The Clinical, Genomic, and Transcriptomic Landscape of BRAF Mutant Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Characteristics of BRAF Mutant Patient Cohort
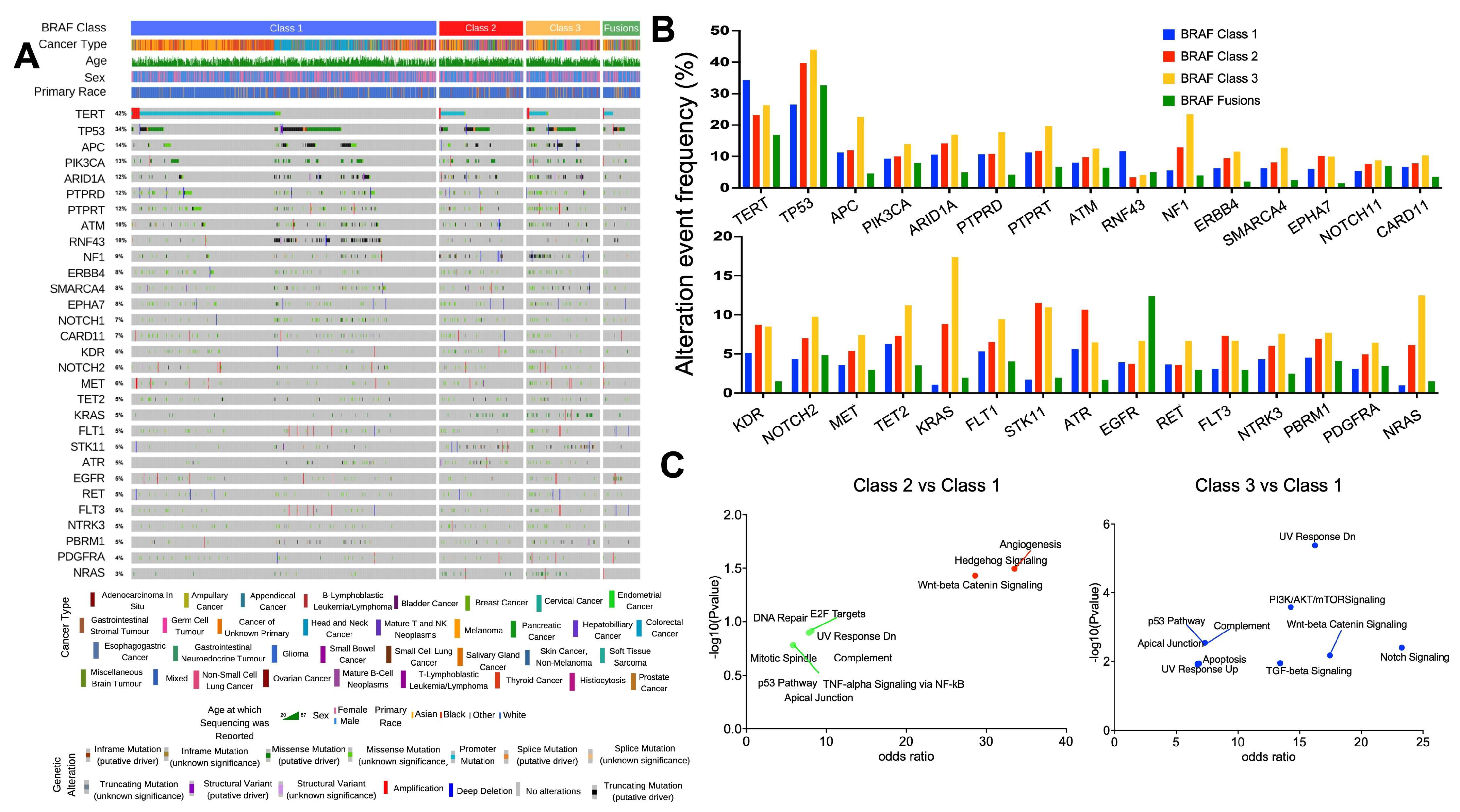
3.2. Relationship between BRAF Mutation Class and Co-Occurring Genomic Alterations
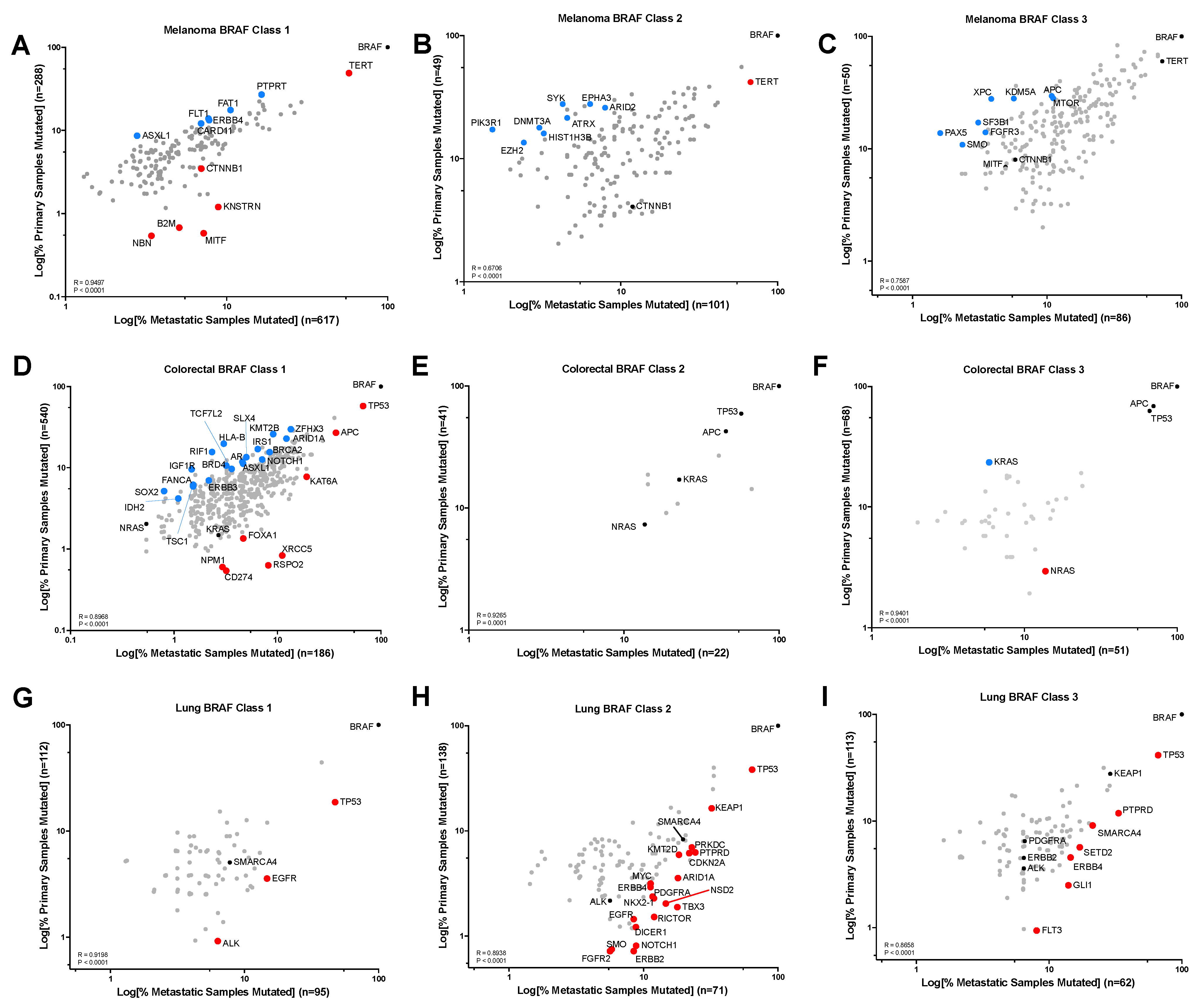
3.3. Gene Alteration Trends within Primary vs. Metastatic Tumors and BRAF Mutation Class
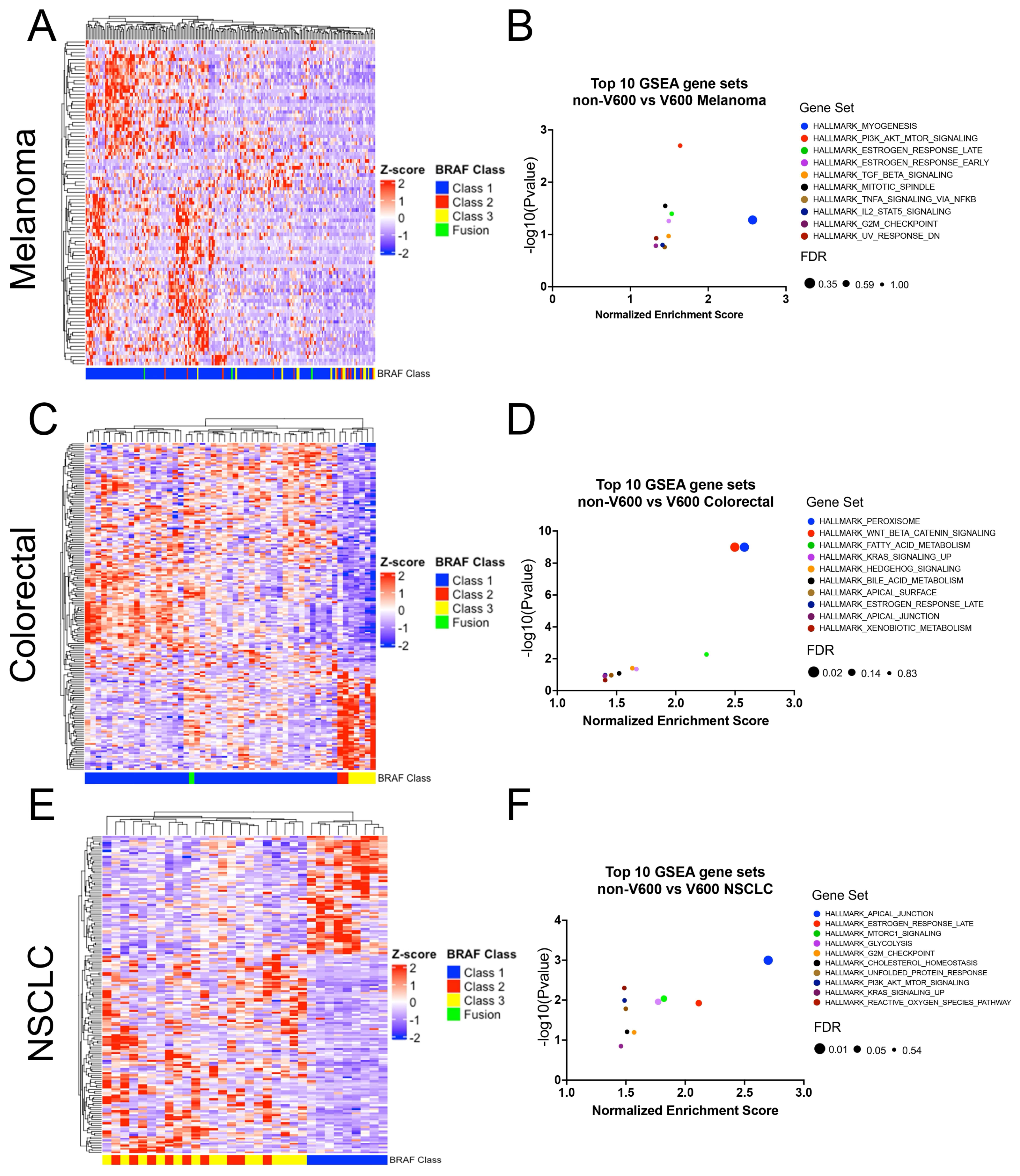
3.4. Relationship between BRAF Mutation Class and Gene Expression
3.5. Relationship between Patients’ Sex and BRAF Mutation Class
3.6. Relationship between Age and BRAF Mutation Class
3.7. Relationship between Primary Race and BRAF Mutation Class
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dankner, M.; Rose, A.A.N.; Rajkumar, S.; Siegel, P.M.; Watson, I.R. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 2018, 37, 3183–3199. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Yaeger, R.; Rodrik-Outmezguine, V.S.; Tao, A.; Torres, N.M.; Chang, M.T.; Drosten, M.; Zhao, H.; Cecchi, F.; Hembrough, T.; et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017, 548, 234–238. [Google Scholar] [CrossRef] [PubMed]
- Wan, P.T.; Garnett, M.J.; Roe, S.M.; Lee, S.; Niculescu-Duvaz, D.; Good, V.M.; Jones, C.M.; Marshall, C.J.; Springer, C.J.; Barford, D.; et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004, 116, 855–867. [Google Scholar] [CrossRef] [PubMed]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Yao, Z.; Torres, N.M.; Tao, A.; Gao, Y.; Luo, L.; Li, Q.; de Stanchina, E.; Abdel-Wahab, O.; Solit, D.B.; Poulikakos, P.I.; et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell 2015, 28, 370–383. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Smit, E.F.; Groen, H.J.M.; Mazieres, J.; Besse, B.; Helland, A.; Giannone, V.; D’Amelio, A.M., Jr.; Zhang, P.; Mookerjee, B.; et al. Dabrafenib plus trametinib in patients with previously untreated BRAF(V600E)-mutant metastatic non-small-cell lung cancer: An open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Dummer, R.; Ascierto, P.A.; Gogas, H.J.; Arance, A.; Mandala, M.; Liszkay, G.; Garbe, C.; Schadendorf, D.; Krajsova, I.; Gutzmer, R.; et al. Encorafenib plus binimetinib versus vemurafenib or encorafenib in patients with BRAF -mutant melanoma (COLUMBUS): A multicentre, open-label, randomised phase 3 trial. Lancet Oncol. 2018, 19, 603–615. [Google Scholar] [CrossRef]
- Kopetz, S.; Grothey, A.; Yaeger, R.; Van Cutsem, E.; Desai, J.; Yoshino, T.; Wasan, H.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib, Binimetinib, and Cetuximab in BRAF V600E–Mutated Colorectal Cancer. N. Engl. J. Med. 2019, 381, 1632–1643. [Google Scholar] [CrossRef]
- Robert, C.; Grob, J.J.; Stroyakovskiy, D.; Karaszewska, B.; Hauschild, A.; Levchenko, E.; Chiarion Sileni, V.; Schachter, J.; Garbe, C.; Bondarenko, I.; et al. Five-Year Outcomes with Dabrafenib plus Trametinib in Metastatic Melanoma. N. Engl. J. Med. 2019, 381, 626–636. [Google Scholar] [CrossRef]
- Larkin, J.; Ascierto, P.A.; Dréno, B.; Atkinson, V.; Liszkay, G.; Maio, M.; Mandalà, M.; Demidov, L.; Stroyakovskiy, D.; Thomas, L.; et al. Combined Vemurafenib and Cobimetinib in BRAF-Mutated Melanoma. N. Engl. J. Med. 2014, 371, 1867–1876. [Google Scholar] [CrossRef]
- Mullard, A. BRAF plus MEK inhibitor combo secures tumour-agnostic FDA approval. Nat. Rev. Drug Discov. 2022, 21, 548. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, N.; Maria, A.; Toumbacaris, N.; Zhao, H.; Kargus, K.; Bryant, M.; Waksmundzki, A.; Aricescu, I.; Lefkowitz, R.A.; Li, B.T.; et al. Combined RAF and MEK Inhibition to Treat Activated Non-V600 BRAF-Altered Advanced Cancers. Oncologist 2023, 29, 15–24. [Google Scholar] [CrossRef]
- Dankner, M.; Wang, Y.; Fazelzad, R.; Johnson, B.; Nebhan, C.A.; Dagogo-Jack, I.; Myall, N.J.; Richtig, G.; Bracht, J.W.P.; Gerlinger, M.; et al. Clinical Activity of Mitogen-Activated Protein Kinase-Targeted Therapies in Patients With Non-V600 BRAF-Mutant Tumors. JCO Precis. Oncol. 2022, 6, e2200107. [Google Scholar] [CrossRef] [PubMed]
- Kakadia, S.; Yarlagadda, N.; Awad, R.; Kundranda, M.; Niu, J.; Naraev, B.; Mina, L.; Dragovich, T.; Gimbel, M.; Mahmoud, F. Mechanisms of resistance to BRAF and MEK inhibitors and clinical update of US Food and Drug Administration-approved targeted therapy in advanced melanoma. OncoTargets Ther. 2018, 11, 7095–7107. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Lokhandwala, P.M.; Tseng, L.H.; Rodriguez, E.; Zheng, G.; Pallavajjalla, A.; Gocke, C.D.; Eshleman, J.R.; Lin, M.T. Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer 2019, 19, 665. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N.; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yu, H.; Ida, C.M.; Halling, K.C.; Kipp, B.R.; Geiersbach, K.; Rumilla, K.M.; Gupta, S.; Lin, M.-T.; Zheng, G. Assessment of RAS Dependency for BRAF Alterations Using Cancer Genomic Databases. JAMA Netw. Open 2021, 4, e2035479. [Google Scholar] [CrossRef] [PubMed]
- Nassar, A.H.; Adib, E.; Kwiatkowski, D.J. Distribution of KRAS (G12C) Somatic Mutations across Race, Sex, and Cancer Type. N. Engl. J. Med. 2021, 384, 185–187. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Jones, J.C.; Renfro, L.A.; Al-Shamsi, H.O.; Schrock, A.B.; Rankin, A.; Zhang, B.Y.; Kasi, P.M.; Voss, J.S.; Leal, A.D.; Sun, J.; et al. (Non-V600) BRAF Mutations Define a Clinically Distinct Molecular Subtype of Metastatic Colorectal Cancer. J. Clin. Oncol. 2017, 35, 2624–2630. [Google Scholar] [CrossRef] [PubMed]
- Wahida, A.; Buschhorn, L.; Fröhling, S.; Jost, P.J.; Schneeweiss, A.; Lichter, P.; Kurzrock, R. The coming decade in precision oncology: Six riddles. Nat. Rev. Cancer 2022, 23, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.; Berry, D.; Heney, K.A.; Strong, C.; Ramsay, L.; Lajoie, M.; Alkallas, R.; Nguyen, T.T.; Thomson, C.; Ahanfeshar-Adams, M.; et al. Melanomas with concurrent BRAF non-p.V600 and NF1 loss-of-function mutations are targetable by BRAF/MEK inhibitor combination therapy. Cell Rep. 2022, 39, 110634. [Google Scholar] [CrossRef] [PubMed]
- Dankner, M.; Lajoie, M.; Moldoveanu, D.; Nguyen, T.T.; Savage, P.; Rajkumar, S.; Huang, X.; Lvova, M.; Protopopov, A.; Vuzman, D.; et al. Dual MAPK Inhibition Is an Effective Therapeutic Strategy for a Subset of Class II BRAF Mutant Melanomas. Clin. Cancer Res. 2018, 24, 6483–6494. [Google Scholar] [CrossRef]
- Pons-Tostivint, E.; Lugat, A.; Fontenau, J.F.; Denis, M.G.; Bennouna, J. STK11/LKB1 Modulation of the Immune Response in Lung Cancer: From Biology to Therapeutic Impact. Cells 2021, 10, 3129. [Google Scholar] [CrossRef]
- Di Federico, A.; De Giglio, A.; Parisi, C.; Gelsomino, F. STK11/LKB1 and KEAP1 mutations in non-small cell lung cancer: Prognostic rather than predictive? Eur. J. Cancer 2021, 157, 108–113. [Google Scholar] [CrossRef]
- Paik, P.K.; Fan, P.D.; Qeriqi, B.; Namakydoust, A.; Daly, B.; Ahn, L.; Kim, R.; Plodkowski, A.; Ni, A.; Chang, J.; et al. Targeting NFE2L2/KEAP1 Mutations in Advanced NSCLC With the TORC1/2 Inhibitor TAK-228. J. Thorac. Oncol. 2023, 18, 516–526. [Google Scholar] [CrossRef] [PubMed]
- Gajos-Michniewicz, A.; Czyz, M. WNT Signaling in Melanoma. Int. J. Mol. Sci. 2020, 21, 4852. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tang, C. The Role of ARID1A in Tumors: Tumor Initiation or Tumor Suppression? Front. Oncol. 2021, 11, 745187. [Google Scholar] [CrossRef] [PubMed]
- Carcamo, S.; Nguyen, C.B.; Grossi, E.; Filipescu, D.; Alpsoy, A.; Dhiman, A.; Sun, D.; Narang, S.; Imig, J.; Martin, T.C.; et al. Altered BAF occupancy and transcription factor dynamics in PBAF-deficient melanoma. Cell Rep. 2022, 39, 110637. [Google Scholar] [CrossRef] [PubMed]
- Mandal, J.; Mandal, P.; Wang, T.L.; Shih, I.M. Treating ARID1A mutated cancers by harnessing synthetic lethality and DNA damage response. J. Biomed. Sci. 2022, 29, 71. [Google Scholar] [CrossRef] [PubMed]
- Wanior, M.; Kramer, A.; Knapp, S.; Joerger, A.C. Exploiting vulnerabilities of SWI/SNF chromatin remodelling complexes for cancer therapy. Oncogene 2021, 40, 3637–3654. [Google Scholar] [CrossRef] [PubMed]
- Molina-Cerrillo, J.; San Roman, M.; Pozas, J.; Alonso-Gordoa, T.; Pozas, M.; Conde, E.; Rosas, M.; Grande, E.; Garcia-Bermejo, M.L.; Carrato, A. BRAF Mutated Colorectal Cancer: New Treatment Approaches. Cancers 2020, 12, 1571. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Yaeger, R.; Kotani, D.; Mondaca, S.; Parikh, A.R.; Bando, H.; Van Seventer, E.E.; Taniguchi, H.; Zhao, H.; Thant, C.N.; de Stanchina, E.; et al. Response to Anti-EGFR Therapy in Patients with BRAF non-V600-Mutant Metastatic Colorectal Cancer. Clin. Cancer Res. 2019, 25, 7089–7097. [Google Scholar] [CrossRef]
- Kotani, D.; Bando, H.; Taniguchi, H.; Masuishi, T.; Komatsu, Y.; Yamaguchi, K.; Nakajima, T.; Satoh, T.; Nishina, T.; Esaki, T.; et al. BIG BANG study (EPOC1703): Multicentre, proof-of-concept, phase II study evaluating the efficacy and safety of combination therapy with binimetinib, encorafenib and cetuximab in patients with BRAF non-V600E mutated metastatic colorectal cancer. ESMO Open 2020, 5, e000624. [Google Scholar] [CrossRef]
- Kamath, S.D.; Torrejon, N.; Wei, W.; Tullio, K.; Nair, K.G.; Liska, D.; Krishnamurthi, S.S.; Khorana, A.A. Racial disparities negatively impact outcomes in early-onset colorectal cancer independent of socioeconomic status. Cancer Med. 2021, 10, 7542–7550. [Google Scholar] [CrossRef]
- Xicola, R.M.; Manojlovic, Z.; Augustus, G.J.; Kupfer, S.S.; Emmadi, R.; Alagiozian-Angelova, V.; Triche, T.; Salhia, B.; Carpten, J.; Llor, X.; et al. Lack of APC somatic mutation is associated with early-onset colorectal cancer in African Americans. Carcinogenesis 2018, 39, 1331–1341. [Google Scholar] [CrossRef] [PubMed]
- Hankey, W.; Frankel, W.L.; Groden, J. Functions of the APC tumor suppressor protein dependent and independent of canonical WNT signaling: Implications for therapeutic targeting. Cancer Metastasis Rev. 2018, 37, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Loree, J.M.; Anand, S.; Dasari, A.; Unger, J.M.; Gothwal, A.; Ellis, L.M.; Varadhachary, G.; Kopetz, S.; Overman, M.J.; Raghav, K. Disparity of Race Reporting and Representation in Clinical Trials Leading to Cancer Drug Approvals From 2008 to 2018. JAMA Oncol 2019, 5, e191870. [Google Scholar] [CrossRef]
- Bellenghi, M.; Puglisi, R.; Pontecorvi, G.; De Feo, A.; Carè, A.; Mattia, G. Sex and Gender Disparities in Melanoma. Cancers 2020, 12, 1819. [Google Scholar] [CrossRef] [PubMed]
- Vellano, C.P.; White, M.G.; Andrews, M.C.; Chelvanambi, M.; Witt, R.G.; Daniele, J.R.; Titus, M.; McQuade, J.L.; Conforti, F.; Burton, E.M.; et al. Androgen receptor blockade promotes response to BRAF/MEK-targeted therapy. Nature 2022, 606, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.K.; Sadar, M.D. Non-Genomic Actions of the Androgen Receptor in Prostate Cancer. Front. Endocrinol. (Lausanne) 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Atkins, M.B.; Lee, S.J.; Chmielowski, B.; Tarhini, A.A.; Cohen, G.I.; Truong, T.G.; Moon, H.H.; Davar, D.; O’Rourke, M.; Stephenson, J.J.; et al. Combination Dabrafenib and Trametinib Versus Combination Nivolumab and Ipilimumab for Patients With Advanced BRAF-Mutant Melanoma: The DREAMseq Trial-ECOG-ACRIN EA6134. J. Clin. Oncol. 2023, 41, 186–197. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Yang, Z.; Algazi, A.P.; Lomeli, S.H.; Wang, Y.; Othus, M.; Hong, A.; Wang, X.; Randolph, C.E.; et al. Anti-PD-1/L1 lead-in before MAPK inhibitor combination maximizes antitumor immunity and efficacy. Cancer Cell 2021, 39, 1375–1387.e6. [Google Scholar] [CrossRef]
- Bailey, Z.D.; Feldman, J.M.; Bassett, M.T. How Structural Racism Work—Racist Policies as a Root Cause of U.S. Racial Health Inequities. N. Engl. J. Med. 2021, 384, 768–773. [Google Scholar] [CrossRef]
- Conforti, C.; Zalaudek, I. Epidemiology and Risk Factors of Melanoma: A Review. Dermatol. Pract. Concept. 2021, 11 (Suppl. S1), e2021161S. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | All Patients (n = 5120) | Class 1 (n = 3358) | Class 2 (n = 782) | Class 3 (n = 759) | Fusion (n = 221) |
|---|---|---|---|---|---|
| Age | |||||
| Median—yr | 62 (20–90) | 62 (20–90) | 65 (20–90) | 64 (22–90) | 59 (20–90) |
| Distribution—No./total No. (%) | |||||
| <60 yr | 2180 (43) | 1532 (70) | 241 (11) | 291 (13) | 116 (5) |
| ≥60 yr | 2940 (57) | 1826 (62) | 541 (18) | 468 (16) | 105 (4) |
| Gender—No./total No. (%) | |||||
| Male | 2537 (50) | 1581 (62) | 400 (16) | 441 (17) | 115 (5) |
| Female | 2583 (50) | 1777 (69) | 382 (15) | 318 (12) | 106 (4) |
| Race—No./total No. (%) | |||||
| Asian | 212 (4) | 138 (65) | 24 (11) | 32 (15) | 18 (9) |
| Black | 192 (4) | 95 (50) | 42 (22) | 41 (21) | 14 (7) |
| White | 4504 (88) | 2977 (66) | 691 (15) | 659 (15) | 177 (4) |
| Other | 212 (4) | 148 (70) | 25 (12) | 27 (13) | 12 (6) |
| Sample type—No./total No. (%) | |||||
| Primary site | 2421 (47) | 1560 (64) | 387 (16) | 369 (15) | 105 (4) |
| Metastatic | 2122 (41) | 1379 (65) | 332 (16) | 304 (14) | 107 (5) |
| Not reported | 577 (11) | 419 (73) | 63 (11) | 86 (15) | 9 (2) |
| Type of Tumour—No./total No. (%) | |||||
| Melanoma | 1591 (31) | 1187 (35) | 189 (24) | 174 (23) | 41 (19) |
| Colorectal cancer | 1061 (21) | 842 (25) | 67 (9) | 136 (18) | 16 (7) |
| Non-small cell lung cancer | 714 (14) | 237 (7) | 243 (31) | 203 (27) | 31 (14) |
| Thyroid cancer | 689 (12) | 656 (20) | 14 (2) | 0 | 19 (9) |
| Glioma | 201 (4) | 118 (4) | 26 (3) | 23 (3) | 34 (15) |
| Unknown primary | 125 (2) | 56 (2) | 25 (3) | 38 (5) | 6 (3) |
| Bladder cancer | 39 (1) | 3 (0.1) | 16 (2) | 18 (2) | 2 (1) |
| Hepatobiliary | 75 (1) | 27 (0.8) | 20 (3) | 23 (3) | 5 (2) |
| Pancreatic cancer | 72 (1) | 25 (0.7) | 31 (4) | 5 (1) | 11 (5) |
| Prostate cancer | 72 (1) | 1 (0.03) | 42 (5) | 6 (1) | 22 (10) |
| Other | 481 (9) | 206 (6) | 108 (14) | 133 (18) | 34 (15) |
| Co-Mutations—No./total No. (%) | |||||
| NF-1 | 234 (5) | 59 (25) | 57 (24) | 114 (49) | 4 (2) |
| RAS (HRAS, NRAS, and/or KRAS) | 390 (8) | 44 (11) | 115 (30) | 225 (58) | 6 (2) |
| Variant Allele Frequency (%) | |||||
| <26% | 2242 (47) | 1446 (64) | 382 (17) | 414 (18) | - |
| ≥26% | 2485 (53) | 1802 (73) | 364 (15) | 319 (13) | - |
| Univariable | Multivariable | |||||
|---|---|---|---|---|---|---|
| OR | 95% CI | p Value | OR | 95% CI | p Value | |
| Gender | ||||||
| Male vs. Female | 1.33 | 1.19–1.50 | <0.0001 | 1.55 | 1.35–1.76 | <0.0001 |
| Age | ||||||
| ≥60 vs. <60 | 1.44 | 1.28–1.62 | <0.0001 | 1.28 | 1.12–1.46 | <0.0001 |
| Primary Race | ||||||
| Black vs. other | 2.00 | 1.50–2.67 | <0.0001 | 1.58 | 1.13–2.20 | 0.007 |
| Primary tumour type | ||||||
| Melanoma | 0.54 | 0.48–0.62 | <0.0001 | 0.50 | 0.43–0.59 | <0.0001 |
| Colorectal | 0.42 | 0.36–0.50 | <0.0001 | 0.38 | 0.31–0.46 | <0.0001 |
| NSCLC | 4.89 | 4.13–5.79 | <0.0001 | 3.08 | 2.53–3.75 | <0.0001 |
| Sample Type | ||||||
| Metastatic vs. primary | 0.97 | 0.86–1.10 | 0.699 | - | - | - |
| Genomic co-mutations | ||||||
| RAS mt vs. wild-type | 18.4 | 13.37–25.34 | <0.0001 | 19.18 | 13.80–26.64 | <0.0001 |
| Variant Allele Frequency | ||||||
| ≥26% vs. <26% | 0.68 | 0.61–0.78 | <0.0001 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kazandjian, S.; Rousselle, E.; Dankner, M.; Cescon, D.W.; Spreafico, A.; Ma, K.; Kavan, P.; Batist, G.; Rose, A.A.N. The Clinical, Genomic, and Transcriptomic Landscape of BRAF Mutant Cancers. Cancers 2024, 16, 445. https://doi.org/10.3390/cancers16020445
Kazandjian S, Rousselle E, Dankner M, Cescon DW, Spreafico A, Ma K, Kavan P, Batist G, Rose AAN. The Clinical, Genomic, and Transcriptomic Landscape of BRAF Mutant Cancers. Cancers. 2024; 16(2):445. https://doi.org/10.3390/cancers16020445
Chicago/Turabian StyleKazandjian, Suzanne, Emmanuelle Rousselle, Matthew Dankner, David W. Cescon, Anna Spreafico, Kim Ma, Petr Kavan, Gerald Batist, and April A. N. Rose. 2024. "The Clinical, Genomic, and Transcriptomic Landscape of BRAF Mutant Cancers" Cancers 16, no. 2: 445. https://doi.org/10.3390/cancers16020445
APA StyleKazandjian, S., Rousselle, E., Dankner, M., Cescon, D. W., Spreafico, A., Ma, K., Kavan, P., Batist, G., & Rose, A. A. N. (2024). The Clinical, Genomic, and Transcriptomic Landscape of BRAF Mutant Cancers. Cancers, 16(2), 445. https://doi.org/10.3390/cancers16020445








