancer. Front. Genet. 2022, 13, 765569. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).