Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Human Skin Reconstructs (HSR)
2.3. Japanese Medaka (Oryzia latipes) Strains and Breeding
2.4. Transient Transfections
2.5. Real-Time RT-qPCR
2.6. Protein Extraction and Western Blotting
2.7. Chromatin Immunoprecipitation
2.8. Cryosections
2.9. Immunohistochemistry
2.10. Optical Microscopy and Fluorescence Acquisition
2.11. Image Analysis
2.12. Atomic Force Microscopy (AFM)
2.13. Atomic Force Microscopy Data Acquisition
2.14. Atomic Force Microscopy Data Analysis
2.15. Statistical Analysis
3. Results
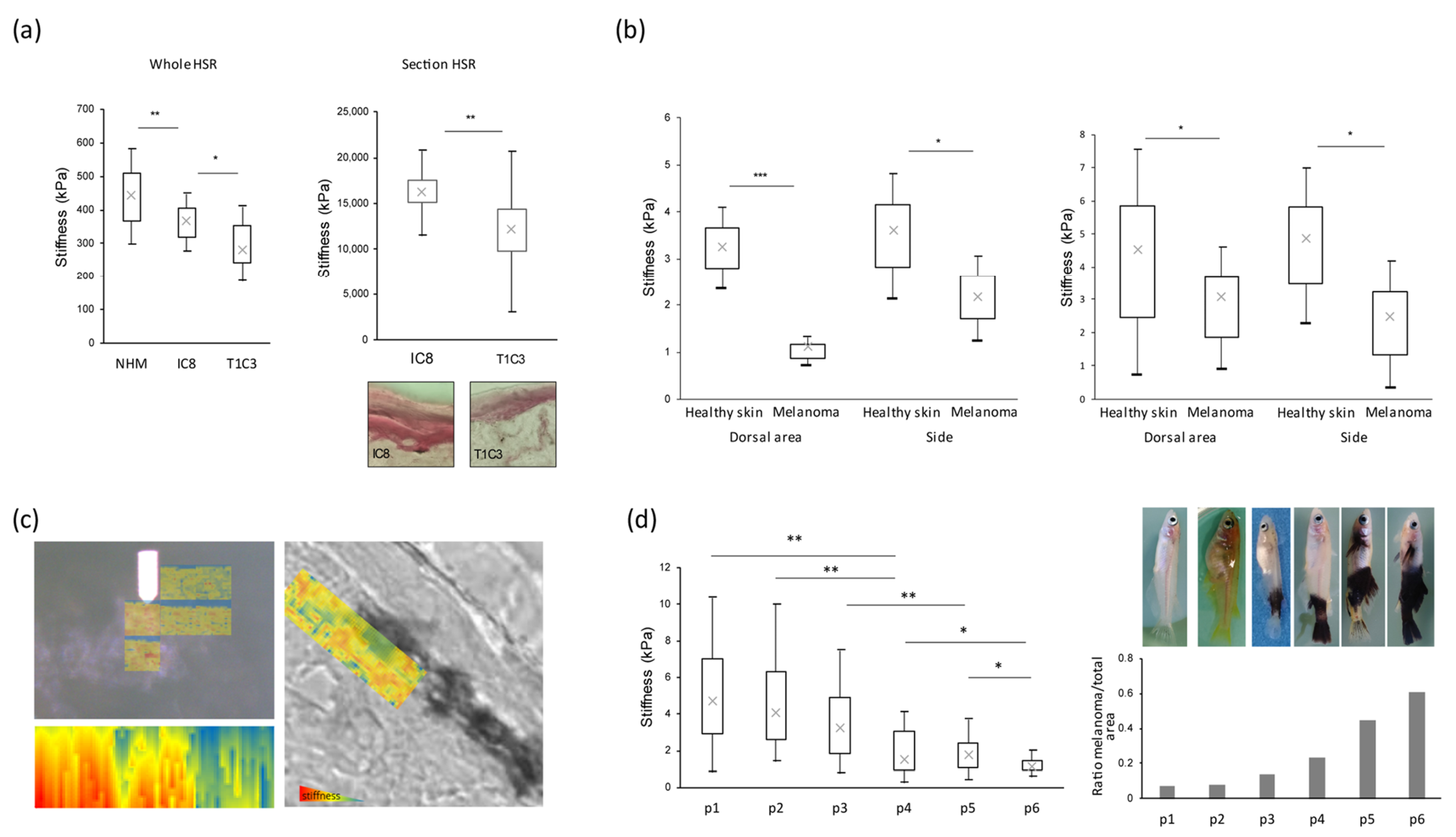
3.1. Stiffness Properties Decrease during Melanoma Transformation and Progression
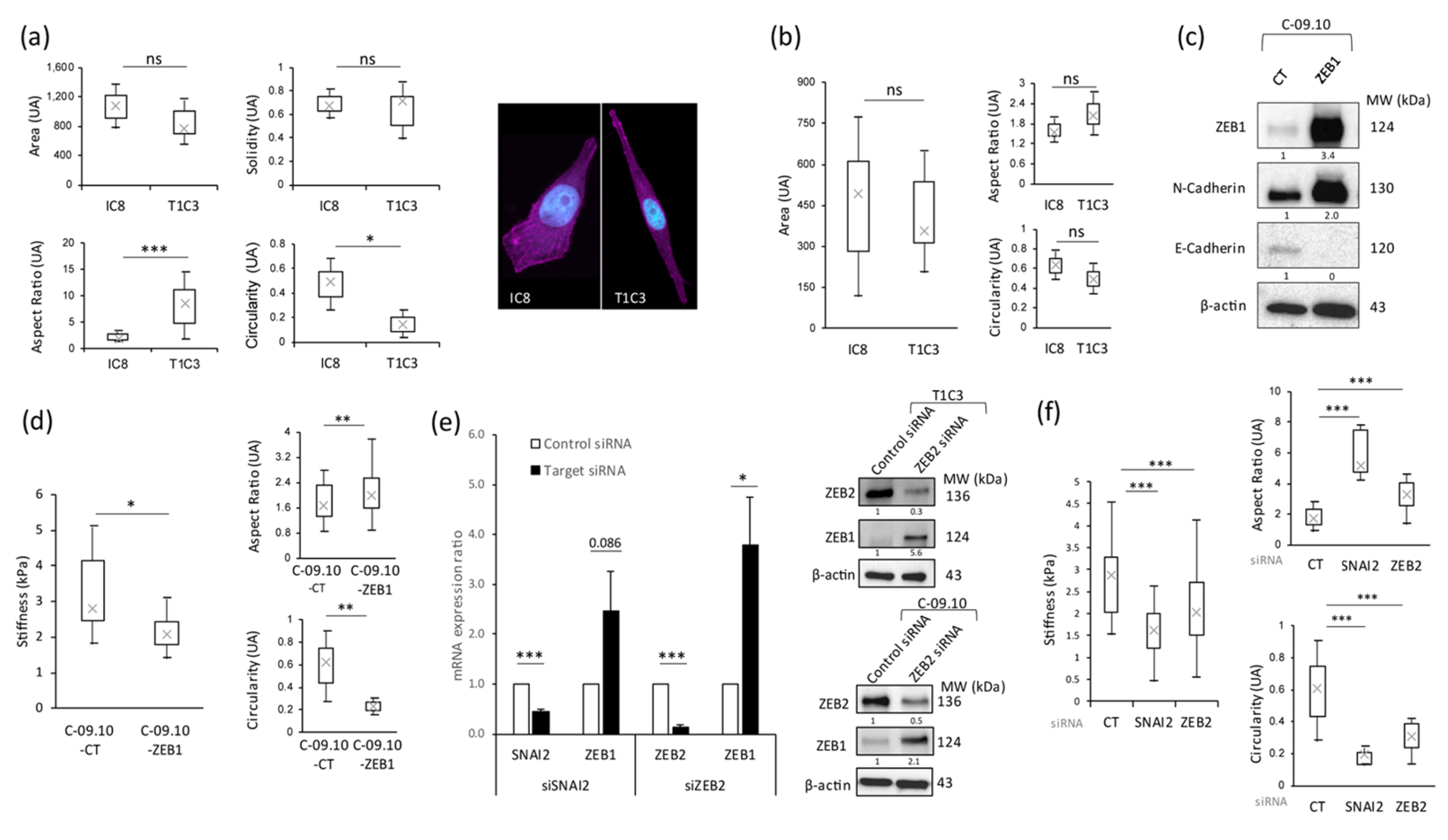
3.2. Stiffness Properties Are Associated with Cell Morphological Changes and with Modulation of Expression of Transcription Factors Regulating Epithelial-to-Mesenchymal Transition
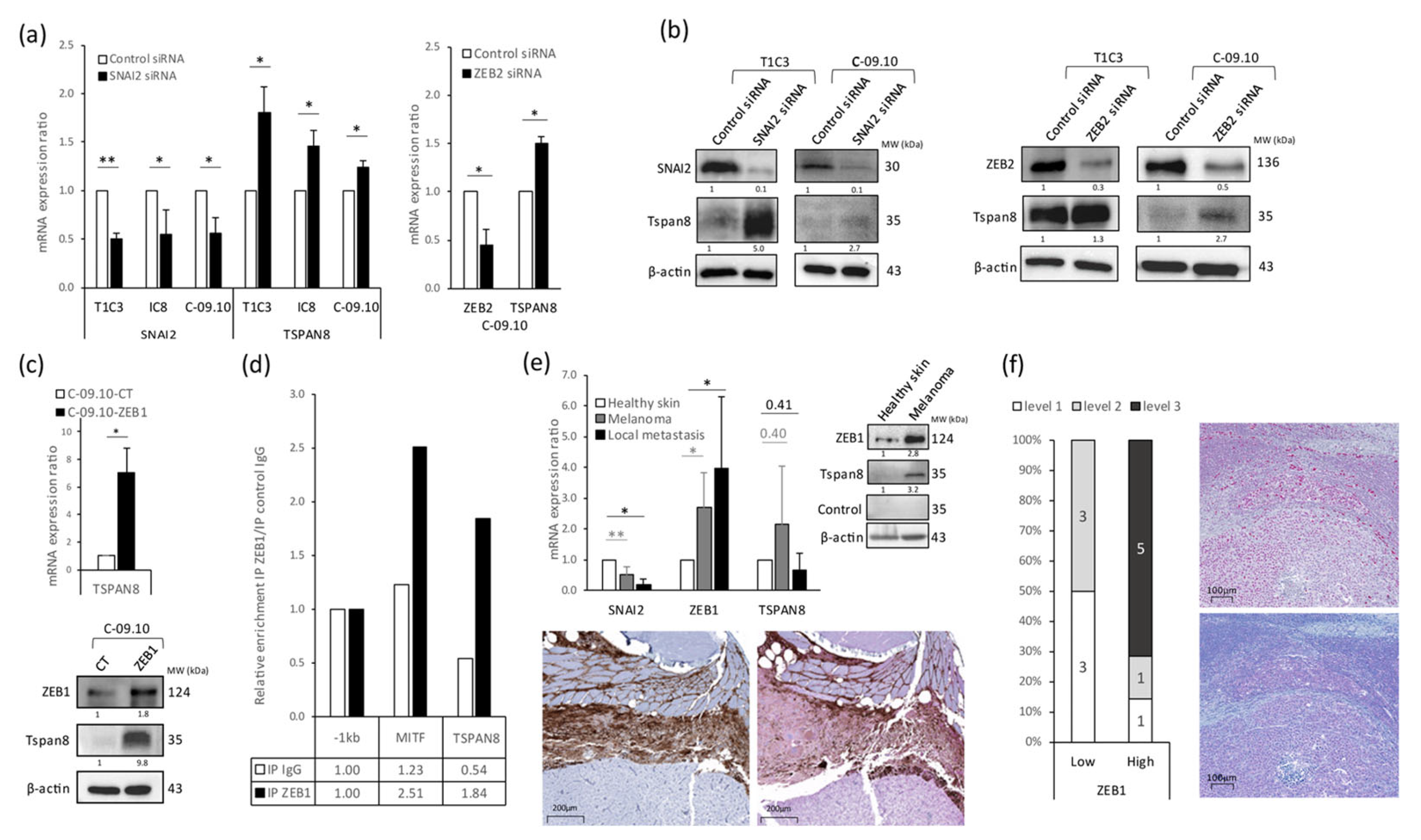
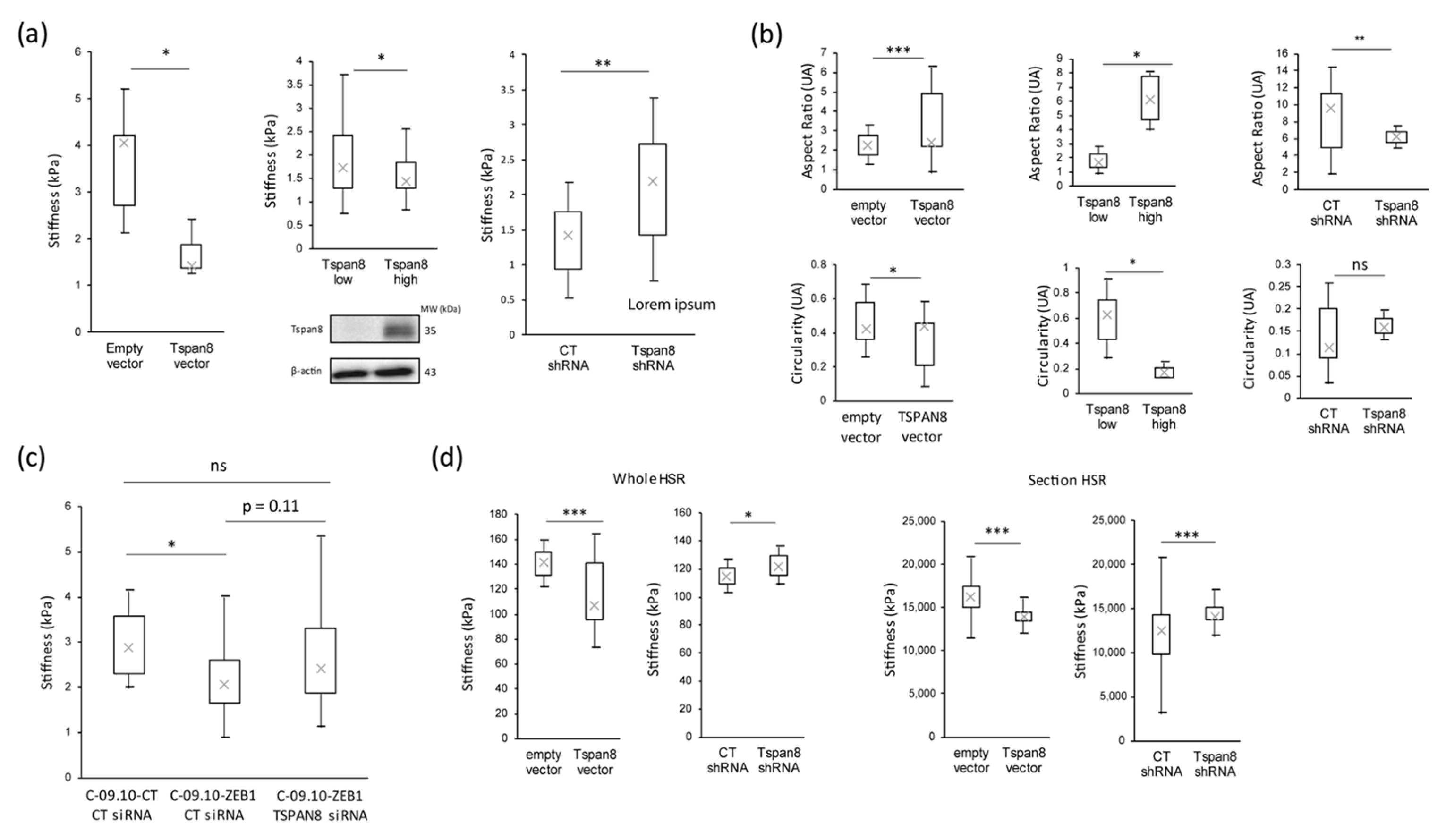
3.3. Tetraspanin 8 (Tspan8) Is the Major Target of EMT-TFs for Cell Stiffness and Morphology Regulation during Melanoma Progression
4. Discussion
5. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shain, A.H.; Bastian, B.C. The Genetic Evolution of Melanoma. N. Engl. J. Med. 2016, 374, 995–996. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the Multicellular Ecosystem of Metastatic Melanoma by Single-Cell RNA-Seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Tsoi, J.; Robert, L.; Paraiso, K.; Galvan, C.; Sheu, K.M.; Lay, J.; Wong, D.J.L.; Atefi, M.; Shirazi, R.; Wang, X.; et al. Multi-Stage Differentiation Defines Melanoma Subtypes with Differential Vulnerability to Drug-Induced Iron-Dependent Oxidative Stress. Cancer Cell 2018, 33, 890–904.e5. [Google Scholar] [CrossRef]
- Hoek, K.S.; Schlegel, N.C.; Brafford, P.; Sucker, A.; Ugurel, S.; Kumar, R.; Weber, B.L.; Nathanson, K.L.; Phillips, D.J.; Herlyn, M.; et al. Metastatic Potential of Melanomas Defined by Specific Gene Expression Profiles with No BRAF Signature. Pigment. Cell Res. 2006, 19, 290–302. [Google Scholar] [CrossRef]
- Widmer, D.S.; Cheng, P.F.; Eichhoff, O.M.; Belloni, B.C.; Zipser, M.C.; Schlegel, N.C.; Javelaud, D.; Mauviel, A.; Dummer, R.; Hoek, K.S. Systematic Classification of Melanoma Cells by Phenotype-Specific Gene Expression Mapping. Pigment. Cell Melanoma Res. 2012, 25, 343–353. [Google Scholar] [CrossRef]
- Marine, J.-C.; Dawson, S.-J.; Dawson, M.A. Non-Genetic Mechanisms of Therapeutic Resistance in Cancer. Nat. Rev. Cancer 2020, 20, 743–756. [Google Scholar] [CrossRef] [PubMed]
- Hugo, W.; Shi, H.; Sun, L.; Piva, M.; Song, C.; Kong, X.; Moriceau, G.; Hong, A.; Dahlman, K.B.; Johnson, D.B.; et al. Non-Genomic and Immune Evolution of Melanoma Acquiring MAPKi Resistance. Cell 2015, 162, 1271–1285. [Google Scholar] [CrossRef] [PubMed]
- Titz, B.; Lomova, A.; Le, A.; Hugo, W.; Kong, X.; ten Hoeve, J.; Friedman, M.; Shi, H.; Moriceau, G.; Song, C.; et al. JUN Dependency in Distinct Early and Late BRAF Inhibition Adaptation States of Melanoma. Cell Discov. 2016, 2, 16028. [Google Scholar] [CrossRef] [PubMed]
- Arozarena, I.; Wellbrock, C. Phenotype Plasticity as Enabler of Melanoma Progression and Therapy Resistance. Nat. Rev. Cancer 2019, 19, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Rambow, F.; Marine, J.-C.; Goding, C.R. Melanoma Plasticity and Phenotypic Diversity: Therapeutic Barriers and Opportunities. Genes Dev. 2019, 33, 1295–1318. [Google Scholar] [CrossRef] [PubMed]
- Goding, C.R.; Arnheiter, H. MITF-the First 25 Years. Genes Dev. 2019, 33, 983–1007. [Google Scholar] [CrossRef]
- Wouters, J.; Kalender-Atak, Z.; Minnoye, L.; Spanier, K.I.; De Waegeneer, M.; Bravo González-Blas, C.; Mauduit, D.; Davie, K.; Hulselmans, G.; Najem, A.; et al. Robust Gene Expression Programs Underlie Recurrent Cell States and Phenotype Switching in Melanoma. Nat. Cell Biol. 2020, 22, 986–998. [Google Scholar] [CrossRef]
- Brabletz, S.; Schuhwerk, H.; Brabletz, T.; Stemmler, M.P. Dynamic EMT: A Multi-tool for Tumor Progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Rashidian, M.; Reinhardt, F.; Bagnato, A.; Keckesova, Z.; Ploegh, H.L.; Weinberg, R.A. Epithelial-to-Mesenchymal Transition Contributes to Immunosuppression in Breast Carcinomas. Cancer Res 2017, 77, 3982–3989. [Google Scholar] [CrossRef] [PubMed]
- Bruneel, K.; Verstappe, J.; Vandamme, N.; Berx, G. Intrinsic Balance between ZEB Family Members Is Important for Melanocyte Homeostasis and Melanoma Progression. Cancers 2020, 12, 2248. [Google Scholar] [CrossRef] [PubMed]
- Caramel, J.; Papadogeorgakis, E.; Hill, L.; Browne, G.J.; Richard, G.; Wierinckx, A.; Saldanha, G.; Osborne, J.; Hutchinson, P.; Tse, G.; et al. A Switch in the Expression of Embryonic EMT-Inducers Drives the Development of Malignant Melanoma. Cancer Cell 2013, 24, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Richard, G.; Dalle, S.; Monet, M.; Ligier, M.; Boespflug, A.; Pommier, R.M.; de la Fouchardière, A.; Perier-Muzet, M.; Depaepe, L.; Barnault, R.; et al. ZEB1-mediated Melanoma Cell Plasticity Enhances Resistance to MAPK Inhibitors. EMBO Mol. Med. 2016, 8, 1143–1161. [Google Scholar] [CrossRef] [PubMed]
- Plaschka, M. ZEB1 Transcription Factor Promotes Immune Escape in Melanoma. J. Immunother. Cancer 2022, 10, e003484. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Tang, Y.; Pommier, R.M.; Benboubker, V.; Grimont, M.; Boivin, F.; Barbollat-Boutrand, L.; Cumunel, E.; Dupeuble, F.; Eberhardt, A.; et al. ZEB1 Controls a Lineage-Specific Transcriptional Program Essential for Melanoma Cell State Transitions. bioRxiv 2023. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-Intrinsic β-Catenin Signalling Prevents Anti-Tumour Immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Diazzi, S.; Tartare-Deckert, S.; Deckert, M. The Mechanical Phenotypic Plasticity of Melanoma Cell: An Emerging Driver of Therapy Cross-Resistance. Oncogenesis 2023, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Frey, K.; Fiechter, M.; Schwager, K.; Belloni, B.; Barysch, M.J.; Neri, D.; Dummer, R. Different Patterns of Fibronectin and Tenascin-C Splice Variants Expression in Primary and Metastatic Melanoma Lesions. Exp. Dermatol. 2011, 20, 685–688. [Google Scholar] [CrossRef]
- Matafora, V.; Farris, F.; Restuccia, U.; Tamburri, S.; Martano, G.; Bernardelli, C.; Sofia, A.; Pisati, F.; Casagrande, F.; Lazzari, L.; et al. Amyloid Aggregates Accumulate in Melanoma Metastasis Modulating YAP Activity. EMBO Rep. 2020, 21, e50446. [Google Scholar] [CrossRef]
- Yu, H.; Mouw, J.K.; Weaver, V.M. Forcing Form and Function: Biomechanical Regulation of Tumor Evolution. Trends Cell Biol. 2011, 21, 47–56. [Google Scholar] [CrossRef]
- Hirata, E.; Girotti, M.R.; Viros, A.; Hooper, S.; Spencer-Dene, B.; Matsuda, M.; Larkin, J.; Marais, R.; Sahai, E. Intravital Imaging Reveals How BRAF Inhibition Generates Drug-Tolerant Microenvironments with High Integrin Β1/FAK Signaling. Cancer Cell 2015, 27, 574–588. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H.; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Weder, G.; Hendriks-Balk, M.C.; Smajda, R.; Rimoldi, D.; Liley, M.; Heinzelmann, H.; Meister, A.; Mariotti, A. Increased Plasticity of the Stiffness of Melanoma Cells Correlates with Their Acquisition of Metastatic Properties. Nanomedicine 2014, 10, 141–148. [Google Scholar] [CrossRef]
- Berthier-Vergnes, O.; El Kharbili, M.; de la Fouchardière, A.; Pointecouteau, T.; Verrando, P.; Wierinckx, A.; Lachuer, J.; Le Naour, F.; Lamartine, J. Gene Expression Profiles of Human Melanoma Cells with Different Invasive Potential Reveal TSPAN8 as a Novel Mediator of Invasion. Br. J. Cancer 2011, 104, 155–165. [Google Scholar] [CrossRef]
- Bailly, M.; Bertrand, S.; Doré, J.-F. Human Tumor Spontaneous Metastasis in Immunosuppressed Newborn Rats. I. Characterization of the Bioassay. Int. J. Cancer 1991, 49, 457–466. [Google Scholar] [CrossRef]
- El Kharbili, M.; Robert, C.; Witkowski, T.; Danty-Berger, E.; Barbollat-Boutrand, L.; Masse, I.; Gadot, N.; de la Fouchardière, A.; McDonald, P.C.; Dedhar, S.; et al. Tetraspanin 8 Is a Novel Regulator of ILK-Driven Β1 Integrin Adhesion and Signaling in Invasive Melanoma Cells. Oncotarget 2017, 8, 17140–17155. [Google Scholar] [CrossRef]
- Agaësse, G.; Barbollat-Boutrand, L.; Sulpice, E.; Bhajun, R.; El Kharbili, M.; Berthier-Vergnes, O.; Degoul, F.; de la Fouchardière, A.; Berger, E.; Voeltzel, T.; et al. A Large-Scale RNAi Screen Identifies LCMR1 as a Critical Regulator of Tspan8-Mediated Melanoma Invasion. Oncogene 2017, 36, 446–457. [Google Scholar] [CrossRef]
- El Kharbili, M.; Cario, M.; Béchetoille, N.; Pain, C.; Boucheix, C.; Degoul, F.; Masse, I.; Berthier-Vergnes, O. Tspan8 Drives Melanoma Dermal Invasion by Promoting ProMMP-9 Activation and Basement Membrane Proteolysis in a Keratinocyte-Dependent Manner. Cancers 2020, 12, 1297. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Wilde, B.; Laisney, J.A.G.C.; Taniguchi, Y.; Takeda, S.; Meierjohann, S. A Mutated EGFR Is Sufficient to Induce Malignant Melanoma with Genetic Background-Dependent Histopathologies. J. Investig. Dermatol. 2010, 130, 249–258. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Kneitz, S.; Wilde, B.; Wagner, T.; Henkel, C.V.; Spaink, H.P.; Meierjohann, S. Conserved Expression Signatures between Medaka and Human Pigment Cell Tumors. PLoS ONE 2012, 7, e37880. [Google Scholar] [CrossRef]
- Schartl, M.; Walter, R.B. Xiphophorus and Medaka Cancer Models. Adv. Exp. Med. Biol. 2016, 916, 531–552. [Google Scholar] [CrossRef]
- Runel, G.; Cario, M.; Lopez-Ramirez, N.; Malbouyres, M.; Ruggiero, F.; Bernard, L.; Puisieux, A.; Caramel, J.; Chlasta, J.; Masse, I. Stiffness Measurement Is a Biomarker of Skin Ageing in Vivo. Exp. Dermatol. 2020, 29, 1233–1237. [Google Scholar] [CrossRef]
- Bechetoille, N.; Haftek, M.; Staquet, M.J.; Cochran, A.J.; Schmitt, D.; Berthier-Vergnes, O. Penetration of Human Metastatic Melanoma Cells through an Authentic Dermal-Epidermal Junction Is Associated with Dissolution of Native Collagen Types IV and VII. Melanoma Res. 2000, 10, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Schartl, M.; Shen, Y.; Maurus, K.; Walter, R.; Tomlinson, C.; Wilson, R.K.; Postlethwait, J.; Warren, W.C. Whole Body Melanoma Transcriptome Response in Medaka. PLoS ONE 2015, 10, e0143057. [Google Scholar] [CrossRef]
- Sugiyama, A.; Schartl, M.; Naruse, K. Histopathologic Features of Melanocytic Tumors in Xiphophorus Melanoma Receptor Kinase (Xmrk)-Transgenic Medaka (Oryzias latipes). J. Toxicol. Pathol. 2019, 32, 111–117. [Google Scholar] [CrossRef]
- Vőfély, R.V.; Gallagher, J.; Pisano, G.D.; Bartlett, M.; Braybrook, S.A. Of Puzzles and Pavements: A Quantitative Exploration of Leaf Epidermal Cell Shape. New Phytol. 2019, 221, 540–552. [Google Scholar] [CrossRef]
- Runel, G.; Lopez-Ramirez, N.; Chlasta, J.; Masse, I. Biomechanical Properties of Cancer Cells. Cells 2021, 10, 887. [Google Scholar] [CrossRef]
- Gladilin, E.; Ohse, S.; Boerries, M.; Busch, H.; Xu, C.; Schneider, M.; Meister, M.; Eils, R. TGFβ-Induced Cytoskeletal Remodeling Mediates Elevation of Cell Stiffness and Invasiveness in NSCLC. Sci. Rep. 2019, 9, 7667. [Google Scholar] [CrossRef]
- Chen, Y.-Q.; Lan, H.-Y.; Wu, Y.-C.; Yang, W.-H.; Chiou, A.; Yang, M.-H. Epithelial-Mesenchymal Transition Softens Head and Neck Cancer Cells to Facilitate Migration in 3D Environments. J. Cell. Mol. Med. 2018, 22, 3837–3846. [Google Scholar] [CrossRef]
- Agaësse, G.; Barbollat-Boutrand, L.; El Kharbili, M.; Berthier-Vergnes, O.; Masse, I. P53 Targets TSPAN8 to Prevent Invasion in Melanoma Cells. Oncogenesis 2017, 6, e309. [Google Scholar] [CrossRef] [PubMed]
- Hemler, M.E. Tetraspanin Proteins Promote Multiple Cancer Stages. Nat. Rev. Cancer 2014, 14, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Zöller, M. Tetraspanins: Push and Pull in Suppressing and Promoting Metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Diaferia, G.R.; Balestrieri, C.; Prosperini, E.; Nicoli, P.; Spaggiari, P.; Zerbi, A.; Natoli, G. Dissection of Transcriptional and Cis-regulatory Control of Differentiation in Human Pancreatic Cancer. EMBO J. 2016, 35, 595–617. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.L.; Pegoraro, A.F.; Li, H.; Li, K.; Yuan, Y.; Xu, G.; Gu, Z.; Sun, J.; Hao, Y.; Gupta, S.K.; et al. Cell Swelling, Softening and Invasion in a Three-Dimensional Breast Cancer Model. Nat. Phys. 2020, 16, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Cross, S.E.; Jin, Y.-S.; Rao, J.; Gimzewski, J.K. Nanomechanical Analysis of Cells from Cancer Patients. Nat. Nanotechnol. 2007, 2, 780–783. [Google Scholar] [CrossRef] [PubMed]
- Suresh, S. Nanomedicine: Elastic Clues in Cancer Detection. Nat. Nanotechnol. 2007, 2, 748–749. [Google Scholar] [CrossRef]
- Cross, S.E.; Jin, Y.-S.; Tondre, J.; Wong, R.; Rao, J.; Gimzewski, J.K. AFM-Based Analysis of Human Metastatic Cancer Cells. Nanotechnology 2008, 19, 384003. [Google Scholar] [CrossRef]
- Tse, H.T.K.; Gossett, D.R.; Moon, Y.S.; Masaeli, M.; Sohsman, M.; Ying, Y.; Mislick, K.; Adams, R.P.; Rao, J.; Di Carlo, D. Quantitative Diagnosis of Malignant Pleural Effusions by Single-Cell Mechanophenotyping. Sci. Transl. Med. 2013, 5, 212ra163. [Google Scholar] [CrossRef] [PubMed]
- Sarna, M.; Zadlo, A.; Pilat, A.; Olchawa, M.; Gkogkolou, P.; Burda, K.; Böhm, M.; Sarna, T. Nanomechanical Analysis of Pigmented Human Melanoma Cells. Pigment. Cell Melanoma Res. 2013, 26, 727–730. [Google Scholar] [CrossRef]
- Watanabe, T.; Kuramochi, H.; Takahashi, A.; Imai, K.; Katsuta, N.; Nakayama, T.; Fujiki, H.; Suganuma, M. Higher Cell Stiffness Indicating Lower Metastatic Potential in B16 Melanoma Cell Variants and in (-)-Epigallocatechin Gallate-Treated Cells. J. Cancer Res. Clin. Oncol. 2012, 138, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Sarna, M.; Krzykawska-Serda, M.; Jakubowska, M.; Zadlo, A.; Urbanska, K. Melanin Presence Inhibits Melanoma Cell Spread in Mice in a Unique Mechanical Fashion. Sci. Rep. 2019, 9, 9280. [Google Scholar] [CrossRef]
- Thomas, N.E.; Kricker, A.; Waxweiler, W.T.; Dillon, P.M.; Busman, K.J.; From, L.; Groben, P.A.; Armstrong, B.K.; Anton-Culver, H.; Gruber, S.B.; et al. Comparison of Clinicopathologic Features and Survival of Histopathologically Amelanotic and Pigmented Melanomas: A Population-Based Study. JAMA Dermatol. 2014, 150, 1306–1314. [Google Scholar] [CrossRef]
- El Kharbili, M.; Agaësse, G.; Barbollat-Boutrand, L.; Pommier, R.M.; de la Fouchardière, A.; Larue, L.; Caramel, J.; Puisieux, A.; Berthier-Vergnes, O.; Masse, I. Tspan8-β-Catenin Positive Feedback Loop Promotes Melanoma Invasion. Oncogene 2019, 38, 3781–3793. [Google Scholar] [CrossRef]
- Berthier-Vergnes, O.; Barbollat-Boutrand, L.; Pommier, R.M.; de la Fouchardière, A.; Combemale, P.; Grimont, M.; Lopez-Ramirez, N.; Caramel, J.; Dalle, S.; Perrot, J.-L.; et al. Tetraspanin8 Expression Predicts an Increased Metastatic Risk and Is Associated with Cancer-Related Death in Human Cutaneous Melanoma. Mol. Cancer 2021, 20, 127. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yang, J.; Xu, B.; Zhang, X. Tumor Metastasis: Mechanistic Insights and Therapeutic Interventions. MedComm 2021, 2, 587–617. [Google Scholar] [CrossRef]
- Er, E.E.; Valiente, M.; Ganesh, K.; Zou, Y.; Agrawal, S.; Hu, J.; Griscom, B.; Rosenblum, M.; Boire, A.; Brogi, E.; et al. Pericyte-like Spreading by Disseminated Cancer Cells Activates YAP and MRTF for Metastatic Colonization. Nat. Cell Biol. 2018, 20, 966–978. [Google Scholar] [CrossRef]
- Tello-Lafoz, M.; Srpan, K.; Sanchez, E.E.; Hu, J.; Remsik, J.; Romin, Y.; Calò, A.; Hoen, D.; Bhanot, U.; Morris, L.; et al. Cytotoxic Lymphocytes Target Characteristic Biophysical Vulnerabilities in Cancer. Immunity 2021, 54, 1037–1054.e7. [Google Scholar] [CrossRef]
- Kim, T.; Hwang, D.; Lee, D.; Kim, J.-H.; Kim, S.-Y.; Lim, D.-S. MRTF Potentiates TEAD-YAP Transcriptional Activity Causing Metastasis. EMBO J. 2017, 36, 520–535. [Google Scholar] [CrossRef]
- Kim, S.I.; Cassella, C.R.; Byrne, K.T. Tumor Burden and Immunotherapy: Impact on Immune Infiltration and Therapeutic Outcomes. Front. Immunol. 2021, 11, 629722. [Google Scholar] [CrossRef] [PubMed]
- Ailane, N.; Greco, C.; Zhu, Y.; Sala-Valdés, M.; Billard, M.; Casal, I.; Bawa, O.; Opolon, P.; Rubinstein, E.; Boucheix, C. Effect of an Anti-Human Co-029/Tspan8 Mouse Monoclonal Antibody on Tumor Growth in a Nude Mouse Model. Front. Physiol. 2014, 5, 364. [Google Scholar] [CrossRef]
- Park, C.S.; Kim, T.-K.; Kim, H.G.; Kim, Y.-J.; Jeoung, M.H.; Lee, W.R.; Go, N.K.; Heo, K.; Lee, S. Therapeutic Targeting of Tetraspanin8 in Epithelial Ovarian Cancer Invasion and Metastasis. Oncogene 2016, 35, 4540–4548. [Google Scholar] [CrossRef] [PubMed]
- Gesierich, S.; Berezovskiy, I.; Ryschich, E.; Zöller, M. Systemic Induction of the Angiogenesis Switch by the Tetraspanin D6.1A/CO-029. Cancer Res. 2006, 66, 7083–7094. [Google Scholar] [CrossRef] [PubMed]
- Heo, K.; Lee, S. TSPAN8 as a Novel Emerging Therapeutic Target in Cancer for Monoclonal Antibody Therapy. Biomolecules 2020, 10, 388. [Google Scholar] [CrossRef]
- Maisonial-Besset, A.; Witkowski, T.; Navarro-Teulon, I.; Berthier-Vergnes, O.; Fois, G.; Zhu, Y.; Besse, S.; Bawa, O.; Briat, A.; Quintana, M.; et al. Tetraspanin 8 (TSPAN 8) as a Potential Target for Radio-Immunotherapy of Colorectal Cancer. Oncotarget 2017, 8, 22034–22047. [Google Scholar] [CrossRef]
- Bonnet, M.; Maisonial-Besset, A.; Zhu, Y.; Witkowski, T.; Roche, G.; Boucheix, C.; Greco, C.; Degoul, F. Targeting the Tetraspanins with Monoclonal Antibodies in Oncology: Focus on Tspan8/Co-029. Cancers 2019, 11, 179. [Google Scholar] [CrossRef]
- Stehn, J.R.; Haass, N.K.; Bonello, T.; Desouza, M.; Kottyan, G.; Treutlein, H.; Zeng, J.; Nascimento, P.R.B.B.; Sequeira, V.B.; Butler, T.L.; et al. A Novel Class of Anticancer Compounds Targets the Actin Cytoskeleton in Tumor Cells. Cancer Res 2013, 73, 5169–5182. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Runel, G.; Lopez-Ramirez, N.; Barbollat-Boutrand, L.; Cario, M.; Durand, S.; Grimont, M.; Schartl, M.; Dalle, S.; Caramel, J.; Chlasta, J.; et al. Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion. Cancers 2024, 16, 694. https://doi.org/10.3390/cancers16040694
Runel G, Lopez-Ramirez N, Barbollat-Boutrand L, Cario M, Durand S, Grimont M, Schartl M, Dalle S, Caramel J, Chlasta J, et al. Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion. Cancers. 2024; 16(4):694. https://doi.org/10.3390/cancers16040694
Chicago/Turabian StyleRunel, Gaël, Noémie Lopez-Ramirez, Laetitia Barbollat-Boutrand, Muriel Cario, Simon Durand, Maxime Grimont, Manfred Schartl, Stéphane Dalle, Julie Caramel, Julien Chlasta, and et al. 2024. "Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion" Cancers 16, no. 4: 694. https://doi.org/10.3390/cancers16040694
APA StyleRunel, G., Lopez-Ramirez, N., Barbollat-Boutrand, L., Cario, M., Durand, S., Grimont, M., Schartl, M., Dalle, S., Caramel, J., Chlasta, J., & Masse, I. (2024). Cancer Cell Biomechanical Properties Accompany Tspan8-Dependent Cutaneous Melanoma Invasion. Cancers, 16(4), 694. https://doi.org/10.3390/cancers16040694






