AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status over Time
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Deep Learning Predictive Modeling
2.1.1. Discrete Time-to-Event Data
2.1.2. Time-Varying Covariates: Proposed Extension
2.2. Experiments
2.2.1. Study Data: SEER-Medicare Linked Dataset
2.2.2. Cohort Selection
2.2.3. Data Cleaning, Standardization, Encoding, and Embedding
2.2.4. Performance Metrics
2.2.5. Model Hyperparameters
2.2.6. Models Validation
3. Results
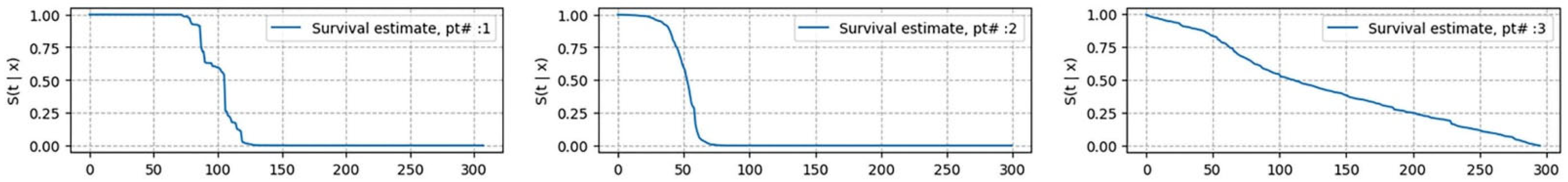
Model Interpretability
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Braun, S.; Vogl, F.D.; Naume, B.; Janni, W.; Osborne, M.P.; Coombes, R.C.; Schlimok, G.; Diel, I.J.; Gerber, B.; Gebauer, G.; et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med. 2005, 353, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Dent, R.; Trudeau, M.; Pritchard, K.I.; Hanna, W.M.; Kahn, H.K.; Sawka, C.A.; Lickley, L.A.; Rawlinson, E.; Sun, P.; Narod, S.A. Triple-negative breast cancer: Clinical features and patterns of recurrence. Clin. Cancer Res. 2007, 13, 4429–4434. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Swartz, M.D.; Zhao, H.; Kapadia, A.S.; Lai, D.; Rowan, P.J.; Buchholz, T.A.; Giordano, S.H. Hazard of recurrence among women after primary breast cancer treatment—A 10-year follow-up using data from SEER-Medicare. Cancer Epidemiol. Biomark. Prev. 2012, 21, 800–809. [Google Scholar] [CrossRef] [PubMed]
- Castellano, I.; Chiusa, L.; Vandone, A.M.; Beatrice, S.; Goia, M.; Donadio, M.; Arisio, R.; Muscarà, F.; Durando, A.; Viale, G.; et al. A simple and reproducible prognostic index in luminal ER-positive breast cancers. Ann. Oncol. 2013, 24, 2292–2297. [Google Scholar] [CrossRef]
- Silber, J.H.; Rosenbaum, P.R.; Clark, A.S.; Giantonio, B.J.; Ross, R.N.; Teng, Y.; Wang, M.; Niknam, B.A.; Ludwig, J.M.; Wang, W.; et al. Characteristics associated with differences in survival among black and white women with breast cancer. JAMA 2013, 310, 389–397. [Google Scholar] [CrossRef]
- Vera-Badillo, F.E.; Templeton, A.J.; de Gouveia, P.; Diaz-Padilla, I.; Bedard, P.L.; Al-Mubarak, M.; Amir, E. Androgen receptor expression and outcomes in early breast cancer: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2014, 106, djt319. [Google Scholar] [CrossRef]
- Daly, B.; Olopade, O.I. A perfect storm: How tumor biology, genomics, and health care delivery patterns collide to create a racial survival disparity in breast cancer and proposed interventions for change. CA Cancer J. Clin. 2015, 65, 221–238. [Google Scholar] [CrossRef]
- Shi, R.; Taylor, H.; McLarty, J.; Liu, L.; Mills, G.; Burton, G. Effects of payer status on breast cancer survival: A retrospective study. BMC Cancer 2015, 15, 211. [Google Scholar] [CrossRef]
- Colleoni, M.; Sun, Z.; Price, K.N.; Karlsson, P.; Forbes, J.F.; Thürlimann, B.; Gianni, L.; Castiglione, M.; Gelber, R.D.; Coates, A.S.; et al. Annual Hazard Rates of Recurrence for Breast Cancer During 24 Years of Follow-Up: Results From the International Breast Cancer Study Group Trials I to V. J. Clin. Oncol. 2016, 34, 927–935. [Google Scholar] [CrossRef]
- Giuliani, J.; Mercanti, A.; Bonetti, A. Late recurrence (more than 10 years) in early (tumors equal to or smaller than 2 cm) breast cancer patients. Clin. Transl. Oncol. 2016, 18, 859–862. [Google Scholar] [CrossRef]
- Miyoshi, Y.; Shien, T.; Ogiya, A.; Ishida, N.; Yamazaki, K.; Horii, R.; Horimoto, Y.; Masuda, N.; Yasojima, H.; Inao, T.; et al. Collaborative Study Group of Scientific Research of the Japanese Breast Cancer Society. Differences in expression of the cancer stem cell marker aldehyde dehydrogenase 1 among estrogen receptor-positive/human epidermal growth factor receptor type 2-negative breast cancer cases with early, late, and no recurrence. Breast Cancer Res. 2016, 18, 73. [Google Scholar] [PubMed]
- Janni, W.J.; Rack, B.; Terstappen, L.W.; Pierga, J.Y.; Taran, F.A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.C.; Friedl, T.W.; et al. Pooled analysis of the prognostic relevance of circulating tumor cells in primary breast cancer. Clin. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Geurts, Y.M.; Witteveen, A.; Bretveld, R.; Poortmans, P.M.; Sonke, G.S.; Strobbe, L.J.A.; Siesling, S. Patterns and predictors of first and subsequent recurrence in women with early breast cancer. Breast Cancer Res. Treat. 2017, 165, 709–720. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. EBCTCG. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef]
- Sestak, I.; Zhang, Y.; Schroeder, B.E.; Schnabel, C.A.; Dowsett, M.; Cuzick, J.; Sgroi, D. Cross-Stratification and Differential Risk by Breast Cancer Index and Recurrence Score in Women with Hormone Receptor-Positive Lymph Node-Negative Early-Stage Breast Cancer. Clin. Cancer Res. 2016, 22, 5043–5048. [Google Scholar] [CrossRef][Green Version]
- Wieder, R.; Shafiq, B.; Adam, N. African American race is an independent risk factor in survival form initially diagnosed localized breast cancer. J. Cancer 2016, 7, 1587–1598. [Google Scholar] [CrossRef][Green Version]
- Tjensvoll, K.; Nordgård, O.; Skjæveland, M.; Oltedal, S.; Janssen, E.A.M.; Gilje, B. Detection of disseminated tumor cells in bone marrow predict late recurrences in operable breast cancer patients. BMC Cancer 2019, 19, 1131. [Google Scholar] [CrossRef]
- Wieder, R.; Shafiq, B.; Adam, N. Greater Survival Improvement in African American vs. Caucasian Women with Hormone Negative Breast Cancer. J. Cancer 2020, 11, 2808–2820. [Google Scholar] [CrossRef]
- Dar, H.; Johansson, A.; Nordenskjöld, A.; Iftimi, A.; Yau, C.; Perez-Tenorio, G.; Benz, C.; Nordenskjöld, B.; Stål, O.; Esserman, L.J.; et al. Assessment of 25-Year Survival of Women With Estrogen Receptor-Positive/ERBB2-Negative Breast Cancer Treated With and Without Tamoxifen Therapy: A Secondary Analysis of Data From the Stockholm Tamoxifen Randomized Clinical Trial. JAMA Netw. Open. 2021, 4, e2114904. [Google Scholar] [CrossRef]
- Prakash, O.; Hossain, F.; Danos, D.; Lassak, A.; Scribner, R.; Miele, L. Racial disparities in triple negative breast cancer: A review of the role of biologic and non-biologic factors. Front. Public Health 2020, 8, 576964. [Google Scholar] [CrossRef]
- Hoskins, K.F.; Danciu, O.C.; Ko, N.Y.; Calip, G.S. Association of Race/Ethnicity and the 21-Gene Recurrence Score With Breast Cancer-Specific Mortality Among US Women. JAMA Oncol. 2021, 7, 370–378. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, K.F.; Calip, G.S.; Huang, H.C.; Ibraheem, A.; Danciu, O.C.; Rauscher, G.H. Association of social determinants and tumor biology with racial disparity in survival from early-stage, hormone-dependent breast cancer. JAMA Oncol. 2023, 9, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Gagliato Dde, M.; Gonzalez-Angulo, A.M.; Lei, X.; Theriault, R.L.; Giordano, S.H.; Valero, V.; Hortobagyi, G.N.; Chavez-Macgregor, M. Clinical impact of delaying initiation of adjuvant chemotherapy in patients with breast cancer. J. Clin. Oncol. 2014, 32, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Sheppard, V.B.; Oppong, B.A.; Hampton, R.; Snead, F.; Horton, S.; Hirpa, F.; Brathwaite, E.J.; Makambi, K.; Onyewu, S.; Boisvert, M.; et al. Disparities in breast cancer surgery delay: The lingering effect of race. Ann. Surg. Oncol. 2015, 22, 2902–2911. [Google Scholar] [CrossRef] [PubMed]
- Wieder, R. Awakening of Dormant Breast Cancer Cells in the Bone Marrow. Cancers 2023, 15, 3021. [Google Scholar] [CrossRef]
- Dillekås, H.; Demicheli, R.; Ardoino, I.; Jensen, S.A.H.; Biganzoli, E.; Straume, O. The recurrence pattern following delayed breast reconstruction after mastectomy for breast cancer suggests a systemic effect of surgery on occult dormant micrometastases. Breast Cancer Res. Treat. 2016, 158, 169–178. [Google Scholar] [CrossRef]
- Janni, W.; Vogl, F.D.; Wiedswang, G.; Synnestvedt, M.; Fehm, T.; Jückstock, J.; Borgen, E.; Rack, B.; Braun, S.; Sommer, H.; et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse—A European pooled analysis. Clin. Cancer Res. 2011, 17, 2967–2976. [Google Scholar] [CrossRef]
- Kwan, M.L.; Kushi, L.H.; Weltzien, E.; Tam, E.K.; Castillo, A.; Sweeney, C.; Caan, B.J. Alcohol consumption and breast cancer recurrence and survival among women with early-stage breast cancer: The life after cancer epidemiology study. J. Clin. Oncol. 2010, 28, 4410–4416. [Google Scholar] [CrossRef]
- Simapivapan, P.; Boltong, A.; Hodge, A. To what extent is alcohol consumption associated with breast cancer recurrence and second primary breast cancer? A systematic review. Cancer Treat. Rev. 2016, 50, 155–167. [Google Scholar] [CrossRef]
- Nechuta, S.; Chen, W.Y.; Cai, H.; Poole, E.M.; Kwan, M.L.; Flatt, S.W.; Patterson, R.E.; Pierce, J.P.; Caan, B.J.; Ou Shu, X. A pooled analysis of post-diagnosis lifestyle factors in association with late estrogen-receptor-positive breast cancer prognosis. Int. J. Cancer 2016, 138, 2088–2097. [Google Scholar] [CrossRef]
- Suresh, K.; Severn, C.; Ghosh, D. Survival prediction models: An introduction to discrete-time modeling. BMC Med. Res. Methodol. 2022, 22, 207. [Google Scholar] [CrossRef] [PubMed]
- Kattan, M.W. Comparison of cox regression with other methods for determining prediction models and nomograms. J. Urol. 2003, 170, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Kleinbaum, D.G.; Klein, M. Survival Analysis; Springer: New York, NY, USA, 2010; Volume 3. [Google Scholar]
- Cox, D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B (Methodol.) 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Meir, T.; Gutman, R.; Gorfine, M. PyDTS: A Python Package for Discrete Time Survival Analysis with Competing Risks. arXiv 2022, arXiv:2204.05731. [Google Scholar]
- Meir, T.; Gorfine, M. Discrete-time Competing-Risks Regression with or without Penalization. arXiv 2023, arXiv:2303.01186. [Google Scholar]
- Gensheimer, M.F.; Narasimhan, B. A scalable discrete-time survival model for neural networks. PeerJ 2019, 7, e6257. [Google Scholar] [CrossRef]
- Faraggi, D.; Simon, R. A neural network model for survival data. Stat. Med. 1995, 14, 73–82. [Google Scholar] [CrossRef]
- Katzman, J.L.; Shaham, U.; Cloninger, A.; Bates, J.; Jiang, T.; Kluger, Y. DeepSurv: Personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 2018, 18, 24. [Google Scholar] [CrossRef]
- Lee, C.; Zame, W.R.; Yoon, J.; van der Schaar, M. DeepHit: A deep learning approach to survival analysis with competing risks. Proc. AAAI Conf. Artif. Intell. 2018, 32, 2314–2321. [Google Scholar] [CrossRef]
- Kvamme, H.; Borgan, Ø.; Scheel, I. Time-to-event prediction with neural networks and Cox regression. J. Mach. Learn. Res. 2019, 20, 1–30. [Google Scholar]
- Enewold, L.; Parsons, H.; Zhao, L.; Bott, D.; Rivera, D.R.; Barrett, M.J.; Virnig, B.A.; Warren, J.L. Updated overview of the SEER-Medicare data: Enhanced content and applications. JNCI Monogr. 2020, 2020, 3–13. [Google Scholar]
- Perez, M.; Murphy, C.C.; Pruitt, S.L.; Rashdan, S.; Rahimi, A.; Gerber, D.E. Potential Impact of Revised NCI Eligibility Criteria Guidance: Prior Malignancy Exclusion in Breast Cancer Clinical Trials. J. Natl. Compr. Cancer Netw. 2022, 20, 792–799. [Google Scholar] [CrossRef] [PubMed]
- Goldvaser, H.; Ribnikar, D.; Majeed, H.; Ocana, A.; Amir, E. Absolute benefit from adjuvant chemotherapy in contemporary clinical trials: A systemic review and meta-analysis. Cancer Treat. Rev. 2018, 71, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Hayes, D.F. Disease related indicators for a proper choice of adjuvant treatments. Breast 2011, 20 (Suppl. S3), S162–S164. [Google Scholar] [CrossRef]
- Guo, C.; Berkhahn, F. Entity embeddings of categorical variables. arXiv 2016, arXiv:1604.06737. [Google Scholar]
- Ishwaran, H.; Kogalur, U.B.; Blackstone, E.H.; Lauer, M.S. Random survival forests. Ann. Appl. Stat. 2008, 2, 841–860. [Google Scholar] [CrossRef]
- Brier, G.W. Verification of forecasts expressed in terms of probability. Mon. Weather Rev. 1950, 78, 1–3. [Google Scholar] [CrossRef]
- Graf, E.; Schmoor, C.; Sauerbrei, W.; Schumacher, M. Assessment and comparison of prognostic classification schemes for survival data. Stat. Med. 1999, 18, 2529–2545. [Google Scholar] [CrossRef]
- Gerds, T.A.; Schumacher, M. Consistent estimation of the expected goldvbrier score in general survival models with right-censored event times. Biom. J. 2006, 48, 1029–1040. [Google Scholar] [CrossRef]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical bayesian optimization of machine learning algorithms. Adv. Neural Inf. Process. Syst. 2012, 25, 1–9. [Google Scholar]
- Mucaki, E.J.; Baranova, K.; Pham, H.Q.; Rezaeian, I.; Angelov, D.; Ngom, A.; Rueda, L.; Rogan, P.K. Predicting Outcomes of Hormone and Chemotherapy in the: Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) Study by Biochemically-inspired Machine Learning. F1000Research 2016, 5, 2124. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Connors, A.F.; Dawson, N.V.; Desbiens, N.A.; Fulkerson, W.J.; Goldman, L.; Knaus, W.A.; Lynn, J.; Oye, R.K.; Bergner, M.; Damiano, A.; et al. A controlled trial to improve care for seriously iII hospitalized patients: The study to understand prognoses and preferences for outcomes and risks of treatments (SUPPORT). JAMA 1995, 274, 1591–1598. [Google Scholar] [CrossRef]
- Kvamme, H.; Borgan, Ø. Continuous and discrete-time survival prediction with neural networks. Lifetime Data Anal. 2021, 27, 710–736. [Google Scholar] [CrossRef] [PubMed]
- Social Security Period Life Table, 2021, As Used in the 2024 Trustees Report. Available online: https://www.ssa.gov/oact/STATS/table4c6.html (accessed on 8 April 2024).
- Ribeiro, M.T.; Singh, S.; Guestrin, C. “Why should I trust you?”: Explaining the predictions of any classifier. In Proceedings of the KDD ‘16: The 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.I. Consistent feature attribution for tree ensembles. arXiv 2017, arXiv:1706.06060. [Google Scholar]
- Miglani, V.; Yang, A.; Markosyan, A.H.; Garcia-Olano, D.; Kokhlikyan, N. Using captum to explain generative language models. arXiv 2023, arXiv:2312.05491. [Google Scholar]
- Russo, A.; Autelitano, M.; Bisanti, L. Re: Frequency and cost of chemotherapy-related serious adverse effects in a population sample of women with breast cancer. J. Natl. Cancer Inst. 2006, 98, 1826–1827. [Google Scholar] [CrossRef]
- Nyrop, K.A.; Damone, E.M.; Deal, A.M.; Wheeler, S.B.; Charlot, M.; Reeve, B.B.; Basch, E.; Shachar, S.S.; Carey, L.A.; Reeder-Hayes, K.E.; et al. Patient-reported treatment toxicity and adverse events in Black and White women receiving chemotherapy for early breast cancer. Breast Cancer Res. Treat. 2022, 191, 409–422. [Google Scholar] [CrossRef]
- Rosenzweig, M.Q.; Mazanec, S.R. Racial differences in breast cancer therapeutic toxicity: Implications for practice. Cancer Epidemiol. Biomark. Prev. 2023, 32, 157–158. [Google Scholar] [CrossRef]
- Barnett, K.; Mercer, S.W.; Norbury, M.; Watt, G.; Wyke, S.; Guthrie, B. Epidemiology of multimorbidity and implications for health care, research, and medical education: A cross-sectional study. Lancet 2012, 380, 37–43. [Google Scholar] [CrossRef]
- Freid, V.M.; Bernstein, A.B.; Bush, M.A. Multiple chronic conditions among adults aged 45 and over: Trends over the past 10 years. NCHS Data Brief 2012, 100, 1–8. Available online: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=med9&NEWS=N&AN=23101759 (accessed on 8 April 2024).
- McGrath, J.J.; Al-Hamzawi, A.; Alonso, J.; Altwaijri, Y.; Andrade, L.H.; Bromet, E.J.; Bruffaerts, R.; de Almeida, J.M.C.; Chardoul, S.; Chiu, W.T.; et al. Age of onset and cumulative risk of mental disorders: A cross-national analysis of population surveys from 29 countries. Lancet Psychiatry 2023, 10, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, N.L.; Blackford, A.L.; Joshu, C.E.; Boyd, C.M.; Varadhan, R. Life expectancy estimates based on comorbidities and frailty to inform preventive care. J. Am. Geriatr. Soc. 2022, 70, 99–109. [Google Scholar] [CrossRef] [PubMed]
- Jatoi, I.; Anderson, W.F.; Jeong, J.H.; Redmond, C.K. Breast cancer adjuvant therapy: Time to consider its time-dependent effects. J. Clin. Oncol. 2011, 29, 2301–2304. [Google Scholar] [CrossRef] [PubMed]
- Hudis, C.A.; Dickler, M. Increasing precision in adjuvant therapy for breast cancer. N. Engl. J. Med. 2016, 375, 790–791. [Google Scholar] [CrossRef] [PubMed]
- Chan, N.; Toppmeyer, D.L. The Final Verdict: Chemotherapy Benefits Estrogen Receptor-Negative Isolated Local Recurrence. J. Clin. Onc. 2018, 36, 1058–1059. [Google Scholar] [CrossRef]
- El Haji, H.; Souadka, A.; Patel, B.N.; Sbihi, N.; Ramasamy, G.; Patel, B.K.; Ghogho, M.; Banerjee, I. Evolution of Breast Cancer Recurrence Risk Prediction: A Systematic Review of Statistical and Machine Learning-Based Models. JCO Clin. Cancer Inform. 2023, 7, e2300049. [Google Scholar] [CrossRef]

| Hyperparameter | Type | Range |
|---|---|---|
| Batch size | Categorical | [32, 64, 128, 256, 512] |
| Epochs | Categorical | [100, 200, 300, 500] |
| Dropout rate | Categorical | [0.001, 0.01, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8] |
| Number of layers | Integer values | [2, 5] |
| Number of nodes | Categorical | [32, 64, 128, 256, 512] |
| Alpha | Categorical | [0.0, 0.001, 0.1, 0.2, 0.5, 0.8, 0.9, 0.99, 1.0] |
| Sigma | Categorical | [0.01, 0.1, 0.25, 0.5, 1.0, 10, 100] |
| Learning rate | Continuous | [0.0001, 0.1] |
| Model | Time-Dependent Concordance | Integrated Brier Score | ||||||
|---|---|---|---|---|---|---|---|---|
| Support | Metabric | Support | Metabric | |||||
| As Reported in the Literature | Our Results | Reported in the Literature | Our Results | Reported in the Literature | Our Results | Reported in the Literature | Our Results | |
| Cox-Time | 0.630 | 0.647 | 0.664 | 0.683 | 0.212 | 0.182 | 0.173 | 0.150 |
| DeepHit | 0.639 | 0.646 | 0.675 | 0.676 | 0.227 | 0.196 | 0.186 | 0.103 |
| DeepSurv | 0.615 | 0.630 | 0.640 | 0.710 | 0.213 | 0.231 | 0.175 | 0.136 |
| Nnet-Survival (Logistic Hazard) | 0.625 | 0.617 | 0.658 | 0.674 | 0.184 | 0.205 | 0.172 | 0.142 |
| Stage | Number of Entries | Number of Patients | Age ± SD | Comorbidity Index ± SD | Number of Entries | Number of Patients | Age ± SD | Comorbidity Index ± SD |
|---|---|---|---|---|---|---|---|---|
| ER/PR+ | ER/PR− | |||||||
| I | 17,400,569 | 92,467 | 74.7 ± 6.8 | 2.9 ± 2.9 | 2,741,780 | 13,880 | 74.0 ± 6.7 | 2.8 ± 2.9 |
| II | 8,828,801 | 47,469 | 75.7 ± 7.5 | 2.9 ± 3.1 | 2,292,617 | 12,560 | 75.2 ± 7.5 | 2.6 ± 3.0 |
| III | 2,604,115 | 14,825 | 75.8 ± 7.6 | 2.5 ± 3.0 | 979,047 | 5966 | 75.7 ± 7.7 | 2.1 ± 2.9 |
| Model | Time-Dependent Concordance | Integrated Brier Score | Time-Dependent Concordance | Integrated Brier Score | ||||
|---|---|---|---|---|---|---|---|---|
| SM_Time-Fixed Patients’ Covariates ± SD | SM_Time-Fixed & Varying Patients’ Covariates ± SD | SM_Time-Fixed Patients’ Covariates ± SD | SM_Time-Fixed & Varying Patients’ Covariates ± SD | SM_Time-Fixed Patients’ Covariates ± SD | SM_Time-Fixed & Varying Patients’ Covariates ± SD | SM_Time-Fixed Patients’ Covariates ± SD | SM_Time-Fixed & Varying Patients’ Covariates ± SD | |
| ER/PR+ | ER/PR− | |||||||
| Stage I | ||||||||
| Cox-Time | 0.679 ± 0.001 | 0.987 ± 0.001 | 0.112 ± 0.002 | 0.009 ± 0.003 | 0.690 ± 0.005 | 0.987 ± 0.002 | 0.120 ± 0.002 | 0.011 ± 0.001 |
| DeepHit | 0.667 ± 0.002 | 0.958 ± 0.001 | 0.110 ± 0.003 | 0.013 ± 0.001 | 0.671 ± 0.003 | 0.960 ± 0.003 | 0.127 ± 0.001 | 0.042 ± 0.003 |
| DeepSurv | 0.682 ± 0.001 | 0.969 ± 0.002 | 0.110 ± 0.003 | 0.030 ± 0.009 | 0.670 ± 0.006 | 0.996 ± 0.001 | 0.117 ± 0.001 | 0.018 ± 0.003 |
| Nnet-Survival (Logistic Hazard) | 0.668 ± 0.001 | 0.976 ± 0.001 | 0.131 ± 0.003 | 0.037 ± 0.002 | 0.642 ± 0.005 | 0.980 ± 0.001 | 0.110 ± 0.002 | 0.042 ± 0.002 |
| Stage II | ||||||||
| Cox-Time | 0.689 ± 0.003 | 0.988 ± 0.001 | 0.106 ± 0.001 | 0.007 ± 0.003 | 0.676 ± 0.006 | 0.978 ± 0.003 | 0.110 ± 0.002 | 0.011 ± 0.001 |
| DeepHit | 0.722 ± 0.001 | 0.988 ± 0.001 | 0.122 ± 0.001 | 0.080 ± 0.007 | 0.724 ± 0.003 | 0.842 ± 0.005 | 0.129 ± 0.002 | 0.001 ± 0.003 |
| DeepSurv | 0.672 ± 0.003 | 0.965 ± 0.003 | 0.105 ± 0.001 | 0.029 ± 0.001 | 0.663 ± 0.006 | 0.993 ± 0.001 | 0.104 ± 0.001 | 0.026 ± 0.002 |
| Nnet-Survival (Logistic Hazard) | 0.661 ± 0.001 | 0.977 ± 0.001 | 0.110 ± 0.001 | 0.038 ± 0.003 | 0.618 ± 0.002 | 0.984 ± 0.001 | 0.118 ± 0.001 | 0.024 ± 0.001 |
| Stage III | ||||||||
| Cox-Time | 0.642 ± 0.001 | 0.981 ± 0.002 | 0.091 ± 0.003 | 0.008 ± 0.001 | 0.709 ± 0.005 | 0.968 ± 0.004 | 0.080 ± 0.001 | 0.011 ± 0.001 |
| DeepHit | 0.621 ± 0.002 | 0.981 ± 0.001 | 0.094 ± 0.001 | 0.052 ± 0.003 | 0.703 ± 0.004 | 0.973 ± 0.001 | 0.085 ± 0.001 | 0.089 ± 0.007 |
| DeepSurv | 0.660 ± 0.004 | 0.984 ± 0.006 | 0.089 ± 0.003 | 0.024 ± 0.002 | 0.666 ± 0.014 | 0.993 ± 0.002 | 0.079 ± 0.002 | 0.024 ± 0.001 |
| Nnet-Survival (Logistic Hazard) | 0.627 ± 0.002 | 0.944 ± 0.009 | 0.091 ± 0.003 | 0.005 ± 0.002 | 0.604 ± 0.005 | 0.944 ± 0.003 | 0.087 ± 0.002 | 0.064 ± 0.007 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, N.; Wieder, R. AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status over Time. Cancers 2024, 16, 3527. https://doi.org/10.3390/cancers16203527
Adam N, Wieder R. AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status over Time. Cancers. 2024; 16(20):3527. https://doi.org/10.3390/cancers16203527
Chicago/Turabian StyleAdam, Nabil, and Robert Wieder. 2024. "AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status over Time" Cancers 16, no. 20: 3527. https://doi.org/10.3390/cancers16203527
APA StyleAdam, N., & Wieder, R. (2024). AI Survival Prediction Modeling: The Importance of Considering Treatments and Changes in Health Status over Time. Cancers, 16(20), 3527. https://doi.org/10.3390/cancers16203527







