Simple Summary
Melanoma is the third most common type of skin cancer. Melanoma is a heterogeneous tumor, composed of genetically divergent subpopulations. These subpopulations exist in the form of cancer stem-like cells (CSCs) and many non-cancer stem cells (non-CSCs). CSCs are characterized by their unique surface proteins and aberrant signaling pathways. The unique characteristics of CSCs are responsible for the promotion of melanoma progression, drug resistance, and recurrence. Beyond unique characteristics of CSCs, melanomas also harbor significant alterations in functional genes (BRAF, CDKN2A, NRAS, TP53, and NF1). Of these, the most common are the BRAF and NRAS oncogenes, with 50% of melanomas demonstrating the BRAF mutation (BRAFV600E). Although the successful targeting of BRAFV600E does improve overall survival, the long-term efficacy of available therapeutic options is limited, and the development of acquired resistance is mostly common.
Abstract
Melanoma is the third most common type of skin cancer, characterized by its heterogeneity and propensity to metastasize to distant organs. Melanoma is a heterogeneous tumor, composed of genetically divergent subpopulations, including a small fraction of melanoma-initiating cancer stem-like cells (CSCs) and many non-cancer stem cells (non-CSCs). CSCs are characterized by their unique surface proteins associated with aberrant signaling pathways with a causal or consequential relationship with tumor progression, drug resistance, and recurrence. Melanomas also harbor significant alterations in functional genes (BRAF, CDKN2A, NRAS, TP53, and NF1). Of these, the most common are the BRAF and NRAS oncogenes, with 50% of melanomas demonstrating the BRAF mutation (BRAFV600E). While the successful targeting of BRAFV600E does improve overall survival, the long-term efficacy of available therapeutic options is limited due to adverse side effects and reduced clinical efficacy. Additionally, drug resistance develops rapidly via mechanisms involving fast feedback re-activation of MAPK signaling pathways. This article updates information relevant to the mechanisms of melanoma progression and resistance and particularly the mechanistic role of CSCs in melanoma progression, drug resistance, and recurrence.
1. Introduction
Despite improved treatment options, the prognosis for patients with advanced malignant melanoma remains poor, as measured by progression-free and overall survival [1]. Traditional therapeutics primarily target rapidly proliferating tumor cells, leaving tumor-initiating cells intact [2,3]. Consequently, tumor-initiating cells stimulate the production of new tumor clones with the propensity to disseminate to distant organs and to confer resistance to anticancer agents [4,5].
Aggressive tumor cells, including melanoma, share many characteristics with embryonic progenitors, which contribute to the mystery of tumor cell plasticity. While the multi-linage differentiation of embryonic stem cells (ESCs) is mainly controlled by a distinct microenvironment milieu leading the specification of the pluripotent ESCs [6,7], the differentiation in the case of the CSC concept refers to the ability of tumor cells to give rise to phenotypically diverse populations that reflect the histological features of the initial tumor in vivo [6]. In both embryonic and adult stem cells, differentiation is controlled by epigenetic mechanisms, and the plasticity of differentiation in these cells is associated with transcription accessibility for genes expressed in different normal tissues [7]. Abnormalities in genetic and/or epigenetic controls can lead to the development of cancer, which can be maintained by self-renewing CSCs [8,9]. Like normal stem cells, CSCs can show plasticity for differentiation [10]. CSC plasticity is mostly associated with transcription accessibility for genes that are normally expressed in different tissues, including tissues other than those from which the cancers originated [11].
In recent years, there has been an increased focus on cancer stem-like cells (CSCs) in experimental models of tumor initiation, progression, recurrence, and treatment resistance [9,12,13]. The working hypothesis suggests that CSCs, representing a small fraction of tumor cells, potentiate neoplastic clones [3]. Like other CSCs, melanoma stem-like cells (MSCs) are characterized by the expression of stemness properties-dependent protein markers and well-defined aberrant signaling pathways.
Most available therapeutics fail to target MSCs. In fact, MSCs are genetically evolved to evade drug toxicity and to promote tumor progression and metastases [14,15]. This article focuses on the molecular mechanisms of melanoma progression and treatment resistance, and particularly the mechanistic role of CSCs in melanoma progression, drug resistance, and recurrence.
2. Melanoma Heterogeneity and Plasticity
Although phenotypic diversity and plasticity in melanoma have been described >40 years ago [16,17], the molecular characterization of specific phenotypic states was first determined after the functional characterization of the gene encoding MITF [18]. Consequently, it is possible to investigate the specific phenotypic states evoked by microenvironmental signals.
In addition to its significant role in the regulation of pigment cell development [19], MITF is widely discussed as a key regulator of genes leading to the regulation of melanogenesis and primary differentiation-associated function of melanocytes [20,21]. Also, accumulated evidence indicated that deregulation of either MITF expression or activity can cause melanocyte dedifferentiation [22,23]. Beyond its role in melanoma and melanocytes differentiation, MITF has been reported to be essential for the regulation of genes implicated in several biological processes such as survival [24], cell cycle control [25], invasion [26], autophagy [27], senescence bypass [28], and DNA damage repair and chromosome stability [28,29].
Tumor heterogeneity is widely documented to play an imperative role in cancer development, evolution, and resistance to therapy. As one of the most heterogeneous human cancers, melanoma demonstrates high levels of biological complexity during disease progression. As result, melanoma cells undergo genetic, epigenetic, and/or phenotypic modification to survive in the human body. In addition to the aforementioned melanoma alterations, the microenvironment of melanoma cells plays a crucial role in the regulation of melanoma initiation, progression, treatment resistance, and recurrence [30,31].
The study of single-cell genotyping demonstrated a complex clonal diversity among tumor cells, a phenomenon that is recognized as tumor heterogeneity; “the same tumor cells exhibit different morphological and phenotypic profiles” [32,33]. Tumor heterogeneity is a tumor phenomenon that refers to the existence of subpopulations of cells with different genotypes and phenotypes that can exhibit different biological behaviors within a primary tumor and its metastases or between tumors of the same histopathological subtype in the form of intra- and inter-tumor phenotypes [34,35,36].
Melanoma has the highest mutation frequency among human cancers, which contributes to the development of significant melanoma heterogeneity [37,38,39,40,41,42].
Intratumoral heterogeneity refers to development of tumor subpopulations with variable genetic traits in the same tumor, intertumoral heterogeneity refers to the differences between lesions in the same patients [32,33,37,43,44], and phenotypic heterogeneity results from irreversible changes in tumor cells within a homogenous population in response to microenvironmental signals without undergoing genetic alterations [45,46,47].
Genetic intratumoral heterogeneity results from genomic instability in the form of frequent mutation of genes encoding for key proteins of the aberrant signaling pathways linked to the development of genetically divergent subpopulations [48,49]. The most common examples of the genetic intratumoral heterogeneity are the melanoma subpopulations bearing mutant (Mut) protein and its wild-type (WT) counterpart such as BRAFMut or BRAFWT [50,51,52,53], KITL576P, KITWT [54], BRAFV600E/NRASWT or BRAFWT/NRASQ61R [48], NRASG13R or NRASWT [49], as well as melanoma subpopulations with heterogenous expression of BRAFV600E [53].
Epigenetic intertumoral heterogeneity refers to tumor subpopulations with epigenetic variation [32]. The most reported examples for epigenetic intratumoral heterogeneity are those describing melanoma subpopulations bearing RASSF1A, CDKN2A, DAPK, MGMT, and RB1 genes with hypermethylated promoters [55,56] or melanoma subpopulations with heterogeneous expression of melanoma-associated antigen A3 (MAGE-A3) as a consequence of differential methylation of the MAGE-A3 promoter [56].
The microphthalmia-associated transcription factor MITF has been widely reported to be the master regulator of melanocyte biology in addition to being one of the key factors that is essential for the regulation of melanoma progression and invasion [32]. Melanoma is one of the most genetically and phenotypically heterogeneous cancers at inter-patient, inter-tumor, and intra-tumor level [57,58]. In addition to its role as key regulator of melanoma progression and invasion, MITF is one of the main determinants of melanoma heterogeneity and therapy resistance [57,58,59]. Consequently, the genetic, epigenetic, and phenotypic heterogeneity of melanoma result from the development of melanoma subpopulations within a tumor and can display remarkable variability in their phenotypic traits [60]. These heterogenous melanoma subpopulations are characterized by the expression of MITFhigh and MITFlow proteins [61,62,63], MITF and BRN2 (non-canonical melanoma tumor-suppressor) proteins [64,65,66], and MITF and PAX3 [65] proteins. Also, phenotypic intertumoral heterogeneity includes subpopulations that can undergo phenotypic transition from one subpopulation to another. This includes the transition from MITFhigh/NF-κBlow to MITFlow/NF-κBhigh/AXLhigh during the development of melanoma resistance [67,68], transition from primary melanoma that expresses ZEB2high/SNAIL2high/ZEB1low/TWIST1low to metastatic melanoma expressing ZEB2low/SNAIL2low/ZEB1high/TWIST1high [69], transition of melanoma of the ABCB5+ subpopulation to the melanoma ABCB5−subpopulation [70,71,72,73], and transition of the melanoma CD133+ subpopulation to the melanoma CD133− subpopulation [73,74,75,76,77]. Accordingly, the development of phenotypic intratumoral heterogeneity, in response to treatment with BRAF inhibitors, is common. The transition of the melanoma MART-1neg/NGFRhigh subpopulation to the MART-1neg/NGFRneg subpopulation [78,79], or the transition of from NRASWT/BRAFV600E to NRASG13R/BRAFV600E [80] following treatment with BRAF inhibitors, has also been reported.
Cancer cells can exhibit a high level of plasticity or the ability to dynamically switch between CSC and non-CSC states and even among different subsets of CSCs. Plasticity gives melanoma cells the ability to dynamically switch between a differentiated state with limited tumorigenic potential and an undifferentiated or cancer stem-like cell state (CSC) that is responsible for long-term tumor growth [81]. In addition to the ability to transit into distinct CSC states with different competence to evade drug toxicity to disseminate to distant organs, cancer cell plasticity has been shown to be linked to the epithelial-to-mesenchymal transition-like program in melanoma that relies not only on cell-autonomous mechanisms but also on signals provided by the tumor microenvironment and/or signals induced in response to therapy [82,83] in response to active mutations in key molecules of both MAPK and PI3K/AKT/PTEN pathways [84,85].
The development of cell cycle heterogeneity and the enhancement of cell differentiation and metabolic reprogramming contribute to the evolution of the phenotypic drug resistance of melanoma [85,86,87]. Figure 1 describes the mechanisms of cellular plasticity that allows for the adaptation of melanoma cells to a variety of environmental stresses.
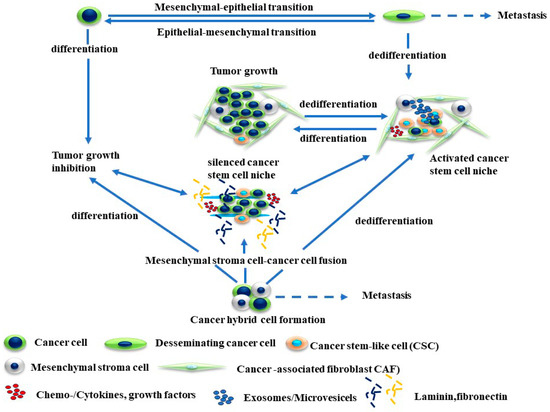
Figure 1.
Mechanisms of melanoma plasticity. The development of melanoma plasticity is mediated by genetic and epigenetic mechanisms that can derive epithelial–mesenchymal transition (EMT), mesenchymal–epithelial transition (MET), and cancer cell fusion. Consequently, the development and accumulation of populations of cancer cell progenitors or cancer stem cells (CSCs) can form a cancer cell niche. The enrichment of CSC populations in an activated niche is the main source for their differentiation or trans- or dedifferentiation, and metastasis of melanoma cells can take place at the same time in distinct compartments of tumor cells.
3. Mechanisms of Tumor Progression and Drug Resistance
Drug resistance is one of the largest challenges to melanoma treatment. Melanoma initiation and progression is mediated via genetic and epigenetic alterations to the key molecules in multiple signaling pathways such as RAS/RAF/MAPK, JNK, PI3K/Akt, and Jak/STAT pathways [86,87]. Likewise, dysregulation of MITF protein results in the development of melanoma progression and drug resistance [88].
The analysis of melanoma circulated tumor DNA (ctDNA) using next-generation sequencing (NGS) has been used as a reliable tool to monitor a driver mutation as a predictor marker for survival high-risk stage III cutaneous melanoma patients [89,90] and drug resistance [91]. The most mutated genes, which are associated with the development of resistance to targeted therapy in melanoma include CDKN2A [92,93], RB1 [92,94], PIK3CA, AKT3, HOXD8 [95], PAX5 [93], MAP3K8 [96], and MITF [95]. All these genes are either involved in the regulation of MAPK and PI3K/AKT signaling pathways in addition to serving as tumor suppressors affecting drug resistance in other cancers including Homeobox protein Hox-D8 (HOXD8) [97] and Paired Box 5 (PAX5) [98].
The most common genetic alterations result from frequent mutations to the DNA sequence of significant genes, while epigenetic alterations are mediated by cytosine methylation of DNA regulatory regions [99,100].
Genetic alterations involve inherited mutation to melanoma development, particularly familial melanomas [101]. These genetic alterations result from familial/inherited and somatic CDKN2A mutations to both p14ARF and p16INK4A, which impact melanoma suppression [102,103]. Although the CDKN2A and CDK4 genes have been primarily linked to familial melanoma, the contribution of these genes only accounts for a small percentage of familial melanoma [101,104]. The main function of p14ARF is to restrict cell proliferation via a p53 stabilization-dependent mechanism, which induces the cyclin-dependent kinase inhibitor p21 [105,106]. The main function of p16INK4A is to control cell proliferation by inhibiting cyclin-dependent kinases 4 and 6 (CDK4/6) and cyclin D1 [107,108]. Accordingly, CDKN2A mutations are common in melanoma and even reported in 8 to 57% of familial melanoma cases [109,110]. Somatic mutations to key genes such as BRAF are common risk factors associated with melanoma development in more than 5% of patients with BRAF mutation [111,112]. The frequent mutation of BRAF, particularly of BRAFV600E, has been reported in benign nevi, the precursors for melanoma genesis [113,114].
Epigenetic-dependent mechanisms such as methylation, chromatin modification, and remodeling are essential for the regulation of melanoma progression and drug resistance [115,116].
In addition to histone modification, noncoding RNA (ncRNA) expression, chromatin remodeling, and nucleosome positioning, most epigenetic changes are mediated by aberrant methylation of DNA-dependent mechanisms via the addition of a methyl group to the fifth carbon position of a cytosine molecule, which leads to generation of 5-methylcytosine (5-mC) molecules that constitute approximately 2–8% of the total cytosines of DNA regulatory sequences of interest [117,118].
Analysis of DNA methylation in melanoma cell lines has revealed a large group of hypermethylated genes, one of which includes the MITF gene [18,119]. MITF is significantly involved in the regulation of multiple biological processes, including melanoma differentiation, proliferation, migration, and senescence [120,121,122]. Likewise, the hypermethylation of the phosphatase and tensin (PTEN) homologue promoter plays a key role in the regulation of melanoma progression and resistance [123,124]. PTEN is the suppressor of PI3K, whose loss is associated with PI3K activation that, in turn, plays an essential role in the development of non-inherited melanomas [125,126].
The resistance of any tumor results from the development of primary and/or acquired resistance mechanisms. As mentioned, the occurrence of primary resistance in melanoma is attributed to the accumulation of both genetic and epigenetic alterations to tumor cells and significant changes in their microenvironment [127,128], while the development of acquired/adaptive resistance results from tumor treatment-induced genetic and epigenetic alterations to key molecules of aberrant signaling pathways in tumor cells [129,130].
Primary resistance development is mediated in great part by tyrosine-kinase-dependent phosphorylation of tyrosine residues that drive various cellular functions, including proliferation, differentiation, migration, and survival. Activation of tyrosine kinases is mediated by receptor tyrosine kinase (RTK) and/or non-receptor tyrosine kinase (NRTK)-dependent mechanisms [131,132,133,134,135]. RTK activation is strictly regulated and well balanced via ligand stimulation, chromosomal rearrangement, point mutations and amplification to RTK/NRTK genes [131,132,133,134,135,136]. RTK activation is mediated by variable mechanisms and via a cascade of phosphorylation events leading to the enhancement of cell growth, migration, differentiation, survival, or apoptosis. The most common RTK activation mechanisms include ligand-stabilized dimerization or oligomerization of their protein monomers [135,136]. Dimerization-dependent activation of RTK is mediated through the phosphorylation of tyrosine residues located in the kinase activation loop or juxta membrane domain of transmembrane receptors [135,136]. Phosphorylation of the tyrosine residues of RTKs is essential to trigger the activation of Ras/Raf/MEK/ERK and PI3K/AKT pathways [131,137].
Acquired resistance for any tumor type occurs via two different biological processes; one is early intrinsic/adaptive tumor resistance, while the other is known as late acquired resistance. Intrinsic resistance of tumors like melanoma results from the re-activation of RTK-dependent pathways (i.e., Ras/Raf/MEK/ERK and PI3K/AKT) as consequence of treatment with their specific inhibitors [138,139,140]. While a proportion of patients are intrinsically resistant to BRAF inhibitors, most patients who initially respond to BRAF inhibitors exhibit acquired resistance once treatment is initiated [139,140]. The development of BRAF inhibitor-associated acquired resistance is mediated by variable mechanisms. These mechanisms are mostly mediated by either upstream or downstream signaling, leading to re-activation of the MAPK pathway via BRAF-dependent and -independent mechanisms [141,142]. The upstream signaling-dependent mechanisms are mediated by the activation of ARAF and CRAF kinases to replace the inhibited BRAFV600E functioning [143,144,145], while downstream signaling-dependent mechanisms are mediated by ERK negative feedback effects on RAS that restore RAS activity and enhance the formation of BRAFV600E dimers. Of note, BRAF inhibitors can only bind one component of each dimer, a mechanism which allows the unbound BRAF monomers to interact with CRAF monomers to form BRAF-CRAF heterodimers that ultimately trigger the re-activation of ERK signaling to reduce long-term BRAF inhibitor efficacy [146,147].
In addition to the above-mentioned mechanisms, other mechanisms such as the amplification of BRAF mutations [148] and/or alternative splicing of BRAF gene, are involved in the re-activation of the MAPK pathway, leading to the development of melanoma acquired resistance to BRAF inhibitors [149]. Also, both insulin growth factor-1 receptor (IGF-1R) and PI3K/AKT pathways have been reported to be involved in the development of melanoma resistance [150,151,152]. The possible mechanisms of melanoma progression and drug resistance are summarized in Figure 2.
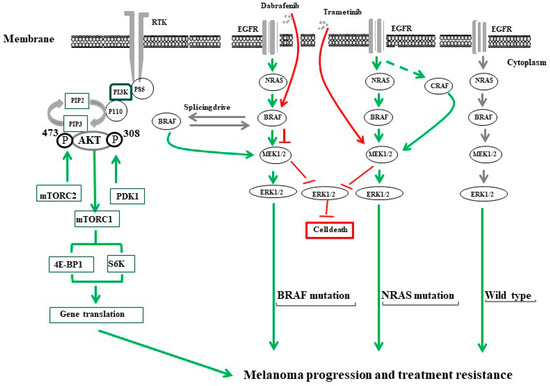
Figure 2.
The common mechanisms of melanoma progression and treatment resistance. PI3K/AKT/mTOR and RAS/RAF/MEK/ERK pathways are the most characterized tumor growth/treatment resistance-driving signals in melanoma. The activation of PI3K/AKT/mTOR is tightly controlled via a multistep process that can be initiated through tyrosine kinase receptors (RTKs) such as IGF-1R. The stimulation of IGF-1R by its ligand triggers the activation of PI3K via their catalytic (p110) and regulatory (p85) subunits leading to the conversion of phosphatidylinositol (3,4)-bisphosphate (PIP2) lipids to phosphatidylinositol (3,4,5)-trisphosphate (PIP3) allowing PDK1 to phosphorylate the tyrosine 308 (T308) residue and mTORC2 to phosphorylate tyrosine 473 (T473) in the activation loop of AKT. Activated AKT then activates mTORC1 that, in turn, enhances the activation of the eukaryotic translation initiation factor 4E binding protein 1 (4EBP1), and ribosomal protein S6 kinase (S6K). The activation of the Ras/RAF/MEK/ERK signaling pathway is mediated by the stimulation of epidermal growth factor (EGF) by its ligand for BRAF wild type or by active mutation in the cells bearing a NRAS mutation. The treatment of melanoma cells bearing BRAFV600E with specific inhibitors (e.g., dabrafenib) stimulates melanoma cells to overcome the inhibitory effect of BRAF through the generation of BRAF isoforms via alternative splicing. Treatment of patients with a NRAS mutation (NRASQ61R) with MEK inhibitors (e.g., trametinib) generates ERK negative feedback effects on RAS, which creates a bypass mechanism through activation of ARAF and/or CRAF to replace functional BRAF.
4. Cancer Stem Cells
Tumors are unique and complex ecosystems in which heterogeneous cell subpopulations with variable molecular profiles, aggressiveness, and proliferation potential coexist and interact in addition to exhibiting some self-renewal properties [153,154]. Two major models have been proposed to explain how tumors grow and progress [155,156]. In one of these models, namely the stochastic model, all the tumor cells are similar in their biological features, but their fates are determined by their intrinsic signals and their microenvironment-dependent signals. Although not all cancer cell progeny has the potential to behave like a cancer stem cell, they have the potential to retain plasticity to go from a non-stem cell to a stem cell-like precursor [156]. While in the hierarchical model, the cancer stem cells are biologically different and can self-renew in addition to giving rise to various progeny cells including those lacking the ability to self-renew [157]. However, the hierarchical model is often considered to be the most common model for sustained tumor propagation rather than the stochastic model [156].
Like normal stem cells, CSCs are hierarchically organized at the cellular level in origin tissues as a small fraction of genetically divergent subpopulations [158,159]. The development of these subpopulations results from the segregation of genetic material of functional genes to intrinsically asymmetric cell division of stem cell lineage to produce two daughter cells, both different in their genetic materials and phenotype [160,161]. The process of asymmetric cell division is genetically programed to produce one cell with stemness properties that is recognized as a CSC and one cell that is recognized as a non-CSC [124]. CSCs are adopted and genetically programed to grow continuously and divide indefinitely, whereas non-CSCs are characterized by their limited cell division [160,161]. While non-CSCs are sensitive to anti-cancer agents, CSCs evade drug toxicity and relocate and metastasize to distant organs [4,14,74].
Many potential biomarkers of CSCs have been identified based on their expression in human solid tumors. Potential CSC markers include neural crest nerve growth factor/neurotrophin receptor CD271 [4,78,79,162]. CD271+ melanoma cells are characterized by their tumorigenicity [4,78,79], and propensity to metastasize to the brain [130]. CD20, the cell surface marker of normal B cells, exhibits elevated expression in melanoma [4,14,163,164,165]. Transcription factors such as Nanog and Oct3/4 transcription factors have been found to be markedly elevated in melanospheres when compared to adherent melanoma cells [166,167]. Likewise, the activation of signaling pathways is common in normal stem cells such as Wnt, and Notch and Hedgehog are also activated in melanoma CSCs [168,169,170,171]. Aldehyde dehydrogenase (ALDH1) has also been identified as a potential marker of CSCs associated with multidrug and/or immunological resistance [170]. Accordingly, genes such as ABCB1, ABCB5, and ABCG2 undergo differential expression in both melanocytes and melanoma cells [4,71,171,172,173]. Finally, Sox10 expression maintains the growth of MSCs to grow as non-adherent tumorigenic spheres [174,175].
The involvement of the stem cell marker CD133 protein in the maintenance of melanoma stemness properties and drug resistance is mediated by its C-terminal domain, which contains tyrosine binding sites located on tyrosine 828 (Tyr828) and tyrosine 852 (Tyr852) residues [74,176]. These two tyrosine residues are phosphorylation targets of the non-receptor tyrosine kinase (NRTK) Fyn [176]. The contribution of CD133 to the regulation of CSC functions such as self-renewal, differentiation, and drug resistance are likely mediated by the NRTK, Fyn-dependent mechanism via the phosphorylation of Tyr828 residue located on the cytoplasmic domain of CD133 [74,176]. Our laboratory has demonstrated that the phosphorylation of Tyr828 is essential to trigger the activation of PI3K and its downstream-dependent signaling pathways in melanoma [74]. Many studies have demonstrated that increased CD133 expression is associated with high tumorigenicity and metastatic potential for melanoma cells [75,177,178,179]. Also, CD133 protein has been implicated in the regulation of tumor resistance [180,181,182,183].
CD133-expressing CSCs have been shown to exhibit resistance to chemotherapy and radiation therapy in addition to being associated with poor prognosis in various cancers [182]. We and others demonstrated that CD133+ cancer cells confer resistance to many chemotherapeutic agents such as caffeic acid phenethyl ester [4], taxol [14], and fotemustine [74]. Accordingly, CD133-dependent mechanisms have been shown to be involved in the development of melanoma resistance to chemotherapy [74].
Thus, understanding the mechanisms which are involved in the regulation of CSC growth and maintenance may help develop innovative therapeutic approaches for melanoma treatment.
The preliminary prevailing hypothesis suggests that tumor-initiating cells are derived from normal stem cells rather than from progenitor cells; the development of CSCs from normal stem cells and from progenitor stem cells has been shown [156,184,185]. Figure 3 outlines the pathways by which MSCs develop from either normal stem cells or cancer progenitor cells. In addition to their origin and unique properties, CSCs serve as an experimental model by which the mechanisms of tumor progression, recurrence, and treatment resistance can be investigated.
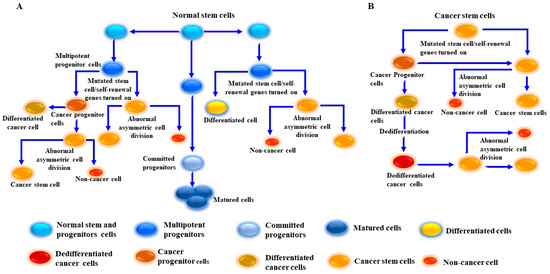
Figure 3.
Proposed model for cancer stem-like cell (CSC)/melanoma stem cell (MSC) generation from normal and cancer cells. (A) Generation of CSCs/MSCs results from the transformation of normal stem/progenitor cells into undifferentiated cancer cells via multiple genetic mutations and dedifferentiation-dependent mechanisms. (B) Generation of MSCs from cancer cells. This model describes the generation of MSCs from tumor cells through the activation of aberrant signaling pathways via driver mutations to tumor growth signaling pathways and activation of self-renewal genes. MSCs become educated to divide into tumor progenitor cells/MSCs. The produced MSCs possess genetic properties which allow for the division into one differentiated cell and one MSC. Once the dedifferentiation process of the differentiated cell is complete, the dedifferentiated cells can be transformed into MSCs. CSCs/MSCs undergo abnormal asymmetric cell division to produce two daughter cells, one CSC and one non-CSC.
5. Mechanisms of Melanoma Treatment Failure and Recurrence
Mechanistically, the role of CSCs in tumor initiation, metastasis, and therapy resistance have been demonstrated [4,14,74,174]. Stemness property-associated proteins like CD133 and CD271 have been purposed to play a functional role in melanoma progression, metastasis, and drug resistance [74,75,79,165,186]. Of note, approximately 50% of melanoma cases involve BRAF mutations [187]. BRAF inhibitors (e.g., vemurafenib, dabrafenib) have become the preferred therapy for melanoma patients with mutated BRAFV600E [188,189,190,191,192]. Unfortunately, while the successful targeting of BRAFV600E improves overall survival [188,193], acquired resistance development is common in most patients. This type of resistance is the direct consequence of secondary NRAS mutations [194,195] in response to BRAF inhibitor induced-ERK negative feedback effects on RAS as well as RTK-dependent RAS activation, which triggers the activation of both ARAF and CRAF, BRAF amplification, and activation of pathways like PI3K [196,197,198].
Melanoma progression, metastasis, treatment failure, and recurrence are attributed to the presence of genetically divergent MSC subpopulations with stemness properties [199,200]. First assessment of melanoma patients treated with BRAF inhibitors revealed that 20% targeted the BRAFV600E mutation [201,202]. Thus, the development of intrinsic resistance in melanoma patients with the BRAFV600E mutation results from the presence of a portion of cells (e.g., tumor-initiating cells/CSCs) that confer drug resistance [201,203].
While treatment with BRAF inhibitors displays initial tumor regression in most melanoma patients, complete tumor regression rarely occurs [204,205]. The main cause of this noted poor prognosis is attributed to BRAF inhibitor-mediated compensatory mechanisms. These compensatory mechanisms include elevation of ARAF and CRAF proteins, a mechanism through which melanoma cells sustain the activation of MAPK activity during BRAF inhibitors treatment [206,207,208]. Although BRAF inhibitor-resistant cells remain sensitive to MEK inhibitors, the resistance of these melanoma cells to various structurally different MEK inhibitors can be mediated by the generation of the compensatory mechanism via activation of RTKs such as IGF-1R [205,206,207,208]. Like other cells with a melanocyte origin, melanoma cells express IGF-1R. Importantly, MEK inhibitor-resistant melanoma cells demonstrate higher surface levels of IGF-1R when compared to BRAF-sensitive melanoma cells [205,206,207,208]. As such, IGF-1R is known to play an important role in tumor progression and drug resistance [209]. IGF-1R-induced effects are mediated via activation of MAPK and PI3K signaling [206,207,208,209]. This crosstalk between the MAPK and IGF-1R/PI3K/AKT signaling pathways promotes the survival and expansion of BRAF-inhibitor-resistant cells.
The most widely recognized challenge of current therapies for solid tumors involves this development of drug resistance and tumor recurrence. Common molecular mechanisms involved in therapeutic resistance include increased drug metabolism and drug efflux; enhanced repair capacity of damaged DNA; re-activation of drug targets; overactivation of growth and survival signaling pathways; amplification of genetic mutations; and impaired activity of apoptosis/autophagy-dependent pathways [142,152]. Tumor heterogeneity is also an important driver of drug resistance [32,80,210,211]. In contrast to non-CSCs, CSCs are characterized by specific features that allow them to escape drug toxicity [211,212]. These specific features include expression of different members of the ATP-binding cassette (ABC) transporters such as ABCB5, ABCG1, ABCG2, and ABCG5, all “efflux pumps”, to extrude drugs from cells into the microenvironment [213,214]. In addition to the ABC transporters, high levels of ALDH activity modulate CSC resistance to anti-cancer agents [160,215,216]. CSCs have been shown to be resistant to radiation therapy, based on their ability to repair damaged DNA [74,217,218]. Finally, inhibition of fotemustine-induced DNA damage in CD133+ melanoma subpopulations is associated with the activation of checkpoint kinases CHK1 and CHK2 [74]. Taken together, these observations strongly support the notation that melanoma CSCs represent a target for melanoma treatment with standard therapies being combined to be directed against the tumor bulk.
Current approaches to cancer immunotherapy include the non-specific stimulation of antitumor immune response through stimulation of endogenous effector cells via cytokine-dependent mechanisms, active immunization, adoptive immunotherapy, and the targeting of immune checkpoints or immune regulatory molecules [219]. The most common cytokine-based therapy involves targeting interleukin-2 (IL-2), which was the first type of immunotherapy approved for melanoma treatment [220,221,222]. Various systemic immune therapies have also been examined for patients with stage II or III melanoma, particularly for patients with a high risk of systemic recurrence [223]. The most common immunotherapeutic of this type involves targeting interferon α-2b and peginterferon α-2b [223,224].
Accordingly, the combination of chemotherapy with either immunotherapy or biotherapy in metastatic melanoma treatment has been explored in multiple trials; notable examples include the combination of cisplatin, vinblastine, and dacarbazine with IL-2 and interferon-α-2b immunotherapy [225,226,227,228].
Tumor vaccination-based therapies for melanoma treatment include multiple approaches. Unfortunately, most protein- or peptide-based vaccines lack significant immunogenicity and are unable to induce a robust immune response in monotherapy [229,230]. Likewise, the administration of customized vaccines, derived from tumor patient whole-cell lysates, does not show any clinical benefit in randomized controlled trials [231]. Other approaches include recombinant vectors, which encode entire genes or the antigenic epitope [232], and active vaccination of dendritic cells pulsed with tumor cell RNA, DNA, or cell lysate [233,234].
The treatment of melanoma with adoptive cell therapy has also been employed. Adoptive cell therapy involves the clinical utilization of autologous engineered T cells expressing receptors that specifically recognize various tumor-associated antigens such as melanocytic protein Melan-A/MART-1-TCG genes or that secrete specific cytokines [232,233,234,235].
Immune checkpoint modulators and immune modulator molecules have also been investigated [234]. Immune checkpoints target cell surface proteins such as cytotoxic T-lymphocyte antigen-4 (CTLA4) and programmed cell death-1 (PD-1) [235,236,237,238]. These proteins are implicated in the regulation of immune response initiation and duration [194]. Inhibition of these immune checkpoint inhibitors by anti-CTLA4 and anti-PD-1 antibodies demonstrated significant tumor regression and long-term durable cancer control when compared to other systematic therapies [238,239].
The expression of CSC makers in human tumor tissues is closely associated with the number of tumor-infiltrating immune cells [240,241]. As such, the CSC immune response is frequently compromised by immune evasion properties. These immune evasion properties are mediated by CSC-secreted immunosuppressive factors as well as the ability of CSCs to recruit immunosuppressive noncancerous cells [242,243,244], enhancing immune evasion strategies.
CSC-mediated immunosuppression both in vitro and in vivo allows CSCs to evade anti-tumor immune-mediated reactions [245]. Melanoma is characterized by their expression of a variety of antigens, including differentiation and cancer testis antigens such as MelanA/Mart1, HMB45, tyrosinase, gp100, and NYESO1 [246,247]. MSCs have the potential to initiate the activation of several mechanisms to maintain tumor survival and escape the patient’s immune reactions [244,248], via their mechanistic role in the suppression of anti-tumor immune reactions. One of these mechanisms involves the expression of multidrug resistance (MDR) genes and related functional transporters. MDR is the ability to mount resistance to multiple, structurally unrelated therapeutic drugs with different mechanisms of action [249,250,251]. MDR pathways in melanoma are responsible for the decrease in intracellular drug accumulation and is mediated by energy-dependent efflux pumps via ABC transporters that translocate solutes across the cellular membrane [252,253]. We and others have shown the involvement of the ABCB5 transporter in the modulation of melanoma resistance to anti-cancer agents like caffeic acid phenethyl ester [4] and doxorubicin [4,254,255]. Accordingly, blockage of ABCB5 enhances intracellular drug accumulation in melanoma cells sensitive to the therapeutic effects of BRAF inhibitors [256].
Resistance of MSCs to BRAF and MEK inhibitor-induced cell death has been reported [257,258]. Increased activity of Stearoyl-CoA-desaturase 1 (SCD1), the rate-limiting enzyme in the formation of monounsaturated fatty acids, has been shown to overcome BRAF and MEK inhibitor-induced death of MSCs [259].
The CD133 protein remains the most important identified stem cell marker. Functional analysis of this protein suggests that CD133 is a key tumor progression and treatment-resistance-driving signaling protein in melanoma [74], via a mechanism mediated by binding of the regulatory subunit of PI3K, p85, to the Tyrosine 828 (Tyr828) residue located on the cytoplasmic domain of the CD133 protein [74]. In addition to its contribution to the regulation of melanoma proliferation and metastasis [260,261,262], the overexpression of CD271 is associated with the induction of stem-like quiescence [160,260,261,262]. CD271 expression is lost in early progression when melanoma cells invade the dermis [263]. The low expression of CD271 is likewise associated with phenotype switching in melanoma [261].
Both tumor autonomous mechanisms and adaptive survival signaling may be related to melanoma progression and drug resistance. Once the processes of melanoma progression have been initiated, tumor cells tend to detach from their natural binding partners, namely keratinocytes, to interact with host cells such as fibroblasts and endothelial cells [2,264], inducing the secretion of the hepatocyte growth factor (HGF), endothelial growth factor (EGF), neuregulin (NRG), and IGF-1R [265,266,267]. Secretion of these tumor-derived growth factors often occurs in response to treatment with BRAF inhibitors, a common mechanism by which melanoma cells confer resistance to BRAF inhibitors [268,269].
Accordingly, melanoma treatment with BRAF inhibitors is theorized to remodel the host environment [151,270]. Clinically utilized BRAF triggers contradictory mitogen-activated protein kinase (MAPK) signaling pathways as well as triggers differentiation in normal skin fibroblasts, particularly those bearing BRAFWT cells [271]. Thus, such bidirectional signaling between tumor and host cells plays an active role in the configuration of these mechanisms, which is essential for the development of adaptive resistance to BRAF inhibitors [197,272].
Treatment with BRAF and/or MEK inhibitors mediates both autocrine and paracrine effects, which in turn trigger a stress-induced senescent phenotype in different melanoma subpopulations [273,274]. Cellular senescence is a phenomenon that occurs in melanoma patients during and/or after melanoma treatment has been initiated [275,276].
Cellular senescence is an autonomous tumor suppressor mechanism associated with the stabilization of cell cycle arrest [277,278]. Senescent cells are characterized by their ability to secrete variable factors that can change tumor cells and their microenvironment, allowing tumor cells to evade the toxicity of anti-cancer agents and subsequently grow and metastasize. Senescence-associated secretory phenotype (SASP) is associated with senescent cells wherein cells secrete high levels of inflammatory cytokines, immune modulators, growth factors, and proteases [279,280,281,282,283,284,285]. SASP may consist of exosomes and ectosomes containing enzymes, microRNA, DNA fragments, chemokines, and other bioactive factors [286]. Previous and current reports indicate that senescent cells are highly secretory cells and drive a range of different functions via SASP-dependent mechanisms [287,288]. The formation of SASP composition is dynamically and spatially regulated; thus, changing SASP composition can determine the beneficial and detrimental aspects of the senescence program, tipping the balance to either an immunosuppressive/pro-fibrotic environment or pro-inflammatory/fibrolytic state. The temporal and spatial regulation of SASP and NOTCH signaling may regulate SASP composition [289,290]. As the composition of SASP is dynamically and spatially regulated, a change in SASP can determine whether the senescence program progresses to an immunosuppressive/profibrotic environment or proinflammatory/fibrolytic state [287,288]. In summary, SASP has the potential to trigger tumor growth through the change in tumor microenvironment composition that ultimately influences treatment outcomes.
Melanoma cells with a senescent phenotype are characterized by the upregulation of two genes, one of which is matrix metalloprotease 2 (MMP2) [291,292]. Vemurafenib-induced MMP-2 has been shown to enhance melanoma invasiveness via an alteration in tumor microenvironment [280]. The other gene encodes monocyte chemoattractant protein 1 (MCP1). Functionally, MCP1 is involved in the initiation and subsequent activation of the poly (ADP-ribose) polymerase-1 (PARP-1)/NF-kB signaling cascade, which is essential for the regulation of tumor progression and dissemination to distant organs [287,291,293].
BRAF, ALK, or EGFR kinase inhibitors induce a complex network of secreted signals (secretome) in most tumor types including melanoma [294,295]. In addition to their role in the development of acquired resistance, the BRAF, ALK, or EGFR inhibitor-induced secretome results in an incomplete tumor regression and the stimulation of tumor growth and metastasis [291,292].
For example, vemurafenib-induced MMP-2 has been reported to enhance melanoma invasiveness and alter the tumor microenvironment, which triggers tumor resistance mechanisms [285,296]. Figure 4 provides a brief description of the possible mechanisms regulating the feedback activation of tumor growth factor (TGF) receptors during BRAF and MEK inhibitor-based therapy in MSCs.
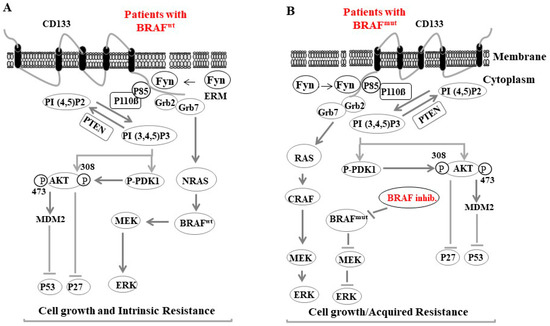
Figure 4.
A proposed model for Fyn-CD133 signaling to the PI3K pathway induces melanoma intrinsic and acquired resistance to anti-cancer agent in melanoma cells. (A) Fyn stimulated CD133 signaling to the PI3K pathway in melanoma population bearing the BRAF wild type (BRAFWT) is mediated by Fyn/CD133/PI3K/PDK1/AKT-induced inhibition of p27, Fyn/CD133/PI3K/AKT/MDM2-induced ubiquitination of p53 and Fyn/CD133/Grb2/Grb7/NRAS7BRAF/MEK/ERK-induced cell growth. (B) Intrinsic resistance of melanoma cells: Fyn stimulated CD133 signaling to the Grb2/Grb7/NRAS7BRAF/MEK/ERK pathway is responsible for acquired resistance of BRAF inhibitors (e.g., dabrafenib).
6. Conclusions
Melanoma progression and treatment resistance are attributed to tumor heterogeneity-dependent mechanisms that are mostly associated with development of genetically divergent subpopulations. These subpopulations exist in the form of CSCs or non-CSCs. Like other CSCs of any solid tumors, MSCs are identifiable via stemness specific markers. Thus, MSCs are central to tumor development, drug resistance, and recurrence. The development of MSCs is attributed to genetic and epigenetic changes, which lead to deregulation of several signal transduction pathways, including MAP kinase and PI3K/AKT. While BRAF/MEK and PI3K/AKT pathway inhibitors represent an intriguing therapeutic option for patients with metastatic melanoma, the success of these therapeutics is reduced by the development of acquired resistance via MSC-dependent mechanisms. These resistance mechanisms result from treatment pressure-induced by either BRAF/MEK or PI3K/AKT pathway inhibition. Both PI3K/AKT/mTOR and RAF/MEK/ERK signaling cascades are derived from numerous feedback loops and are interconnected at multiple points of crosstalk. Inhibition of one of these pathways can result in the activation of the other signaling cascade. The re-activation of the MAP kinase pathway occurs either via bypass-dependent activation of MEK by c-RAF/RAF1 or amplification and relative splicing of BRAF. Conversely, activation of the PI3K/AKT pathway results from the inhibition of the MAP kinase pathway. Thus, dual targeting of both pathways may improve melanoma treatment efficacy and lead to better clinical outcomes.
Author Contributions
Conceptualization, Y.A.H., R.T.B., M.H., N.K., T.W.F., A.A.A. and Y.H.; methodology, S.-Y.H., S.-L.H., S.S., M.H., H.S., S.-Y.H., T.W.F., A.A.A. and Y.H.; software, H.S., N.K., M.H. and T.W.F.; validation, M.H., Y.H., R.T.B., A.A.A. and S.S.; formal analysis, N.K., M.H. and T.W.F.; investigation, M.H., Y.H., Y.A.H., R.T.B., H.S., A.A.A. and S.S.; resources, Y.H., M.H., Y.A.H. and R.T.B.; data curation, M.H., N.K., A.A.A., H.S., Y.A.H., R.T.B. and S.S.; writing M.H., T.W.F., N.K., A.A.A. and Y.H.; writing—review and editing, M.H., Y.H. and S.S.; visualization, M.H., S.-Y.H., A.A.A. and S.-L.H.; supervision, M.H., S.S. and Y.H.; project administration, M.H., S.-Y.H., S.S., A.A.A. and S.-L.H.; funding acquisition, M.H., Y.H. and M.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest. Disclosures: Robert T. Brodell is a principal investigator for clinical trials (Novartis and Sanofi) and the Corevitas psoriasis biologic registry and owns stock in Veradermics, Inc. He has served on advisory boards for Amgen and Novan. The remaining authors have no conflicts of interest to disclose.
References
- Weiss, S.A.; Hanniford, D.; Hernando, E.; Osman, I. Revisiting determinants of prognosis in cutaneous melanoma. Cancer 2015, 121, 4108–4123. [Google Scholar] [CrossRef] [PubMed]
- Czyz, M.; Koprowska, K.; Sztiller-Sikorska, M. Parthenolide reduces the frequency of ABCB5-positive cells and clonogenic capacity of melanoma cells from anchorage independent melanospheres. Cancer Biol. Ther. 2013, 14, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- El-Khattouti, A.; Sheehan, N.T.; Monico, J.; Drummond, H.A.; Haikel, Y.; Brodell, R.T.; Megahed, M.; Hassan, M. CD133+ melanoma subpopulation acquired resistance to caffeic acid phenethyl ester-induced apoptosis is attributed to the elevated expression of ABCB5: Significance for melanoma treatment. Cancer Lett. 2015, 357, 83–104. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, M.C.; Fumagalli, M.R.; La Porta, C.A.M. Cancer stem cells, plasticity, and drug resistance. Cancer Drug Resist. 2020, 3, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, J.; Jiang, L.; James Kang, Y. Cancer and stem cells. Exp. Biol. Med. 2021, 246, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Lotem, J.; Sachs, L. Epigenetics and the plasticity of differentiation in normal and cancer stem cells. Oncogene 2006, 25, 7663–7672. [Google Scholar] [CrossRef]
- Toh, T.B.; Lim, J.J.; Chow, E.K. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 29. [Google Scholar] [CrossRef]
- Schatton, T.; Frank, M.H. Cancer stem cells and human malignant melanoma. Pigment. Cell Melanoma Res. 2008, 21, 39–55. [Google Scholar] [CrossRef]
- Warrier, N.M.; Kelkar, N.; Johnson, C.T.; Govindarajan, T.; Prabhu, V.; Kumar, P. Understanding cancer stem cells and plasticity: Towards better therapeutics. Eur. J. Cell Biol. 2023, 102, 151321. [Google Scholar] [CrossRef]
- Tang, D.G. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012, 22, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Raimondi, M.; Fontana, F.; Montagnani Marelli, M.; Moretti, R.M.; Limonta, P. Cellular and molecular biology of cancer stem cells in melanoma: Possible therapeutic implications. Semin. Cancer Biol. 2019, 59, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Diener, J.; Sommer, L. Reemergence of neural crest stem cell-like states in melanoma during disease progression and treatment. Stem Cells Transl. Med. 2021, 10, 522–533. [Google Scholar] [CrossRef] [PubMed]
- El-Khattouti, A.; Selimovic, D.; Haïkel, Y.; Megahed, M.; Gomez, C.R.; Hassan, M. Identification and analysis of CD133+ melanoma stem-like cells conferring resistance to taxol: An insight into the mechanisms of their resistance and response. Cancer Lett. 2014, 343, 123–133. [Google Scholar] [CrossRef]
- Fattore, L.; Mancini, R.; Ciliberto, G. Cancer Stem Cells and the Slow Cycling Phenotype: How to Cut the Gordian Knot Driving Resistance to Therapy in Melanoma. Cancers 2020, 12, 3368. [Google Scholar] [CrossRef]
- Rambow, F.; Marine, J.C.; Goding, C.R. Melanoma plasticity and phenotypic diversity: Therapeutic barriers and opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef]
- Diazzi, S.; Tartare-Deckert, S.; Deckert, M. The mechanical phenotypic plasticity of melanoma cell: An emerging driver of therapy cross-resistance. Oncogenesis 2023, 12, 7. [Google Scholar] [CrossRef]
- Hartman, M.L.; Czyz, M. MITF in melanoma: Mechanisms behind its expression and activity. Cell. Mol. Life Sci. 2015, 72, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, J.J.; Fisher, D.E. The roles of microphthalmia-associated transcription factor and pigmentation in melanoma. Arch. Biochem. Biophys. 2014, 563, 28–34. [Google Scholar] [CrossRef]
- Gelmi, M.C.; Houtzagers, L.E.; Strub, T.; Krossa, I.; Jager, M.J. MITF in Normal Melanocytes, Cutaneous and Uveal Melanoma: A Delicate Balance. Int. J. Mol. Sci. 2022, 23, 6001. [Google Scholar] [CrossRef]
- Chauhan, J.S.; Hölzel, M.; Lambert, J.P.; Buffa, F.M.; Goding, C.R. The MITF regulatory network in melanoma. Pigment. Cell Melanoma Res. 2022, 35, 517–533. [Google Scholar] [CrossRef]
- Fernández-Barral, A.; Orgaz, J.L.; Baquero, P.; Ali, Z.; Moreno, A.; Tiana, M.; Gómez, V.; Riveiro-Falkenbach, E.; Cañadas, C.; Zazo, S.; et al. Regulatory and functional connection of microphthalmia-associated transcription factor and anti-metastatic pigment epithelium derived factor in melanoma. Neoplasia 2014, 16, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Selzer, E.; Wacheck, V.; Lucas, T.; Heere-Ress, E.; Wu, M.; Weilbaecher, K.N.; Schlegel, W.; Valent, P.; Wrba, F.; Pehamberger, H.; et al. The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res. 2002, 62, 2098–2103. [Google Scholar]
- Kawakami, A.; Fisher, D.E. The master role of microphthalmia-associated transcription factor in melanocyte and melanoma biology. Lab. Investig. 2017, 97, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Loercher, A.E.; Tank, E.M.; Delston, R.B.; Harbour, J.W. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 2005, 168, 35–40. [Google Scholar] [CrossRef] [PubMed]
- Dilshat, R.; Fock, V.; Kenny, C.; Gerritsen, I.; Lasseur, R.M.J.; Travnickova, J.; Eichhoff, O.M.; Cerny, P.; Möller, K.; Sigurbjörnsdóttir, S.; et al. MITF reprograms the extracellular matrix and focal adhesion in melanoma. eLife 2021, 10, e63093. [Google Scholar] [CrossRef]
- Möller, K.; Sigurbjornsdottir, S.; Arnthorsson, A.O.; Pogenberg, V.; Dilshat, R.; Fock, V.; Brynjolfsdottir, S.H.; Bindesboll, C.; Bessadottir, M.; Ogmundsdottir, H.M.; et al. MITF has a central role in regulating starvation-induced autophagy in melanoma. Sci. Rep. 2019, 9, 1055. [Google Scholar] [CrossRef]
- Leclerc, J.; Ballotti, R.; Bertolotto, C. Pathways from senescence to melanoma: Focus on MITF sumoylation. Oncogene 2017, 36, 6659–6667. [Google Scholar] [CrossRef] [PubMed]
- Binet, R.; Lambert, J.P.; Tomkova, M.; Tischfield, S.; Baggiolini, A.; Picaud, S.; Sarkar, S.; Louphrasitthiphol, P.; Dias, D.; Carreira, S.; et al. DNA damage-induced interaction between a lineage addiction oncogenic transcription factor and the MRN complex shapes a tissue-specific DNA Damage Response and cancer predisposition. bioRxiv 2023. [Google Scholar] [CrossRef]
- Qin, S.S.; Han, B.J.; Williams, A.; Jackson, K.M.; Jewell, R.; Chacon, A.C.; Lord, E.M.; Linehan, D.C.; Kim, M.; Reuben, A.; et al. Intertumoral Genetic Heterogeneity Generates Distinct Tumor Microenvironments in a Novel Murine Synchronous Melanoma Model. Cancers 2021, 13, 2293. [Google Scholar] [CrossRef]
- Menaa, F.; Houben, R.; Eyrich, M.; Broecker, E.B.; Becker, J.C.; Wischhusen, J. Stem cells, melanoma and cancer stem cells: The good, the bad and the evil? G. Ital. Dermatol. Venereol. 2009, 144, 287–296. [Google Scholar] [PubMed]
- Ng, M.F.; Simmons, J.L.; Boyle, G.M. Heterogeneity in Melanoma. Cancers 2022, 14, 3030. [Google Scholar] [CrossRef] [PubMed]
- Grzywa, T.M.; Paskal, W.; Włodarski, P.K. Intratumor and Intertumor Heterogeneity in Melanoma. Transl. Oncol. 2017, 10, 956–975. [Google Scholar] [CrossRef] [PubMed]
- Khoja, L.; Shenjere, P.; Hodgson, C.; Hodgetts, J.; Clack, G.; Hughes, A.; Lorigan, P.; Dive, C. Prevalence and heterogeneity of circulating tumour cells in metastatic cutaneous melanoma. Melanoma Res. 2014, 24, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Keall, P.J.; Webb, S. Optimum parameters in a model for tumour control probability, including interpatient heterogeneity: Evaluation of the log-normal distribution. Phys. Med. Biol. 2007, 52, 291–302. [Google Scholar] [CrossRef]
- Cornelius, E.A.; Neumann, R.D.; Zoghbi, S.S.; Kirkwood, J.M.; Ernstoff, M.S.; Unger, M.M. Intrapatient and interpatient comparison of tumor size and monoclonal antibody uptake in melanoma. Clin. Nucl. Med. 1988, 13, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Anaka, M.; Hudson, C.; Lo, P.H.; Do, H.; Caballero, O.L.; Davis, I.D.; Dobrovic, A.; Cebon, J.; Behren, A. Intratumoral genetic heterogeneity in metastatic melanoma is accompanied by variation in malignant behaviors. BMC Med. Genom. 2013, 6, 40. [Google Scholar] [CrossRef]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Béranger, G.; Lassalle, S.; et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef]
- Thrane, K.; Eriksson, H.; Maaskola, J.; Hansson, J.; Lundeberg, J. Spatially Resolved Transcriptomics Enables Dissection of Genetic Heterogeneity in Stage III Cutaneous Malignant Melanoma. Cancer Res. 2018, 78, 5970–5979. [Google Scholar] [CrossRef]
- Gerber, T.; Willscher, E.; Loeffler-Wirth, H.; Hopp, L.; Schadendorf, D.; Schartl, M.; Anderegg, U.; Camp, G.; Treutlein, B.; Binder, H.; et al. Mapping heterogeneity in patient-derived melanoma cultures by single-cell RNA-seq. Oncotarget 2017, 8, 846–862. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, L.; Meyer, N.; Tournier, E.; Grand, D.; Uro-Coste, E.; Rochaix, P.; Brousset, P.; Lamant, L. Highly Concordant Results Between Immunohistochemistry and Molecular Testing of Mutated V600E BRAF in Primary and Metastatic Melanoma. Acta Derm. Venereol. 2016, 96, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Kemper, K.; Krijgsman, O.; Cornelissen-Steijger, P.; Shahrabi, A.; Weeber, F.; Song, J.Y.; Kuilman, T.; Vis, D.J.; Wessels, L.F.; Voest, E.E.; et al. Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol. Med. 2015, 7, 1104–1118. [Google Scholar] [CrossRef] [PubMed]
- Sakaizawa, K.; Ashida, A.; Kiniwa, Y.; Okuyama, R. BRAF Mutation Heterogeneity in Melanoma Lesions. Acta Derm.-Venereol. 2020, 100, adv00045. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Wang, J.; Huang, W.; Liu, J.; Yan, W. Identification of Transcriptional Heterogeneity and Construction of a Prognostic Model for Melanoma Based on Single-Cell and Bulk Transcriptome Analysis. Front. Cell Dev. Biol. 2022, 10, 874429. [Google Scholar] [CrossRef]
- Karras, P.; Bordeu, I.; Pozniak, J.; Nowosad, A.; Pazzi, C.; Van Raemdonck, N.; Landeloos, E.; Van Herck, Y.; Pedri, D.; Bervoets, G.; et al. A cellular hierarchy in melanoma uncouples growth and metastasis. Nature 2022, 610, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.M.; Eccles, M.R. Phenotype Switching and the Melanoma Microenvironment; Impact on Immunotherapy and Drug Resistance. Int. J. Mol. Sci. 2023, 24, 1601. [Google Scholar] [CrossRef] [PubMed]
- Avagliano, A.; Fiume, G.; Pelagalli, A.; Sanità, G.; Ruocco, M.R.; Montagnani, S.; Arcucci, A. Metabolic Plasticity of Melanoma Cells and Their Crosstalk with Tumor Microenvironment. Front. Oncol. 2020, 10, 722. [Google Scholar] [CrossRef]
- Sensi, M.; Nicolini, G.; Petti, C.; Bersani, I.; Lozupone, F.; Molla, A.; Vegetti, C.; Nonaka, D.; Mortarini, R.; Parmiani, G.; et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene 2006, 25, 3357–3364. [Google Scholar] [CrossRef]
- Wilmott, J.S.; Tembe, V.; Howle, J.R.; Sharma, R.; Thompson, J.F.; Rizos, H.; Lo, R.S.; Kefford, R.F.; Scolyer, R.A.; Long, G.V. Intratumoral molecular heterogeneity in a BRAF-mutant, BRAF inhibitor-resistant melanoma: A case illustrating the challenges for personalized medicine. Mol. Cancer Ther. 2012, 11, 2704–2708. [Google Scholar] [CrossRef]
- Yancovitz, M.; Litterman, A.; Yoon, J.; Ng, E.; Shapiro, R.L.; Berman, R.S.; Pavlick, A.C.; Darvishian, F.; Christos, P.; Mazumdar, M.; et al. Intra- and inter-tumor heterogeneity of BRAF(V600E) mutations in primary and metastatic melanoma. PLoS ONE 2012, 7, e29336. [Google Scholar] [CrossRef]
- Lin, J.; Goto, Y.; Murata, H.; Sakaizawa, K.; Uchiyama, A.; Saida, T.; Takata, M. Polyclonality of BRAF mutations in primary melanoma and the selection of mutant alleles during progression. Br. J. Cancer 2011, 104, 464–468. [Google Scholar] [CrossRef] [PubMed]
- Bruno, W.; Martinuzzi, C.; Andreotti, V.; Pastorino, L.; Spagnolo, F.; Dalmasso, B.; Cabiddu, F.; Gualco, M.; Ballestrero, A.; Bianchi-Scarrà, G.; et al. Heterogeneity and frequency of BRAF mutations in primary melanoma: Comparison between molecular methods and immunohistochemistry. Oncotarget 2017, 8, 8069–8082. [Google Scholar] [CrossRef] [PubMed]
- Busam, K.J.; Hedvat, C.; Pulitzer, M.; von Deimling, A.; Jungbluth, A.A. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am. J. Surg. Pathol. 2013, 37, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Gremel, G.; Lee, R.J.; Girotti, M.R.; Mandal, A.K.; Valpione, S.; Garner, G.; Ayub, M.; Wood, S.; Rothwell, D.G.; Fusi, A.; et al. Distinct subclonal tumour responses to therapy revealed by circulating cell-free DNA. Ann. Oncol. 2016, 27, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Rastetter, M.; Schagdarsurengin, U.; Lahtz, C.; Fiedler, E.; Marsch, W.C.; Dammann, R.; Helmbold, P. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol. Histopathol. 2007, 22, 1005–1015. [Google Scholar] [CrossRef]
- Sigalotti, L.; Fratta, E.; Coral, S.; Tanzarella, S.; Danielli, R.; Colizzi, F.; Fonsatti, E.; Traversari, C.; Altomonte, M.; Maio, M. Intratumoralalal heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation-regulated and functionally reverted by 5-aza-2’-deoxycytidine. Cancer Res. 2004, 64, 9167–9171. [Google Scholar] [CrossRef] [PubMed]
- Ennen, M.; Keime, C.; Gambi, G.; Kieny, A.; Coassolo, S.; Thibault-Carpentier, C.; Margerin-Schaller, F.; Davidson, G.; Vagne, C.; Lipsker, D.; et al. MITF-High and MITF-Low Cells and a Novel Subpopulation Expressing Genes of Both Cell States Contribute to Intra- and Intertumoral Heterogeneity of Primary Melanoma. Clin. Cancer Res. 2017, 23, 7097–7107. [Google Scholar] [CrossRef] [PubMed]
- Eichhoff, O.M.; Zipser, M.C.; Xu, M.; Weeraratna, A.T.; Mihic, D.; Dummer, R.; Hoek, K.S. The immunohistochemistry of invasive and proliferative phenotype switching in melanoma: A case report. Melanoma Res. 2010, 20, 349–355. [Google Scholar] [CrossRef]
- Ballesteros-Álvarez, J.; Dilshat, R.; Fock, V.; Möller, K.; Karl, L.; Larue, L.; Ögmundsdóttir, M.H.; Steingrímsson, E. MITF and TFEB cross-regulation in melanoma cells. PLoS ONE 2020, 15, e0238546. [Google Scholar] [CrossRef]
- Pillai, M.; Rajaram, G.; Thakur, P.; Agarwal, N.; Muralidharan, S.; Ray, A.; Barbhaya, D.; Somarelli, J.A.; Jolly, M.K. Mapping phenotypic heterogeneity in melanoma onto the epithelial-hybrid-mesenchymal axis. Front. Oncol. 2022, 12, 913803. [Google Scholar] [CrossRef]
- Salti, G.I.; Manougian, T.; Farolan, M.; Shilkaitis, A.; Majumdar, D.; Das Gupta, T.K. Micropthalmia transcription factor: A new prognostic marker in intermediate-thickness cutaneous malignant melanoma. Cancer Res. 2000, 60, 5012–5016. [Google Scholar] [PubMed]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Lister, J.A.; Capper, A.; Zeng, Z.; Mathers, M.E.; Richardson, J.; Paranthaman, K.; Jackson, I.J.; Elizabeth Patton, E. A conditional zebrafish MITF mutation reveals MITF levels are critical for melanoma promotion vs. regression in vivo. J. Investig. Dermatol. 2014, 134, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Simmons, J.L.; Pierce, C.J.; Al-Ejeh, F.; Boyle, G.M. MITF and BRN2 contribute to metastatic growth after dissemination of melanoma. Sci. Rep. 2017, 7, 10909. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.P.; Rana, S.; Ferguson, J.; Rowling, E.J.; Flaherty, K.T.; Wargo, J.A.; Marais, R.; Wellbrock, C. A PAX3/BRN2 rheostat controls the dynamics of BRAF mediated MITF regulation in MITF. Pigment. Cell Melanoma Res. 2019, 32, 280–291. [Google Scholar] [CrossRef]
- Golan, T.; Messer, A.R.; Amitai-Lange, A.; Melamed, Z.; Ohana, R.; Bell, R.E.; Kapitansky, O.; Lerman, G.; Greenberger, S.; Khaled, M.; et al. Interactions of Melanoma Cells with Distal Keratinocytes Trigger Metastasis via Notch Signaling Inhibition of MITF. Mol. Cell 2015, 59, 664–676. [Google Scholar] [CrossRef]
- Müller, J.; Krijgsman, O.; Tsoi, J.; Robert, L.; Hugo, W.; Song, C.; Kong, X.; Possik, P.A.; Cornelissen-Steijger, P.D.; Geukes Foppen, M.H.; et al. Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun. 2014, 5, 5712. [Google Scholar] [CrossRef]
- Konieczkowski, D.J.; Johannessen, C.M.; Abudayyeh, O.; Kim, J.W.; Cooper, Z.A.; Piris, A.; Frederick, D.T.; Barzily-Rokni, M.; Straussman, R.; Haq, R.; et al. A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors. Cancer Discov. 2014, 4, 816–827. [Google Scholar] [CrossRef]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A switch in the expression of embryonic EMT-inducers drives the development of malignant melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef]
- Wang, S.; Tang, L.; Lin, J.; Shen, Z.; Yao, Y.; Wang, W.; Tao, S.; Gu, C.; Ma, J.; Xie, Y.; et al. ABCB5 promotes melanoma metastasis through enhancing NF-κB p65 protein stability. Biochem. Biophys. Res. Commun. 2017, 492, 18–26. [Google Scholar] [CrossRef]
- Tuccitto, A.; Tazzari, M.; Beretta, V.; Rini, F.; Miranda, C.; Greco, A.; Santinami, M.; Patuzzo, R.; Vergani, B.; Villa, A.; et al. Immunomodulatory Factors Control the Fate of Melanoma Tumor Initiating Cells. Stem Cells 2016, 34, 2449–2460. [Google Scholar] [CrossRef] [PubMed]
- Schatton, T.; Murphy, G.F.; Frank, N.Y.; Yamaura, K.; Waaga-Gasser, A.M.; Gasser, M.; Zhan, Q.; Jordan, S.; Duncan, L.M.; Weishaupt, C.; et al. Identification of cells initiating human melanomas. Nature 2008, 451, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.Y.; Schwartz, B.E.; Hsu, M.Y. CD133+ melanoma subpopulations contribute to perivascular niche morphogenesis and tumorigenicity through vasculogenic mimicry. Cancer Res. 2012, 72, 5111–5118. [Google Scholar] [CrossRef]
- Jamal, S.M.E.; Alamodi, A.; Wahl, R.U.; Grada, Z.; Shareef, M.A.; Hassan, S.Y.; Murad, F.; Hassan, S.L.; Santourlidis, S.; Gomez, C.R.; et al. Melanoma stem cell maintenance and chemo-resistance are mediated by CD133 signal to PI3K-dependent pathways. Oncogene 2020, 39, 5468–5478. [Google Scholar] [CrossRef] [PubMed]
- Simbulan-Rosenthal, C.M.; Dougherty, R.; Vakili, S.; Ferraro, A.M.; Kuo, L.W.; Alobaidi, R.; Aljehane, L.; Gaur, A.; Sykora, P.; Glasgow, E.; et al. CRISPR-Cas9 Knockdown and Induced Expression of CD133 Reveal Essential Roles in Melanoma Invasion and Metastasis. Cancers 2019, 11, 1490. [Google Scholar] [CrossRef] [PubMed]
- Monzani, E.; Facchetti, F.; Galmozzi, E.; Corsini, E.; Benetti, A.; Cavazzin, C.; Gritti, A.; Piccinini, A.; Porro, D.; Santinami, M.; et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer 2007, 43, 935–946. [Google Scholar] [CrossRef]
- Zimmerer, R.M.; Korn, P.; Demougin, P.; Kampmann, A.; Kokemüller, H.; Eckardt, A.M.; Gellrich, N.C.; Tavassol, F. Functional features of cancer stem cells in melanoma cell lines. Cancer Cell Int. 2013, 13, 78. [Google Scholar] [CrossRef]
- Boiko, A.D.; Razorenova, O.V.; van de Rijn, M.; Swetter, S.M.; Johnson, D.L.; Ly, D.P.; Butler, P.D.; Yang, G.P.; Joshua, B.; Kaplan, M.J.; et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature 2010, 466, 133–137. [Google Scholar] [CrossRef]
- Civenni, G.; Walter, A.; Kobert, N.; Mihic-Probst, D.; Zipser, M.; Belloni, B.; Seifert, B.; Moch, H.; Dummer, R.; van den Broek, M.; et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011, 71, 3098–3109. [Google Scholar] [CrossRef]
- Su, Y.; Bintz, M.; Yang, Y.; Robert, L.; Ng, A.H.C.; Liu, V.; Ribas, A.; Heath, J.R.; Wei, W. Phenotypic heterogeneity and evolution of melanoma cells associated with targeted therapy resistance. PLoS Comput. Biol. 2019, 15, e1007034. [Google Scholar] [CrossRef]
- da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Muñoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef] [PubMed]
- Vachtenheim, J.; Ondrušová, L. Many Distinct Ways Lead to Drug Resistance in BRAF- and NRAS-Mutated Melanomas. Life 2021, 11, 424. [Google Scholar] [CrossRef]
- Huang, F.; Santinon, F.; Flores González, R.E.; Del Rincón, S.V. Melanoma Plasticity: Promoter of Metastasis and Resistance to Therapy. Front. Oncol. 2021, 11, 756001. [Google Scholar] [CrossRef] [PubMed]
- Roesch, A. Tumor heterogeneity and plasticity as elusive drivers for resistance to MAPK pathway inhibition in melanoma. Oncogene 2015, 34, 2951–2957. [Google Scholar] [CrossRef] [PubMed]
- Corrales, E.; Levit-Zerdoun, E.; Metzger, P.; Mertes, R.; Lehmann, A.; Münch, J.; Lemke, S.; Kowar, S.; Boerries, M. PI3K/AKT signaling allows for MAPK/ERK pathway independence mediating dedifferentiation-driven treatment resistance in melanoma. Cell Commun. Signal. 2022, 20, 187. [Google Scholar] [CrossRef]
- Sarkar, D.; Leung, E.Y.; Baguley, B.C.; Finlay, G.J.; Askarian-Amiri, M.E. Epigenetic regulation in human melanoma: Past and future. Epigenetics 2015, 10, 103–121. [Google Scholar] [CrossRef] [PubMed]
- Karami Fath, M.; Azargoonjahromi, A.; Soofi, A.; Almasi, F.; Hosseinzadeh, S.; Khalili, S.; Sheikhi, K.; Ferdousmakan, S.; Owrangi, S.; Fahimi, M.; et al. Current understanding of epigenetics role in melanoma treatment and resistance. Cancer Cell Int. 2022, 22, 313. [Google Scholar] [CrossRef]
- Lavelle, T.J.; Alver, T.N.; Heintz, K.M.; Wernhoff, P.; Nygaard, V.; Nakken, S.; Øy, G.F.; Bøe, S.L.; Urbanucci, A.; Hovig, E. Dysregulation of MITF Leads to Transformation in MC1R-Defective Melanocytes. Cancers 2020, 12, 1719. [Google Scholar] [CrossRef]
- Diefenbach, R.J.; Lee, J.H.; Rizos, H. Monitoring Melanoma Using Circulating Free DNA. Am. J. Clin. Dermatol. 2019, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Saw, R.P.; Thompson, J.F.; Lo, S.; Spillane, A.J.; Shannon, K.F.; Stretch, J.R.; Howle, J.; Menzies, A.M.; Carlino, M.S.; et al. Pre-operative ctDNA predicts survival in high-risk stage III cutaneous melanoma patients. Ann. Oncol. 2019, 30, 815–822. [Google Scholar] [CrossRef]
- Olbryt, M.; Pigłowski, W.; Rajczykowski, M.; Pfeifer, A.; Student, S.; Fiszer-Kierzkowska, A. Genetic Profiling of Advanced Melanoma: Candidate Mutations for Predicting Sensitivity and Resistance to Targeted Therapy. Target. Oncol. 2020, 15, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Long, G.V.; Fung, C.; Menzies, A.M.; Pupo, G.M.; Carlino, M.S.; Hyman, J.; Shahheydari, H.; Tembe, V.; Thompson, J.F.; Saw, R.P.; et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014, 5, 5694. [Google Scholar] [CrossRef] [PubMed]
- Wheler, J.; Yelensky, R.; Falchook, G.; Kim, K.B.; Hwu, P.; Tsimberidou, A.M.; Stephens, P.J.; Hong, D.; Cronin, M.T.; Kurzrock, R. Next generation sequencing of exceptional responders with BRAF-mutant melanoma: Implications for sensitivity and resistance. BMC Cancer 2015, 15, 61. [Google Scholar] [CrossRef] [PubMed][Green Version]
- LoRusso, P.M.; Boerner, S.A.; Pilat, M.J.; Forman, K.M.; Zuccaro, C.Y.; Kiefer, J.A.; Liang, W.S.; Hunsberger, S.; Redman, B.G.; Markovic, S.N.; et al. Pilot Trial of Selecting Molecularly Guided Therapy for Patients with Non-V600 BRAF-Mutant Metastatic Melanoma: Experience of the SU2C/MRA Melanoma Dream Team. Mol. Cancer Ther. 2015, 14, 1962–1971. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Wagle, N.; Sucker, A.; Treacy, D.J.; Johannessen, C.M.; Goetz, E.M.; Place, C.S.; Taylor-Weiner, A.; Whittaker, S.; Kryukov, G.V.; et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014, 4, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, C.M.; Boehm, J.S.; Kim, S.Y.; Thomas, S.R.; Wardwell, L.; Johnson, L.A.; Emery, C.M.; Stransky, N.; Cogdill, A.P.; Barretina, J.; et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 2010, 468, 968–972. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Song, Y.; Liu, D.; Liu, G.; Mao, X.; Dong, B.; Braicu, E.I.; Sehouli, J. Potential role of the HOXD8 transcription factor in cisplatin resistance and tumour metastasis in advanced epithelial ovarian cancer. Sci. Rep. 2018, 8, 13483. [Google Scholar] [CrossRef] [PubMed]
- Kurimoto, K.; Hayashi, M.; Guerrero-Preston, R.; Koike, M.; Kanda, M.; Hirabayashi, S.; Tanabe, H.; Takano, N.; Iwata, N.; Niwa, Y.; et al. PAX5 gene as a novel methylation marker that predicts both clinical outcome and cisplatin sensitivity in esophageal squamous cell carcinoma. Epigenetics 2017, 12, 865–874. [Google Scholar] [CrossRef]
- Jurmeister, P.; Wrede, N.; Hoffmann, I.; Vollbrecht, C.; Heim, D.; Hummel, M.; Wolkenstein, P.; Koch, I.; Heynol, V.; Schmitt, W.D.; et al. Mucosal melanomas of different anatomic sites share a common global DNA methylation profile with cutaneous melanoma but show location-dependent patterns of genetic and epigenetic alterations. J. Pathol. 2022, 256, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Marzese, D.M.; Scolyer, R.A.; Roqué, M.; Vargas-Roig, L.M.; Huynh, J.L.; Wilmott, J.S.; Murali, R.; Buckland, M.E.; Barkhoudarian, G.; Thompson, J.F.; et al. DNA methylation and gene deletion analysis of brain metastases in melanoma patients identifies mutually exclusive molecular alterations. Neuro Oncol. 2014, 16, 1499–1509. [Google Scholar] [CrossRef]
- Zocchi, L.; Lontano, A.; Merli, M.; Dika, E.; Nagore, E.; Quaglino, P.; Puig, S.; Ribero, S. Familial Melanoma and Susceptibility Genes: A Review of the Most Common Clinical and Dermoscopic Phenotypic Aspect, Associated Malignancies and Practical Tips for Management. J. Clin. Med. 2021, 10, 3760. [Google Scholar] [CrossRef]
- Ming, Z.; Lim, S.Y.; Rizos, H. Genetic Alterations in the INK4a/ARF Locus: Effects on Melanoma Development and Progression. Biomolecules 2020, 10, 1447. [Google Scholar] [CrossRef]
- Leachman, S.A.; Lucero, O.M.; Sampson, J.E.; Cassidy, P.; Bruno, W.; Queirolo, P.; Ghiorzo, P. Identification, genetic testing, and management of hereditary melanoma. Cancer Metastasis Rev. 2017, 36, 77–90. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Pellegrini, C.; Cardelli, L.; Ciciarelli, V.; Di Nardo, L.; Fargnoli, M.C. Familial Melanoma: Diagnostic and Management Implications. Dermatol. Pract. Concept. 2019, 9, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.; Fry, E.A. Aberrant Expression of p14ARF in human cancers: A new biomarker? Tumor Microenviron. 2018, 1, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Hemmer, R.M.; Sedivy, J.M. Role of p14(ARF) in replicative and induced senescence of human fibroblasts. Mol. Cell. Biol. 2001, 21, 6748–6757. [Google Scholar] [CrossRef]
- McConnell, B.B.; Gregory, F.J.; Stott, F.J.; Hara, E.; Peters, G. Induced expression of p16(INK4a) inhibits both CDK4- and CDK2-associated kinase activity by reassortment of cyclin-CDK-inhibitor complexes. Mol. Cell Biol. 1999, 19, 1981–1989. [Google Scholar] [CrossRef] [PubMed]
- Marzuka-Alcalá, A.; Gabree, M.J.; Tsao, H. Melanoma susceptibility genes and risk assessment. Methods Mol. Biol. 2014, 1102, 381–393. [Google Scholar] [CrossRef]
- Helgadottir, H.; Höiom, V.; Tuominen, R.; Jönsson, G.; Månsson-Brahme, E.; Olsson, H.; Hansson, J. CDKN2a mutation-negative melanoma families have increased risk exclusively for skin cancers but not for other malignancies. Int. J. Cancer 2015, 137, 2220–2226. [Google Scholar] [CrossRef]
- Burgstaller-Muehlbacher, S.; Marko, M.; Müller, C.; Wendt, J.; Pehamberger, H.; Okamoto, I. Novel CDKN2A mutations in Austrian melanoma patients. Melanoma Res. 2015, 25, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.; Baritaki, S.; Militello, L.; Malaponte, G.; Bevelacqua, Y.; Bonavida, B. The Role of B-RAF Mutations in Melanoma and the Induction of EMT via Dysregulation of the NF-κB/Snail/RKIP/PTEN Circuit. Genes Cancer 2010, 1, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Kiuru, M.; Tartar, D.M.; Qi, L.; Chen, D.; Yu, L.; Konia, T.; McPherson, J.D.; Murphy, W.J.; Fung, M.A. Improving classification of melanocytic nevi: Association of BRAF V600E expression with distinct histomorphologic features. J. Am. Acad. Dermatol. 2018, 79, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Ichii-Nakato, N.; Takata, M.; Takayanagi, S.; Takashima, S.; Lin, J.; Murata, H.; Fujimoto, A.; Hatta, N.; Saida, T. High frequency of BRAFV600E mutation in acquired nevi and small congenital nevi, but low frequency of mutation in medium-sized congenital nevi. J. Investig. Dermatol. 2006, 126, 2111–2118. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Berghoff, A.S.; Preusser, M.; Burgstaller-Muehlbacher, S.; Pehamberger, H.; Okamoto, I.; Kittler, H. NRAS and BRAF mutations in melanoma-associated nevi and uninvolved nevi. PLoS ONE 2013, 8, e69639. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.L.; Grant, P.A. The role of DNA methylation and histone modifications in transcriptional regulation in humans. Subcell. Biochem. 2013, 61, 289–317. [Google Scholar] [CrossRef]
- Garcia-Peterson, L.M.; Ndiaye, M.A.; Singh, C.K.; Chhabra, G.; Huang, W.; Ahmad, N. SIRT6 histone deacetylase functions as a potential oncogene in human melanoma. Genes. Cancer 2017, 8, 701–712. [Google Scholar] [CrossRef]
- Hanniford, D.; Ulloa-Morales, A.; Karz, A.; Berzoti-Coelho, M.G.; Moubarak, R.S.; Sánchez-Sendra, B.; Kloetgen, A.; Davalos, V.; Imig, J.; Wu, P.; et al. Epigenetic Silencing of CDR1as Drives IGF2BP3-Mediated Melanoma Invasion and Metastasis. Cancer Cell 2020, 37, 55–70.e15. [Google Scholar] [CrossRef]
- Santourlidis, S.; Schulz, W.A.; Araúzo-Bravo, M.J.; Gerovska, D.; Ott, P.; Bendhack, M.L.; Hassan, M.; Erichsen, L. Epigenetics in the Diagnosis and Therapy of Malignant Melanoma. Int. J. Mol. Sci. 2022, 23, 1531. [Google Scholar] [CrossRef] [PubMed]
- Sanna, A.; Phung, B.; Mitra, S.; Lauss, M.; Choi, J.; Zhang, T.; Njauw, C.N.; Cordero, E.; Harbst, K.; Rosengren, F.; et al. DNA promoter hypermethylation of melanocyte lineage genes determines melanoma phenotype. JCI Insight 2022, 7, e156577. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Eichhoff, O.M.; Widmer, D.S.; Praetorius, C.; Einarsson, S.O.; Valgeirsdottir, S.; Bergsteinsdottir, K.; Schepsky, A.; Dummer, R.; et al. Novel MITF targets identified using a two-step DNA microarray strategy. Pigment. Cell Melanoma Res. 2008, 21, 665–676. [Google Scholar] [CrossRef]
- Widmer, D.S.; Cheng, P.F.; Eichhoff, O.M.; Belloni, B.C.; Zipser, M.C.; Schlegel, N.C.; Javelaud, D.; Mauviel, A.; Dummer, R.; Hoek, K.S. Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment. Cell Melanoma Res. 2012, 25, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Giuliano, S.; Cheli, Y.; Ohanna, M.; Bonet, C.; Beuret, L.; Bille, K.; Loubat, A.; Hofman, V.; Hofman, P.; Ponzio, G.; et al. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res. 2010, 70, 3813–3822. [Google Scholar] [CrossRef] [PubMed]
- Mirmohammadsadegh, A.; Marini, A.; Nambiar, S.; Hassan, M.; Tannapfel, A.; Ruzicka, T.; Hengge, U.R. Epigenetic silencing of the PTEN gene in melanoma. Cancer Res. 2006, 66, 6546–6552. [Google Scholar] [CrossRef]
- Stahl, J.M.; Cheung, M.; Sharma, A.; Trivedi, N.R.; Shanmugam, S.; Robertson, G.P. Loss of PTEN promotes tumor development in malignant melanoma. Cancer Res. 2003, 63, 2881–2890. [Google Scholar] [PubMed]
- Yajima, I.; Kumasaka, M.Y.; Thang, N.D.; Goto, Y.; Takeda, K.; Yamanoshita, O.; Iida, M.; Ohgami, N.; Tamura, H.; Kawamoto, Y.; et al. RAS/RAF/MEK/ERK and PI3K/PTEN/AKT Signaling in Malignant Melanoma Progression and Therapy. Dermatol. Res. Pract. 2012, 2012, 354191. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Ahn, H.J.; Kim, Y.I. Methylation and mutation of the inhibin-α gene in human melanoma cells and regulation of PTEN expression and AKT/PI3K signaling by a demethylating agent. Oncol. Rep. 2022, 47, 37. [Google Scholar] [CrossRef] [PubMed]
- Easty, D.J.; Gray, S.G.; O’Byrne, K.J.; O’Donnell, D.; Bennett, D.C. Receptor tyrosine kinases and their activation in melanoma. Pigment. Cell Melanoma Res. 2011, 24, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Bello, D.M.; Ariyan, C.E.; Carvajal, R.D. Melanoma mutagenesis and aberrant cell signaling. Cancer Control 2013, 20, 261–281. [Google Scholar] [CrossRef]
- Sun, C.; Wang, L.; Huang, S.; Heynen, G.J.; Prahallad, A.; Robert, C.; Haanen, J.; Blank, C.; Wesseling, J.; Willems, S.M.; et al. Reversible and adaptive resistance to BRAF(V600E) inhibition in melanoma. Nature 2014, 508, 118–122. [Google Scholar] [CrossRef]
- McKenna, S.; García-Gutiérrez, L. Resistance to Targeted Therapy and RASSF1A Loss in Melanoma: What Are We Missing? Int. J. Mol. Sci. 2021, 22, 5115. [Google Scholar] [CrossRef]
- Verstraete, K.; Savvides, S.N. Extracellular assembly and activation principles of oncogenic class III receptor tyrosine kinases. Nat. Rev. Cancer 2012, 12, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Elias, D.; Ditzel, H.J. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol. Res. 2015, 100, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Meyers, B.A.; Czako, B.; Leonard, P.; Mseeh, F.; Harris, A.L.; Wu, Q.; Johnson, S.; Parker, C.A.; Cross, J.B.; et al. Allosteric SHP2 Inhibitor, IACS-13909, Overcomes EGFR-Dependent and EGFR-Independent Resistance Mechanisms toward Osimertinib. Cancer Res. 2020, 80, 4840–4853. [Google Scholar] [CrossRef] [PubMed]
- Sini, P.; Cannas, A.; Koleske, A.J.; Di Fiore, P.P.; Scita, G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 2004, 6, 268–274. [Google Scholar] [CrossRef]
- Du, Z.; Lovly, C.M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 2018, 17, 58. [Google Scholar] [CrossRef] [PubMed]
- Rea, K.; Sensi, M.; Anichini, A.; Canevari, S.; Tomassetti, A. EGFR/MEK/ERK/CDK5-dependent integrin-independent FAK phosphorylated on serine 732 contributes to microtubule depolymerization and mitosis in tumor cells. Cell Death Dis. 2013, 4, e815. [Google Scholar] [CrossRef] [PubMed]
- Chappell, W.H.; Steelman, L.S.; Long, J.M.; Kempf, R.C.; Abrams, S.L.; Franklin, R.A.; Bäsecke, J.; Stivala, F.; Donia, M.; Fagone, P.; et al. Ras/Raf/MEK/ERK and PI3K/PTEN/Akt/mTOR inhibitors: Rationale and importance to inhibiting these pathways in human health. Oncotarget 2011, 2, 135–164. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Maachani, U.B.; Schweitzer, M.; Singh, R.; Wang, M.; Chang, R.; Souweidane, M.M. Dual Inhibition of PI3K/AKT and MEK/ERK Pathways Induces Synergistic Antitumor Effects in Diffuse Intrinsic Pontine Glioma Cells. Transl. Oncol. 2017, 10, 221–228. [Google Scholar] [CrossRef]
- Savoia, P.; Zavattaro, E.; Cremona, O. Clinical Implications of Acquired BRAF Inhibitors Resistance in Melanoma. Int. J. Mol. Sci. 2020, 21, 9730. [Google Scholar] [CrossRef]
- Proietti, I.; Skroza, N.; Bernardini, N.; Tolino, E.; Balduzzi, V.; Marchesiello, A.; Michelini, S.; Volpe, S.; Mambrin, A.; Mangino, G.; et al. Mechanisms of Acquired BRAF Inhibitor Resistance in Melanoma: A Systematic Review. Cancers 2020, 12, 2801. [Google Scholar] [CrossRef]
- Yadav, V.; Zhang, X.; Liu, J.; Estrem, S.; Li, S.; Gong, X.Q.; Buchanan, S.; Henry, J.R.; Starling, J.J.; Peng, S.B. Reactivation of mitogen-activated protein kinase (MAPK) pathway by FGF receptor 3 (FGFR3)/Ras mediates resistance to vemurafenib in human B-RAF V600E mutant melanoma. J. Biol. Chem. 2012, 287, 28087–28098. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, P.I.; Sullivan, R.J.; Yaeger, R. Molecular Pathways and Mechanisms of BRAF in Cancer Therapy. Clin. Cancer Res. 2022, 28, 4618–4628. [Google Scholar] [CrossRef] [PubMed]
- McCain, J. The MAPK (ERK) Pathway: Investigational Combinations for the Treatment Of BRAF-Mutated Metastatic Melanoma. Pharm. Ther. 2013, 38, 96–108. [Google Scholar]
- Brummer, T.; Naegele, H.; Reth, M.; Misawa, Y. Identification of novel ERK-mediated feedback phosphorylation sites at the C-terminus of B-Raf. Oncogene 2003, 22, 8823–8834. [Google Scholar] [CrossRef] [PubMed]
- Ritt, D.A.; Monson, D.M.; Specht, S.I.; Morrison, D.K. Impact of feedback phosphorylation and Raf heterodimerization on normal and mutant B-Raf signaling. Mol. Cell. Biol. 2010, 30, 806–819. [Google Scholar] [CrossRef] [PubMed]
- Shao, C.; Dai, W.; Li, H.; Tang, W.; Jia, S.; Wu, X.; Luo, Y. The relationship between RASSF1A gene promoter methylation and the susceptibility and prognosis of melanoma: A meta-analysis and bioinformatics. PLoS ONE 2017, 12, e0171676. [Google Scholar] [CrossRef] [PubMed]
- Fröhlich, F.; Gerosa, L.; Muhlich, J.; Sorger, P.K. Mechanistic model of MAPK signaling reveals how allostery and rewiring contribute to drug resistance. Mol. Syst. Biol. 2023, 19, e10988. [Google Scholar] [CrossRef]
- Song, K.; Minami, J.K.; Huang, A.; Dehkordi, S.R.; Lomeli, S.H.; Luebeck, J.; Goodman, M.H.; Moriceau, G.; Krijgsman, O.; Dharanipragada, P.; et al. Plasticity of Extrachromosomal and Intrachromosomal BRAF Amplifications in Overcoming Targeted Therapy Dosage Challenges. Cancer Discov. 2022, 12, 1046–1069. [Google Scholar] [CrossRef]
- Bokharaie, H.; Kolch, W.; Krstic, A. Analysis of Alternative mRNA Splicing in Vemurafenib-Resistant Melanoma Cells. Biomolecules 2022, 12, 993. [Google Scholar] [CrossRef]
- Capoluongo, E. Insulin-like growth factor system and sporadic malignant melanoma. Am. J. Pathol. 2011, 178, 26–31. [Google Scholar] [CrossRef]
- Jiang, Y.Y.; Huang, H.; Wang, H.J.; Wu, D.; Yang, R.; Tashiro, S.; Onodera, S.; Ikejima, T. Interruption of mitochondrial complex IV activity and cytochrome c expression activated O2−-mediated cell survival in silibinin-treated human melanoma A375-S2 cells via IGF-1R-PI3K-Akt and IGF-1R-PLC γ-PKC pathways. Eur. J. Pharmacol. 2011, 668, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Nazarian, R.; Shi, H.; Wang, Q.; Kong, X.; Koya, R.C.; Lee, H.; Chen, Z.; Lee, M.K.; Attar, N.; Sazegar, H.; et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature 2010, 468, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, A.; Rapsomaniki, M.A.; Barros, V.; Fomitcheva-Khartchenko, A.; Martinelli, A.L.; Rodriguez, A.F.; Gabrani, M.; Rosen-Zvi, M.; Kaigala, G. Quantification of tumor heterogeneity: From data acquisition to metric generation. Trends Biotechnol. 2022, 40, 647–676. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtina, Y.; Moran, K.L.; Portal, M.M. Genetic and Non-Genetic Mechanisms Underlying Cancer Evolution. Cancers 2021, 13, 1380. [Google Scholar] [CrossRef] [PubMed]
- Morrison, S.J.; Kimble, J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature 2006, 441, 1068–1074. [Google Scholar] [CrossRef]
- Li, L.; Clevers, H. Coexistence of quiescent and active adult stem cells in mammals. Science 2010, 327, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Lang, D.; Mascarenhas, J.B.; Shea, C.R. Melanocytes, melanocyte stem cells, and melanoma stem cells. Clin. Dermatol. 2013, 31, 166–178. [Google Scholar] [CrossRef]
- Cabrera, M.C.; Hollingsworth, R.E.; Hurt, E.M. Cancer stem cell plasticity and tumor hierarchy. World J. Stem Cells 2015, 7, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Magee, J.A.; Piskounova, E.; Morrison, S.J. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell 2012, 21, 283–296. [Google Scholar] [CrossRef]
- Wiggans, M.; Pearson, B.J. One stem cell program to rule them all? FEBS J. 2021, 288, 3394–3406. [Google Scholar] [CrossRef] [PubMed]
- Gómez-López, S.; Lerner, R.G.; Petritsch, C. Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell. Mol. Life Sci. 2014, 71, 575–597. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Kong, J.; Brat, D.J. Cancer stem cell division: When the rules of asymmetry are broken. Stem Cells Dev. 2015, 24, 405–416. [Google Scholar] [CrossRef] [PubMed]
- Shahoumi, L.A. Oral Cancer Stem Cells: Therapeutic Implications and Challenges. Front. Oral Health 2021, 2, 685236. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Pestell, T.G.; Lisanti, M.P.; Pestell, R.G. Cancer stem cells. Int. J. Biochem. Cell Biol. 2012, 44, 2144–2151. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. Tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar] [CrossRef]
- Guo, R.; Fierro-Fine, A.; Goddard, L.; Russell, M.; Chen, J.; Liu, C.Z.; Fung, K.M.; Hassell, L.A. Increased expression of melanoma stem cell marker CD271 in metastatic melanoma to the brain. Int. J. Clin. Exp. Pathol. 2014, 7, 8947–8951. [Google Scholar] [PubMed]
- Parmiani, G. Melanoma Cancer Stem Cells: Markers and Functions. Cancers 2016, 8, 34. [Google Scholar] [CrossRef]
- Perego, M.; Tortoreto, M.; Tragni, G.; Mariani, L.; Deho, P.; Carbone, A.; Santinami, M.; Patuzzo, R.; Mina, P.D.; Villa, A.; et al. Heterogeneous phenotype of human melanoma cells with in vitro and in vivo features of tumor-initiating cells. J. Investig. Dermatol. 2010, 130, 1877–1886. [Google Scholar] [CrossRef]
- Kumar, V.; Vashishta, M.; Kong, L.; Wu, X.; Lu, J.J.; Guha, C.; Dwarakanath, B.S. The Role of Notch, Hedgehog, and Wnt Signaling Pathways in the Resistance of Tumors to Anticancer Therapies. Front. Cell Dev. Biol. 2021, 9, 650772. [Google Scholar] [CrossRef]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Nakahata, K.; Uehara, S.; Nishikawa, S.; Kawatsu, M.; Zenitani, M.; Oue, T.; Okuyama, H. Aldehyde Dehydrogenase 1 (ALDH1) Is a Potential Marker for Cancer Stem Cells in Embryonal Rhabdomyosarcoma. PLoS ONE 2015, 10, e0125454. [Google Scholar] [CrossRef]
- Gerard, L.; Duviver, L.; Fourrez, M.; Salazar, P.; Sprimont, L.; Xia, D.; Ambudkar, S.V.; Gottesman, M.M.; Gillet, J.P. Identification of two novel heterodimeric ABC transporters in melanoma: ABCB5β/B6 and ABCB5β/B9. J. Biol. Chem. 2023, 300, 105594. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.J.; Saab, K.R.; Ma, J.; Schatton, T.; Pütz, P.; Zhan, Q.; Murphy, G.F.; Gasser, M.; Waaga-Gasser, A.M.; Frank, N.Y.; et al. ABCB5 maintains melanoma-initiating cells through a proinflammatory cytokine signaling circuit. Cancer Res. 2014, 74, 4196–4207. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.G.; Valencia, J.C.; Gillet, J.P.; Hearing, V.J.; Gottesman, M.M. Involvement of ABC transporters in melanogenesis and the development of multidrug resistance of melanoma. Pigment. Cell Melanoma Res. 2009, 22, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Tudrej, K.B.; Czepielewska, E.; Kozłowska-Wojciechowska, M. SOX10-MITF pathway activity in melanoma cells. Arch. Med. Sci. 2017, 13, 1493–1503. [Google Scholar] [CrossRef]
- Boivin, D.; Labbé, D.; Fontaine, N.; Lamy, S.; Beaulieu, E.; Gingras, D.; Béliveau, R. The stem cell marker CD133 (prominin-1) is phosphorylated on cytoplasmic tyrosine-828 and tyrosine-852 by Src and Fyn tyrosine kinases. Biochemistry 2009, 48, 3998–4007. [Google Scholar] [CrossRef]
- González-Herrero, I.; Romero-Camarero, I.; Cañueto, J.; Cardeñoso-Álvarez, E.; Fernández-López, E.; Pérez-Losada, J.; Sánchez-García, I.; Román-Curto, C. CD133+ cell content correlates with tumour growth in melanomas from skin with chronic sun-induced damage. Br. J. Dermatol. 2013, 169, 830–837. [Google Scholar] [CrossRef]
- Madjd, Z.; Erfani, E.; Gheytanchi, E.; Moradi-Lakeh, M.; Shariftabrizi, A.; Asadi-Lari, M. Expression of CD133 cancer stem cell marker in melanoma: A systematic review and meta-analysis. Int. J. Biol. Markers 2016, 31, e118–e125. [Google Scholar] [CrossRef]
- Liou, G.Y. CD133 as a regulator of cancer metastasis through the cancer stem cells. Int. J. Biochem. Cell Biol. 2019, 106, 1–7. [Google Scholar] [CrossRef]
- Lai, I.C.; Shih, P.H.; Yao, C.J.; Yeh, C.T.; Wang-Peng, J.; Lui, T.N.; Chuang, S.E.; Hu, T.S.; Lai, T.Y.; Lai, G.M. Elimination of cancer stem-like cells and potentiation of temozolomide sensitivity by Honokiol in glioblastoma multiforme cells. PLoS ONE 2015, 10, e0114830. [Google Scholar] [CrossRef]
- Yang, Y.P.; Chien, Y.; Chiou, G.Y.; Cherng, J.Y.; Wang, M.L.; Lo, W.L.; Chang, Y.L.; Huang, P.I.; Chen, Y.W.; Shih, Y.H.; et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microRNA145 with cationic polyurethane-short branch PEI. Biomaterials 2012, 33, 1462–1476. [Google Scholar] [CrossRef] [PubMed]
- Tseng, L.M.; Huang, P.I.; Chen, Y.R.; Chen, Y.C.; Chou, Y.C.; Chen, Y.W.; Chang, Y.L.; Hsu, H.S.; Lan, Y.T.; Chen, K.H.; et al. Targeting signal transducer and activator of transcription 3 pathway by cucurbitacin I diminishes self-renewing and radiochemoresistant abilities in thyroid cancer-derived CD133+ cells. J. Pharmacol. Exp. Ther. 2012, 341, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Barzegar Behrooz, A.; Syahir, A.; Ahmad, S. CD133: Beyond a cancer stem cell biomarker. J. Drug Target. 2019, 27, 257–269. [Google Scholar] [CrossRef] [PubMed]
- Afify, S.M.; Seno, M. Conversion of Stem Cells to Cancer Stem Cells: Undercurrent of Cancer Initiation. Cancers 2019, 11, 345. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.Z. Origin of cancer stem cells: The role of self-renewal and differentiation. Ann. Surg. Oncol. 2008, 15, 407–414. [Google Scholar] [CrossRef]
- Redmer, T.; Walz, I.; Klinger, B.; Khouja, S.; Welte, Y.; Schäfer, R.; Regenbrecht, C. The role of the cancer stem cell marker CD271 in DNA damage response and drug resistance of melanoma cells. Oncogenesis 2017, 6, e291. [Google Scholar] [CrossRef]
- Alqathama, A. BRAF in malignant melanoma progression and metastasis: Potentials and challenges. Am. J. Cancer Res. 2020, 10, 1103–1114. [Google Scholar]
- Tanda, E.T.; Vanni, I.; Boutros, A.; Andreotti, V.; Bruno, W.; Ghiorzo, P.; Spagnolo, F. Current State of Target Treatment in BRAF Mutated Melanoma. Front. Mol. Biosci. 2020, 7, 154. [Google Scholar] [CrossRef] [PubMed]
- Yushak, M.; Chapman, P.; Robert, C.; Kudchadkar, R. Systemic Therapy Options for Patients with Unresectable Melanoma. Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 661–672. [Google Scholar] [CrossRef]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): A multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 1315–1327. [Google Scholar] [CrossRef]
- Sullivan, R.J.; Flaherty, K.T. Resistance to BRAF-targeted therapy in melanoma. Eur. J. Cancer 2013, 49, 1297–1304. [Google Scholar] [CrossRef]
- Trinh, V.A.; You, Y.; Hwu, W.J. Treatment of BRAF-mutated advanced cutaneous melanoma. Chin. Clin. Oncol. 2014, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Chapman, P.B.; Hauschild, A.; Robert, C.; Haanen, J.B.; Ascierto, P.; Larkin, J.; Dummer, R.; Garbe, C.; Testori, A.; Maio, M.; et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N. Engl. J. Med. 2011, 364, 2507–2516. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, M.I.; Widmer, D.S.; Narechania, A.; Eichhoff, O.; Freiberger, S.N.; Wenzina, J.; Cheng, P.F.; Mihic-Probst, D.; Desalle, R.; Dummer, R.; et al. Co-existence of BRAF and NRAS driver mutations in the same melanoma cells results in heterogeneity of targeted therapy resistance. Oncotarget 2016, 7, 77163–77174. [Google Scholar] [CrossRef] [PubMed]
- Randic, T.; Kozar, I.; Margue, C.; Utikal, J.; Kreis, S. NRAS mutant melanoma: Towards better therapies. Cancer Treat. Rev. 2021, 99, 102238. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Bi, Z.; Liu, Y.; Qin, F.; Wei, Y.; Wei, X. Targeting RAS-RAF-MEK-ERK signaling pathway in human cancer: Current status in clinical trials. Genes. Dis. 2023, 10, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Czarnecka, A.M.; Bartnik, E.; Fiedorowicz, M.; Rutkowski, P. Targeted Therapy in Melanoma and Mechanisms of Resistance. Int. J. Mol. Sci. 2020, 21, 4576. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Carvajal, R.; Kim, M.; Yang, H.W. Kinetics of RTK activation determine ERK reactivation and resistance to dual BRAF/MEK inhibition in melanoma. Cell Rep. 2023, 42, 112570. [Google Scholar] [CrossRef]
- Atefi, M.; Titz, B.; Tsoi, J.; Avramis, E.; Le, A.; Ng, C.; Lomova, A.; Lassen, A.; Friedman, M.; Chmielowski, B.; et al. CRAF R391W is a melanoma driver oncogene. Sci. Rep. 2016, 6, 27454. [Google Scholar] [CrossRef]
- Yin, Q.; Shi, X.; Lan, S.; Jin, H.; Wu, D. Effect of melanoma stem cells on melanoma metastasis. Oncol. Lett. 2021, 22, 566. [Google Scholar] [CrossRef]
- Giunta, E.F.; De Falco, V.; Napolitano, S.; Argenziano, G.; Brancaccio, G.; Moscarella, E.; Ciardiello, D.; Ciardiello, F.; Troiani, T. Optimal treatment strategy for metastatic melanoma patients harboring. Ther. Adv. Med. Oncol. 2020, 12, 1758835920925219. [Google Scholar] [CrossRef]
- Griffin, M.; Scotto, D.; Josephs, D.H.; Mele, S.; Crescioli, S.; Bax, H.J.; Pellizzari, G.; Wynne, M.D.; Nakamura, M.; Hoffmann, R.M.; et al. BRAF inhibitors: Resistance and the promise of combination treatments for melanoma. Oncotarget 2017, 8, 78174–78192. [Google Scholar] [CrossRef] [PubMed]
- Fisher, M.L.; Grun, D.; Adhikary, G.; Xu, W.; Eckert, R.L. Inhibition of YAP function overcomes BRAF inhibitor resistance in melanoma cancer stem cells. Oncotarget 2017, 8, 110257–110272. [Google Scholar] [CrossRef] [PubMed]
- Grigore, F.; Yang, H.; Hanson, N.D.; VanBrocklin, M.W.; Sarver, A.L.; Robinson, J.P. BRAF inhibition in melanoma is associated with the dysregulation of histone methylation and histone methyltransferases. Neoplasia 2020, 22, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Vultur, A.; Villanueva, J.; Herlyn, M. BRAF inhibitor unveils its potential against advanced melanoma. Cancer Cell 2010, 18, 301–302. [Google Scholar] [CrossRef] [PubMed]
- Neudauer, C.L.; McCarthy, J.B. Insulin-like growth factor I-stimulated melanoma cell migration requires phosphoinositide 3-kinase but not extracellular-regulated kinase activation. Exp. Cell Res. 2003, 286, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Osrodek, M.; Rozanski, M.; Czyz, M. Insulin Reduces the Efficacy of Vemurafenib and Trametinib in Melanoma Cells. Cancer Manag. Res. 2020, 12, 7231–7250. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Ye, Y.; Zhang, X.D.; Chen, J. Insulin induces drug resistance in melanoma through activation of the PI3K/Akt pathway. Drug Dev. Ther. 2014, 8, 255–262. [Google Scholar] [CrossRef]
- Hilmi, C.; Larribere, L.; Giuliano, S.; Bille, K.; Ortonne, J.P.; Ballotti, R.; Bertolotto, C. IGF1 promotes resistance to apoptosis in melanoma cells through an increased expression of BCL2, BCL-X(L), and survivin. J. Investig. Dermatol. 2008, 128, 1499–1505. [Google Scholar] [CrossRef]
- Winder, M.; Virós, A. Mechanisms of Drug Resistance in Melanoma. Handb. Exp. Pharmacol. 2018, 249, 91–108. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Z.; Qi, F. Aldehyde dehydrogenase-positive melanoma stem cells in tumorigenesis, drug resistance and anti-neoplastic immunotherapy. Mol. Biol. Rep. 2020, 47, 1435–1443. [Google Scholar] [CrossRef] [PubMed]
- Xanthis, V.; Mantso, T.; Dimtsi, A.; Pappa, A.; Fadouloglou, V.E. Human Aldehyde Dehydrogenases: A Superfamily of Similar Yet Different Proteins Highly Related to Cancer. Cancers 2023, 15, 4419. [Google Scholar] [CrossRef] [PubMed]
- Meszaros, P.; Hummel, I.; Klappe, K.; Draghiciu, O.; Hoekstra, D.; Kok, J.W. The function of the ATP-binding cassette (ABC) transporter ABCB1 is not susceptible to actin disruption. Biochim. Biophys. Acta 2013, 1828, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Begicevic, R.R.; Falasca, M. ABC Transporters in Cancer Stem Cells: Beyond Chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef] [PubMed]
- Clark, D.W.; Palle, K. Aldehyde dehydrogenases in cancer stem cells: Potential as therapeutic targets. Ann. Transl. Med. 2016, 4, 518. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, M.; Bravaccini, S.; Fabbri, F.; Arienti, C. Emerging Roles of Aldehyde Dehydrogenase Isoforms in Anti-cancer Therapy Resistance. Front. Med. 2022, 9, 795762. [Google Scholar] [CrossRef] [PubMed]
- Arnold, C.R.; Mangesius, J.; Skvortsova, I.I.; Ganswindt, U. The Role of Cancer Stem Cells in Radiation Resistance. Front. Oncol. 2020, 10, 164. [Google Scholar] [CrossRef]
- Gómez-Millán, J.; Katz, I.S.S.; de Araujo Farias, V.; Linares-Fernández, J.-L.; López-Peñalver, J.; Ortiz-Ferrón, G.; Ruiz-Ruiz, C.; Oliver, F.J.; Ruiz de Almodóvar, J.M. The importance of bystander effects in radiation therapy in melanoma skin-cancer cells and umbilical-cord stromal stem cells. Radiother. Oncol. 2012, 102, 450–458. [Google Scholar] [CrossRef]
- Sanlorenzo, M.; Vujic, I.; Posch, C.; Dajee, A.; Yen, A.; Kim, S.; Ashworth, M.; Rosenblum, M.D.; Algazi, A.; Osella-Abate, S.; et al. Melanoma immunotherapy. Cancer Biol. Ther. 2014, 15, 665–674. [Google Scholar] [CrossRef]
- Atkins, M.B.; Kunkel, L.; Sznol, M.; Rosenberg, S.A. High-dose recombinant interleukin-2 therapy in patients with metastatic melanoma: Long-term survival update. Cancer J. Sci. Am. 2000, 6 (Suppl. S1), S11–S14. [Google Scholar]
- Kobeissi, I.; Tarhini, A.A. Systemic adjuvant therapy for high-risk cutaneous melanoma. Ther. Adv. Med. Oncol. 2022, 14, 17588359221134087. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Kirkwood, J.M.; Hodi, F.S.; Agarwala, S.; Amatruda, T.; Bines, S.D.; Clark, J.I.; Curti, B.; Ernstoff, M.S.; Gajewski, T.; et al. The Society for Immunotherapy of Cancer consensus statement on tumour immunotherapy for the treatment of cutaneous melanoma. Nat. Rev. Clin. Oncol. 2013, 10, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Payne, M.J.; Argyropoulou, K.; Lorigan, P.; McAleer, J.J.; Farrugia, D.; Davidson, N.; Kelly, C.; Chao, D.; Marshall, E.; Han, C.; et al. Phase II pilot study of intravenous high-dose interferon with or without maintenance treatment in melanoma at high risk of recurrence. J. Clin. Oncol. 2014, 32, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.B.; Legha, S.S.; Gonzalez, R.; Anderson, C.M.; Johnson, M.M.; Liu, P.; Papadopoulos, N.E.; Eton, O.; Plager, C.; Buzaid, A.C.; et al. A randomized phase III trial of biochemotherapy versus interferon-alpha-2b for adjuvant therapy in patients at high risk for melanoma recurrence. Melanoma Res. 2009, 19, 42–49. [Google Scholar] [CrossRef]
- Di Trolio, R.; Simeone, E.; Di Lorenzo, G.; Grimaldi, A.M.; Romano, A.; Ayala, F.; Caracò, C.; Mozzillo, N.; Ascierto, P.A. Update on PEG-interferon α-2b as adjuvant therapy in melanoma. Anticancer Res. 2012, 32, 3901–3909. [Google Scholar]
- Hwu, W.J.; Panageas, K.S.; Menell, J.H.; Lamb, L.A.; Aird, S.; Krown, S.E.; Williams, L.J.; Chapman, P.B.; Livingston, P.O.; Wolchok, J.D.; et al. Phase II study of temozolomide plus pegylated interferon-alpha-2b for metastatic melanoma. Cancer 2006, 106, 2445–2451. [Google Scholar] [CrossRef] [PubMed]
- Spieth, K.; Kaufmann, R.; Dummer, R.; Garbe, C.; Becker, J.C.; Hauschild, A.; Tilgen, W.; Ugurel, S.; Beyeler, M.; Bröcker, E.B.; et al. Temozolomide plus pegylated interferon alfa-2b as first-line treatment for stage IV melanoma: A multicenter phase II trial of the Dermatologic Cooperative Oncology Group (DeCOG). Ann. Oncol. 2008, 19, 801–806. [Google Scholar] [CrossRef]
- Addeo, R.; Zappavigna, S.; Luce, A.; Facchini, S.; Caraglia, M. Chemotherapy in the management of brain metastases: The emerging role of fotemustine for patients with melanoma and NSCLC. Expert. Opin. Drug Saf. 2013, 12, 729–740. [Google Scholar] [CrossRef]
- Harari, A.; Graciotti, M.; Bassani-Sternberg, M.; Kandalaft, L.E. Antitumour dendritic cell vaccination in a priming and boosting approach. Nat. Rev. Drug Discov. 2020, 19, 635–652. [Google Scholar] [CrossRef]
- Thurner, B.; Haendle, I.; Röder, C.; Dieckmann, D.; Keikavoussi, P.; Jonuleit, H.; Bender, A.; Maczek, C.; Schreiner, D.; von dem Driesch, P.; et al. Vaccination with mage-3A1 peptide-pulsed mature, monocyte-derived dendritic cells expands specific cytotoxic T cells and induces regression of some metastases in advanced stage IV melanoma. J. Exp. Med. 1999, 190, 1669–1678. [Google Scholar] [CrossRef]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef]
- Rosenberg, S.A.; Zhai, Y.; Yang, J.C.; Schwartzentruber, D.J.; Hwu, P.; Marincola, F.M.; Topalian, S.L.; Restifo, N.P.; Seipp, C.A.; Einhorn, J.H.; et al. Immunizing patients with metastatic melanoma using recombinant adenoviruses encoding MART-1 or gp100 melanoma antigens. J. Natl. Cancer Inst. 1998, 90, 1894–1900. [Google Scholar] [CrossRef] [PubMed]
- Shang, N.; Figini, M.; Shangguan, J.; Wang, B.; Sun, C.; Pan, L.; Ma, Q.; Zhang, Z. Dendritic cells based immunotherapy. Am. J. Cancer Res. 2017, 7, 2091–2102. [Google Scholar] [PubMed]
- Toh, H.C.; Wang, W.W.; Chia, W.K.; Kvistborg, P.; Sun, L.; Teo, K.; Phoon, Y.P.; Soe, Y.; Tan, S.H.; Hee, S.W.; et al. Clinical Benefit of Allogeneic Melanoma Cell Lysate-Pulsed Autologous Dendritic Cell Vaccine in MAGE-Positive Colorectal Cancer Patients. Clin. Cancer Res. 2009, 15, 7726–7736. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.J.; Rosenberg, S.A.; Dudley, M.E.; Yang, J.C.; White, D.E.; Butman, J.A.; Sherry, R.M. Successful treatment of melanoma brain metastases with adoptive cell therapy. Clin. Cancer Res. 2010, 16, 4892–4898. [Google Scholar] [CrossRef]
- Zhang, Y.; Zheng, J. Functions of Immune Checkpoint Molecules Beyond Immune Evasion. Adv. Exp. Med. Biol. 2020, 1248, 201–226. [Google Scholar] [CrossRef]
- Seidel, J.A.; Otsuka, A.; Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front. Oncol. 2018, 8, 86. [Google Scholar] [CrossRef]
- Buchbinder, E.I.; Desai, A. CTLA-4 and PD-1 Pathways: Similarities, Differences, and Implications of Their Inhibition. Am. J. Clin. Oncol. 2016, 39, 98–106. [Google Scholar] [CrossRef]
- Shiravand, Y.; Khodadadi, F.; Kashani, S.M.A.; Hosseini-Fard, S.R.; Hosseini, S.; Sadeghirad, H.; Ladwa, R.; O’Byrne, K.; Kulasinghe, A. Immune Checkpoint Inhibitors in Cancer Therapy. Curr. Oncol. 2022, 29, 3044–3060. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, H.; Shiota, G. Immune evasion by cancer stem cells. Regen. Ther. 2021, 17, 20–33. [Google Scholar] [CrossRef]
- Wu, B.; Shi, X.; Jiang, M.; Liu, H. Cross-talk between cancer stem cells and immune cells: Potential therapeutic targets in the tumor immune microenvironment. Mol. Cancer 2023, 22, 38. [Google Scholar] [CrossRef] [PubMed]
- Bruttel, V.S.; Wischhusen, J. Cancer stem cell immunology: Key to understanding tumorigenesis and tumor immune escape? Front. Immunol. 2014, 5, 360. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Volontè, A.; Cimminiello, C.; Parmiani, G. Immunology of cancer stem cells in solid tumours. A review. Eur. J. Cancer 2014, 50, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, F.; Shields, B.; Makhoul, I.; Avaritt, N.; Wong, H.K.; Hutchins, L.F.; Shalin, S.; Tackett, A.J. Immune surveillance in melanoma: From immune attack to melanoma escape and even counterattack. Cancer Biol. Ther. 2017, 18, 451–469. [Google Scholar] [CrossRef]
- Passarelli, A.; Mannavola, F.; Stucci, L.S.; Tucci, M.; Silvestris, F. Immune system and melanoma biology: A balance between immunosurveillance and immune escape. Oncotarget 2017, 8, 106132–106142. [Google Scholar] [CrossRef] [PubMed]
- Errington, J.A.; Conway, R.M.; Walsh-Conway, N.; Browning, J.; Freyer, C.; Cebon, J.; Madigan, M.C. Expression of cancer-testis antigens (MAGE-A1, MAGE-A3/6, MAGE-A4, MAGE-C1 and NY-ESO-1) in primary human uveal and conjunctival melanoma. Br. J. Ophthalmol. 2012, 96, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Aris, M.; Zubieta, M.R.; Colombo, M.; Arriaga, J.M.; Bianchini, M.; Alperovich, M.; Bravo, A.I.; Barrio, M.M.; Mordoh, J. MART-1- and gp100-expressing and -non-expressing melanoma cells are equally proliferative in tumors and clonogenic in vitro. J. Investig. Dermatol. 2012, 132, 365–374. [Google Scholar] [CrossRef]
- Gonzalez, H.; Hagerling, C.; Werb, Z. Roles of the immune system in cancer: From tumor initiation to metastatic progression. Genes Dev. 2018, 32, 1267–1284. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef]
- Elliott, A.M.; Al-Hajj, M.A. ABCB8 mediates doxorubicin resistance in melanoma cells by protecting the mitochondrial genome. Mol. Cancer Res. 2009, 7, 79–87. [Google Scholar] [CrossRef]
- Fukunaga-Kalabis, M.; Herlyn, M. Beyond ABC: Another mechanism of drug resistance in melanoma side population. J. Investig. Dermatol. 2012, 132, 2317–2319. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.H.; Yu, A.M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr. Pharm. Des. 2014, 20, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Catalano, A.; Iacopetta, D.; Ceramella, J.; Scumaci, D.; Giuzio, F.; Saturnino, C.; Aquaro, S.; Rosano, C.; Sinicropi, M.S. Multidrug Resistance (MDR): A Widespread Phenomenon in Pharmacological Therapies. Molecules 2022, 27, 616. [Google Scholar] [CrossRef]
- Frank, N.Y.; Margaryan, A.; Huang, Y.; Schatton, T.; Waaga-Gasser, A.M.; Gasser, M.; Sayegh, M.H.; Sadee, W.; Frank, M.H. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005, 65, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Keniya, M.V.; Holmes, A.R.; Niimi, M.; Lamping, E.; Gillet, J.P.; Gottesman, M.M.; Cannon, R.D. Drug resistance is conferred on the model yeast Saccharomyces cerevisiae by expression of full-length melanoma-associated human ATP-binding cassette transporter ABCB5. Mol. Pharm. 2014, 11, 3452–3462. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Egger, M.E.; McMasters, K.M.; Hao, H. Differential expression of ABCB5 in BRAF inhibitor-resistant melanoma cell lines. BMC Cancer 2018, 18, 675. [Google Scholar] [CrossRef]
- Schmitt, M.; Sinnberg, T.; Nalpas, N.C.; Maass, A.; Schittek, B.; Macek, B. Quantitative Proteomics Links the Intermediate Filament Nestin to Resistance to Targeted BRAF Inhibition in Melanoma Cells. Mol. Cell. Proteomics 2019, 18, 1096–1109. [Google Scholar] [CrossRef]
- Beretti, F.; Gatti, M.; Zavatti, M.; Bassoli, S.; Pellacani, G.; Maraldi, T. Reactive Oxygen Species Regulation of Chemoresistance and Metastatic Capacity of Melanoma: Role of the Cancer Stem Cell Marker CD271. Biomedicines 2023, 11, 1229. [Google Scholar] [CrossRef]
- Pisanu, M.E.; Maugeri-Saccà, M.; Fattore, L.; Bruschini, S.; De Vitis, C.; Tabbì, E.; Bellei, B.; Migliano, E.; Kovacs, D.; Camera, E.; et al. Inhibition of Stearoyl-CoA desaturase 1 reverts BRAF and MEK inhibition-induced selection of cancer stem cells in BRAF-mutated melanoma. J. Exp. Clin. Cancer Res. 2018, 37, 318. [Google Scholar] [CrossRef]
- Vidal, A.; Redmer, T. Decoding the Role of CD271 in Melanoma. Cancers 2020, 12, 2460. [Google Scholar] [CrossRef]
- Restivo, G.; Diener, J.; Cheng, P.F.; Kiowski, G.; Bonalli, M.; Biedermann, T.; Reichmann, E.; Levesque, M.P.; Dummer, R.; Sommer, L. low neurotrophin receptor CD271 regulates phenotype switching in melanoma. Nat. Commun. 2017, 8, 1988. [Google Scholar] [CrossRef] [PubMed]
- Radke, J.; Roßner, F.; Redmer, T. CD271 determines migratory properties of melanoma cells. Sci. Rep. 2017, 7, 9834. [Google Scholar] [CrossRef] [PubMed]
- Saltari, A.; Truzzi, F.; Quadri, M.; Lotti, R.; Palazzo, E.; Grisendi, G.; Tiso, N.; Marconi, A.; Pincelli, C. CD271 Down-Regulation Promotes Melanoma Progression and Invasion in Three-Dimensional Models and in Zebrafish. J. Investig. Dermatol. 2016, 136, 2049–2058. [Google Scholar] [CrossRef] [PubMed]
- Neagu, M.; Constantin, C.; Caruntu, C.; Dumitru, C.; Surcel, M.; Zurac, S. Inflammation: A key process in skin tumorigenesis. Oncol. Lett. 2019, 17, 4068–4084. [Google Scholar] [CrossRef] [PubMed]
- Nataraj, N.B.; Marrocco, I.; Yarden, Y. Roles for growth factors and mutations in metastatic dissemination. Biochem. Soc. Trans. 2021, 49, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Straussman, R.; Morikawa, T.; Shee, K.; Barzily-Rokni, M.; Qian, Z.R.; Du, J.; Davis, A.; Mongare, M.M.; Gould, J.; Frederick, D.T.; et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature 2012, 487, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Cools-Lartigue, J.; Marshall, J.C.; Caissie, A.L.; Saraiva, V.S.; Burnier, M.N. Secretion of hepatocyte growth factor and vascular endothelial growth factor during uveal melanoma-monocyte in vitro interactions. Melanoma Res. 2005, 15, 141–145. [Google Scholar] [CrossRef]
- Fedorenko, I.V.; Wargo, J.A.; Flaherty, K.T.; Messina, J.L.; Smalley, K.S.M. BRAF Inhibition Generates a Host-Tumor Niche that Mediates Therapeutic Escape. J. Investig. Dermatol. 2015, 135, 3115–3124. [Google Scholar] [CrossRef]
- Luebker, S.A.; Koepsell, S.A. Diverse Mechanisms of BRAF Inhibitor Resistance in Melanoma Identified in Clinical and Preclinical Studies. Front. Oncol. 2019, 9, 268. [Google Scholar] [CrossRef]
- Manzano, J.L.; Layos, L.; Bugés, C.; de Los Llanos Gil, M.; Vila, L.; Martínez-Balibrea, E.; Martínez-Cardús, A. Resistant mechanisms to BRAF inhibitors in melanoma. Ann. Transl. Med. 2016, 4, 237. [Google Scholar] [CrossRef]
- Liu, T.; Zhou, L.; Xiao, Y.; Andl, T.; Zhang, Y. BRAF Inhibitors Reprogram Cancer-Associated Fibroblasts to Drive Matrix Remodeling and Therapeutic Escape in Melanoma. Cancer Res. 2022, 82, 419–432. [Google Scholar] [CrossRef] [PubMed]
- Flach, E.H.; Rebecca, V.W.; Herlyn, M.; Smalley, K.S.; Anderson, A.R. Fibroblasts contribute to melanoma tumor growth and drug resistance. Mol. Pharm. 2011, 8, 2039–2049. [Google Scholar] [CrossRef] [PubMed]
- Pretti, M.A.M.; Bernardes, S.S.; da Cruz, J.G.V.; Boroni, M.; Possik, P.A. Extracellular vesicle-mediated crosstalk between melanoma and the immune system: Impact on tumor progression and therapy response. J. Leukoc. Biol. 2020, 108, 1101–1115. [Google Scholar] [CrossRef] [PubMed]
- Castellani, G.; Buccarelli, M.; Arasi, M.B.; Rossi, S.; Pisanu, M.E.; Bellenghi, M.; Lintas, C.; Tabolacci, C. BRAF Mutations in Melanoma: Biological Aspects, Therapeutic Implications, and Circulating Biomarkers. Cancers 2023, 15, 4026. [Google Scholar] [CrossRef] [PubMed]
- Thompson, E.L.; Hu, J.J.; Niedernhofer, L.J. The Role of Senescent Cells in Acquired Drug Resistance and Secondary Cancer in BRAFi-Treated Melanoma. Cancers 2021, 13, 2241. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front. Cell Dev. Biol. 2021, 9, 645593. [Google Scholar] [CrossRef]
- Herranz, N.; Gil, J. Mechanisms and functions of cellular senescence. J. Clin. Investig. 2018, 128, 1238–1246. [Google Scholar] [CrossRef]
- Peng, J.; Lin, Z.; Chen, W.; Ruan, J.; Deng, F.; Yao, L.; Rao, M.; Xiong, X.; Xu, S.; Zhang, X.; et al. Vemurafenib induces a noncanonical senescence-associated secretory phenotype in melanoma cells which promotes vemurafenib resistance. Heliyon 2023, 9, e17714. [Google Scholar] [CrossRef]
- Paskaš, S.; Krajnović, T.; Basile, M.S.; Dunđerović, D.; Cavalli, E.; Mangano, K.; Mammana, S.; Al-Abed, Y.; Nicoletti, F.; Mijatović, S.; et al. Senescence as a main mechanism of Ritonavir and Ritonavir-NO action against melanoma. Mol. Carcinog. 2019, 58, 1362–1375. [Google Scholar] [CrossRef]
- Ou, H.L.; Hoffmann, R.; González-López, C.; Doherty, G.J.; Korkola, J.E.; Muñoz-Espín, D. Cellular senescence in cancer: From mechanisms to detection. Mol. Oncol. 2021, 15, 2634–2671. [Google Scholar] [CrossRef]
- Laberge, R.M.; Sun, Y.; Orjalo, A.V.; Patil, C.K.; Freund, A.; Zhou, L.; Curran, S.C.; Davalos, A.R.; Wilson-Edell, K.A.; Liu, S.; et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 2015, 17, 1049–1061. [Google Scholar] [CrossRef]
- Homann, L.; Rentschler, M.; Brenner, E.; Böhm, K.; Röcken, M.; Wieder, T. IFN-γ and TNF Induce Senescence and a Distinct Senescence-Associated Secretory Phenotype in Melanoma. Cells 2022, 11, 1514. [Google Scholar] [CrossRef] [PubMed]
- Jochems, F.; Thijssen, B.; De Conti, G.; Jansen, R.; Pogacar, Z.; Groot, K.; Wang, L.; Schepers, A.; Wang, C.; Jin, H.; et al. The Cancer SENESCopedia: A delineation of cancer cell senescence. Cell Rep. 2021, 36, 109441. [Google Scholar] [CrossRef] [PubMed]
- Malaquin, N.; Olivier, M.A.; Martinez, A.; Nadeau, S.; Sawchyn, C.; Coppé, J.P.; Cardin, G.; Mallette, F.A.; Campisi, J.; Rodier, F. Non-canonical ATM/MRN activities temporally define the senescence secretory program. EMBO Rep. 2020, 21, e50718. [Google Scholar] [CrossRef] [PubMed]
- Birch, J.; Gil, J. Senescence and the SASP: Many therapeutic avenues. Genes Dev. 2020, 34, 1565–1576. [Google Scholar] [CrossRef]
- Rodriguez-Ruiz, M.E.; Vitale, I.; Harrington, K.J.; Melero, I.; Galluzzi, L. Immunological impact of cell death signaling driven by radiation on the tumor microenvironment. Nat. Immunol. 2020, 21, 120–134. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef]
- Ohtani, N. The roles and mechanisms of senescence-associated secretory phenotype (SASP): Can it be controlled by senolysis? Inflamm. Regen. 2022, 42, 11. [Google Scholar] [CrossRef]
- Ito, Y.; Hoare, M.; Narita, M. Spatial and Temporal Control of Senescence. Trends Cell Biol. 2017, 27, 820–832. [Google Scholar] [CrossRef]
- Cuollo, L.; Antonangeli, F.; Santoni, A.; Soriani, A. The Senescence-Associated Secretory Phenotype (SASP) in the Challenging Future of Cancer Therapy and Age-Related Diseases. Biology 2020, 9, 485. [Google Scholar] [CrossRef]
- Hanna, S.C.; Krishnan, B.; Bailey, S.T.; Moschos, S.J.; Kuan, P.F.; Shimamura, T.; Osborne, L.D.; Siegel, M.B.; Duncan, L.M.; O’Brien, E.T.; et al. HIF1α and HIF2α independently activate SRC to promote melanoma metastases. J. Clin. Investig. 2013, 123, 2078–2093. [Google Scholar] [CrossRef] [PubMed]
- Shaverdashvili, K.; Wong, P.; Ma, J.; Zhang, K.; Osman, I.; Bedogni, B. MT1-MMP modulates melanoma cell dissemination and metastasis through activation of MMP2 and RAC1. Pigment. Cell Melanoma Res. 2014, 27, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Ohanna, M.; Giuliano, S.; Bonet, C.; Imbert, V.; Hofman, V.; Zangari, J.; Bille, K.; Robert, C.; Bressac-de Paillerets, B.; Hofman, P.; et al. Senescent cells develop a PARP-1 and nuclear factor-{kappa}B-associated secretome (PNAS). Genes Dev. 2011, 25, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Obenauf, A.C.; Zou, Y.; Ji, A.L.; Vanharanta, S.; Shu, W.; Shi, H.; Kong, X.; Bosenberg, M.C.; Wiesner, T.; Rosen, N.; et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature 2015, 520, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, E.; Zimmer, L.; Vaubel, J.; Schadendorf, D. BRAF, MEK and KIT inhibitors for melanoma: Adverse events and their management. Chin. Clin. Oncol. 2014, 3, 29. [Google Scholar] [CrossRef]
- Sandri, S.; Faião-Flores, F.; Tiago, M.; Pennacchi, P.C.; Massaro, R.R.; Alves-Fernandes, D.K.; Berardinelli, G.N.; Evangelista, A.F.; de Lima Vazquez, V.; Reis, R.M.; et al. Vemurafenib resistance increases melanoma invasiveness and modulates the tumor microenvironment by MMP-2 upregulation. Pharmacol. Res. 2016, 111, 523–533. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).