Cellular Transformation by Human Cytomegalovirus
Abstract
Simple Summary
Abstract
1. Introduction
2. HCMV Displays Oncogenic Traits Similar to Human Oncoviruses
3. HCMV, like Human Oncoviruses, Favors an Immunosuppressive Tissue Microenvironment
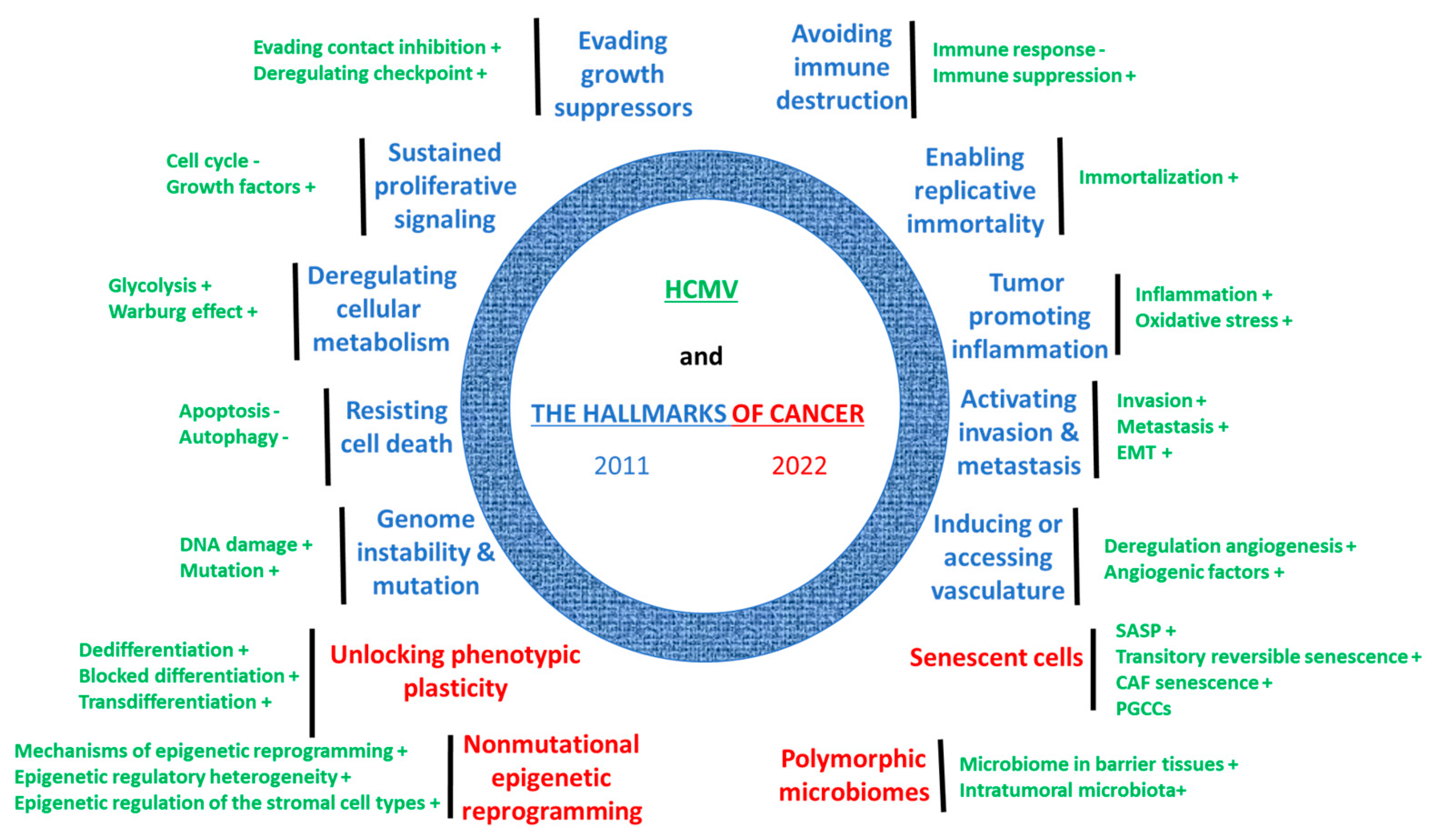
4. HCMV Fulfills Previous and Current Hallmarks of Cancer
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fulkerson, H.L.; Nogalski, M.T.; Collins-McMillen, D.; Yurochko, A.D. Overview of Human Cytomegalovirus Pathogenesis. Methods Mol. Biol. 2021, 2244, 1–18. [Google Scholar] [CrossRef]
- Aldè, M.; Binda, S.; Primache, V.; Pellegrinelli, L.; Pariani, E.; Pregliasco, F.; Di Berardino, F.; Cantarella, G.; Ambrosetti, U. Congenital Cytomegalovirus and Hearing Loss: The State of the Art. JCM 2023, 12, 4465. [Google Scholar] [CrossRef]
- Cobbs, C. Cytomegalovirus Is a Tumor-Associated Virus: Armed and Dangerous. Curr. Opin. Virol. 2019, 39, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Herbein, G. The Human Cytomegalovirus, from Oncomodulation to Oncogenesis. Viruses 2018, 10, 408. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer Weltgesundheitsorganisation (Ed.) IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Volume 100 B, Biological Agents: This Publication Represents the Views and Expert Opinions of an IARC Working Group on the Evaluation of Carcinogenic Risks to Humans, Which Met in Lyon. 2012; 24. [Google Scholar]
- Bouvard, V.; Baan, R.; Straif, K.; Grosse, Y.; Secretan, B.; Ghissassi, F.E.; Benbrahim-Tallaa, L.; Guha, N.; Freeman, C.; Galichet, L.; et al. A Review of Human Carcinogens—Part B: Biological Agents. Lancet Oncol. 2009, 10, 321–322. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Mesri, E.A.; Feitelson, M.A.; Munger, K. Human Viral Oncogenesis: A Cancer Hallmarks Analysis. Cell Host Microbe 2014, 15, 266–282. [Google Scholar] [CrossRef]
- Herbein, G. Tumors and Cytomegalovirus: An Intimate Interplay. Viruses 2022, 14, 812. [Google Scholar] [CrossRef]
- Söderberg-Nauclér, C. New Mechanistic Insights of the Pathogenicity of High-Risk Cytomegalovirus (CMV) Strains Derived from Breast Cancer: Hope for New Cancer Therapy Options. eBioMedicine 2022, 81, 104103. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhu, H.; Shenk, T. Human Cytomegalovirus IE1 and IE2 Proteins Are Mutagenic and Mediate “Hit-and-Run” Oncogenic Transformation in Cooperation with the Adenovirus E1A Proteins. Proc. Natl. Acad. Sci. USA 1997, 94, 3341–3345. [Google Scholar] [CrossRef] [PubMed]
- Soroceanu, L.; Cobbs, C.S. Is HCMV a Tumor Promoter? Virus Res. 2011, 157, 193–203. [Google Scholar] [CrossRef] [PubMed]
- Geisler, J.; Touma, J.; Rahbar, A.; Söderberg-Nauclér, C.; Vetvik, K. A Review of the Potential Role of Human Cytomegalovirus (HCMV) Infections in Breast Cancer Carcinogenesis and Abnormal Immunity. Cancers 2019, 11, 1842. [Google Scholar] [CrossRef] [PubMed]
- Peredo-Harvey, I.; Rahbar, A.; Söderberg-Nauclér, C. Presence of the Human Cytomegalovirus in Glioblastomas—A Systematic Review. Cancers 2021, 13, 5051. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wilkie, A.R.; Weller, M.; Liu, X.; Cohen, J.I. THY-1 Cell Surface Antigen (CD90) Has an Important Role in the Initial Stage of Human Cytomegalovirus Infection. PLoS Pathog. 2015, 11, e1004999. [Google Scholar] [CrossRef] [PubMed]
- Soroceanu, L.; Akhavan, A.; Cobbs, C.S. Platelet-Derived Growth Factor-α Receptor Activation Is Required for Human Cytomegalovirus Infection. Nature 2008, 455, 391–395. [Google Scholar] [CrossRef]
- Stegmann, C.; Hochdorfer, D.; Lieber, D.; Subramanian, N.; Stöhr, D.; Laib Sampaio, K.; Sinzger, C. A Derivative of Platelet-Derived Growth Factor Receptor Alpha Binds to the Trimer of Human Cytomegalovirus and Inhibits Entry into Fibroblasts and Endothelial Cells. PLoS Pathog. 2017, 13, e1006273. [Google Scholar] [CrossRef]
- Wu, Y.; Prager, A.; Boos, S.; Resch, M.; Brizic, I.; Mach, M.; Wildner, S.; Scrivano, L.; Adler, B. Human Cytomegalovirus Glycoprotein Complex gH/gL/gO Uses PDGFR-α as a Key for Entry. PLoS Pathog. 2017, 13, e1006281. [Google Scholar] [CrossRef]
- Li, J.-W.; Yang, D.; Yang, D.; Chen, Z.; Miao, J.; Liu, W.; Wang, X.; Qiu, Z.; Jin, M.; Shen, Z. Tumors Arise from the Excessive Repair of Damaged Stem Cells. Med. Hypotheses 2017, 102, 112–122. [Google Scholar] [CrossRef]
- Lilley, C.E.; Schwartz, R.A.; Weitzman, M.D. Using or Abusing: Viruses and the Cellular DNA Damage Response. Trends Microbiol. 2007, 15, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Cobbs, C.S.; Soroceanu, L.; Denham, S.; Zhang, W.; Kraus, M.H. Modulation of Oncogenic Phenotype in Human Glioma Cells by Cytomegalovirus IE1–Mediated Mitogenicity. Cancer Res. 2008, 68, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Soroceanu, L.; Matlaf, L.; Khan, S.; Akhavan, A.; Singer, E.; Bezrookove, V.; Decker, S.; Ghanny, S.; Hadaczek, P.; Bengtsson, H.; et al. Cytomegalovirus Immediate-Early Proteins Promote Stemness Properties in Glioblastoma. Cancer Res. 2015, 75, 3065–3076. [Google Scholar] [CrossRef] [PubMed]
- Fornara, O.; Bartek Jr, J.; Rahbar, A.; Odeberg, J.; Khan, Z.; Peredo, I.; Hamerlik, P.; Bartek, J.; Stragliotto, G.; Landázuri, N.; et al. Cytomegalovirus Infection Induces a Stem Cell Phenotype in Human Primary Glioblastoma Cells: Prognostic Significance and Biological Impact. Cell Death Differ. 2016, 23, 261–269. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Herbein, G. EZH2-Myc Hallmark in Oncovirus/Cytomegalovirus Infections and Cytomegalovirus’ Resemblance to Oncoviruses. Cells 2024, 13, 541. [Google Scholar] [CrossRef] [PubMed]
- Herbein, G.; Nehme, Z. Polyploid Giant Cancer Cells, a Hallmark of Oncoviruses and a New Therapeutic Challenge. Front. Oncol. 2020, 10, 567116. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human Papillomavirus and Cervical Cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Tomaić, V. Functional Roles of E6 and E7 Oncoproteins in HPV-Induced Malignancies at Diverse Anatomical Sites. Cancers 2016, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, Y.; Zhao, N.; Chen, H.; Qiao, L.; Zhao, W.; Chen, J.J. Role of Cdk1 in the P53-Independent Abrogation of the Postmitotic Checkpoint by Human Papillomavirus E6. J. Virol. 2015, 89, 2553–2562. [Google Scholar] [CrossRef]
- Fan, X.; Liu, Y.; Heilman, S.A.; Chen, J.J. Human Papillomavirus E7 Induces Rereplication in Response to DNA Damage. J. Virol. 2013, 87, 1200–1210. [Google Scholar] [CrossRef]
- Pal, A.; Kundu, R. Human Papillomavirus E6 and E7: The Cervical Cancer Hallmarks and Targets for Therapy. Front. Microbiol. 2020, 10, 3116. [Google Scholar] [CrossRef] [PubMed]
- Kew, M.C. Hepatitis B Virus x Protein in the Pathogenesis of Hepatitis B Virus-induced Hepatocellular Carcinoma. J. Gastro Hepatol. 2011, 26, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wu, Y.; Tsai, H.; Sun, C.; Wu, Y.; Wu, H.; Pei, Y.; Lu, K.; Yen, T.T.; Chang, C.; et al. Intrahepatic Hepatitis B Virus Large Surface Antigen Induces Hepatocyte Hyperploidy via Failure of Cytokinesis. J. Pathol. 2018, 245, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Musa, J.; Li, J.; Grünewald, T.G. Hepatitis B Virus Large Surface Protein Is Priming for Hepatocellular Carcinoma Development via Induction of Cytokinesis Failure. J. Pathol. 2019, 247, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Moosavy, S.H.; Dvoodian, P.; Nazarnezhad, M.A.; Nejatizaheh, A.; Ephtekhar, E.; Mahboobi, H. Epidemiology, Transmission, Diagnosis, and Outcome of Hepatitis C Virus Infection. Electron. Physician 2017, 9, 5646–5656. [Google Scholar] [CrossRef] [PubMed]
- Machida, K.; Liu, J.-C.; McNamara, G.; Levine, A.; Duan, L.; Lai, M.M.C. Hepatitis C Virus Causes Uncoupling of Mitotic Checkpoint and Chromosomal Polyploidy through the Rb Pathway. J. Virol. 2009, 83, 12590–12600. [Google Scholar] [CrossRef] [PubMed]
- Moriya, K.; Fujie, H.; Shintani, Y.; Yotsuyanagi, H.; Tsutsumi, T.; Ishibashi, K.; Matsuura, Y.; Kimura, S.; Miyamura, T.; Koike, K. The Core Protein of Hepatitis C Virus Induces Hepatocellular Carcinoma in Transgenic Mice. Nat. Med. 1998, 4, 1065–1067. [Google Scholar] [CrossRef] [PubMed]
- Grassmann, R.; Aboud, M.; Jeang, K.-T. Molecular Mechanisms of Cellular Transformation by HTLV-1 Tax. Oncogene 2005, 24, 5976–5985. [Google Scholar] [CrossRef]
- Liang, M.-H.; Geisbert, T.; Yao, Y.; Hinrichs, S.H.; Giam, C.-Z. Human T-Lymphotropic Virus Type 1 Oncoprotein Tax Promotes S-Phase Entry but Blocks Mitosis. J. Virol. 2002, 76, 4022–4033. [Google Scholar] [CrossRef]
- Jin, D.-Y.; Spencer, F.; Jeang, K.-T. Human T Cell Leukemia Virus Type 1 Oncoprotein Tax Targets the Human Mitotic Checkpoint Protein MAD1. Cell 1998, 93, 81–91. [Google Scholar] [CrossRef]
- Malu, A.; Hutchison, T.; Yapindi, L.; Smith, K.; Nelson, K.; Bergeson, R.; Pope, J.; Romeo, M.; Harrod, C.; Ratner, L.; et al. The Human T-Cell Leukemia Virus Type-1 Tax Oncoprotein Dissociates NF-κB p65RelA-Stathmin Complexes and Causes Catastrophic Mitotic Spindle Damage and Genomic Instability. Virology 2019, 535, 83–101. [Google Scholar] [CrossRef]
- Mohanty, S.; Harhaj, E.W. Mechanisms of Oncogenesis by HTLV-1 Tax. Pathogens 2020, 9, 543. [Google Scholar] [CrossRef]
- Tsang, S.H.; Wang, R.; Nakamaru-Ogiso, E.; Knight, S.A.B.; Buck, C.B.; You, J. The Oncogenic Small Tumor Antigen of Merkel Cell Polyomavirus Is an Iron-Sulfur Cluster Protein That Enhances Viral DNA Replication. J. Virol. 2016, 90, 1544–1556. [Google Scholar] [CrossRef]
- Kwun, H.J.; Wendzicki, J.A.; Shuda, Y.; Moore, P.S.; Chang, Y. Merkel Cell Polyomavirus Small T Antigen Induces Genome Instability by E3 Ubiquitin Ligase Targeting. Oncogene 2017, 36, 6784–6792. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Shuda, M.; Chang, Y.; Moore, P.S. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science 2008, 319, 1096–1100. [Google Scholar] [CrossRef]
- Yang, J.F.; Liu, W.; You, J. Characterization of Molecular Mechanisms Driving Merkel Cell Polyomavirus Oncogene Transcription and Tumorigenic Potential. PLoS Pathog. 2023, 19, e1011598. [Google Scholar] [CrossRef]
- Lee, J.M.; Lee, K.-H.; Farrell, C.J.; Ling, P.D.; Kempkes, B.; Park, J.H.; Hayward, S.D. EBNA2 Is Required for Protection of Latently Epstein-Barr Virus-Infected B Cells against Specific Apoptotic Stimuli. J. Virol. 2004, 78, 12694–12697. [Google Scholar] [CrossRef]
- Pan, S.-H.; Tai, C.-C.; Lin, C.-S.; Hsu, W.-B.; Chou, S.-F.; Lai, C.-C.; Chen, J.-Y.; Tien, H.-F.; Lee, F.-Y.; Wang, W.-B. Epstein-Barr Virus Nuclear Antigen 2 Disrupts Mitotic Checkpoint and Causes Chromosomal Instability. Carcinogenesis 2009, 30, 366–375. [Google Scholar] [CrossRef]
- Parker, G.A.; Touitou, R.; Allday, M.J. Epstein-Barr Virus EBNA3C Can Disrupt Multiple Cell Cycle Checkpoints and Induce Nuclear Division Divorced from Cytokinesis. Oncogene 2000, 19, 700–709. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.A.; Crook, T.; Bain, M.; Sara, E.A.; Farrell, P.J.; Allday, M.J. Epstein-Barr Virus Nuclear Antigen (EBNA)3C Is an Immortalizing Oncoprotein with Similar Properties to Adenovirus E1A and Papillomavirus E7. Oncogene 1996, 13, 2541–2549. [Google Scholar]
- Lajoie, V.; Lemieux, B.; Sawan, B.; Lichtensztejn, D.; Lichtensztejn, Z.; Wellinger, R.; Mai, S.; Knecht, H. LMP1 Mediates Multinuclearity through Downregulation of Shelterin Proteins and Formation of Telomeric Aggregates. Blood 2015, 125, 2101–2110. [Google Scholar] [CrossRef] [PubMed]
- Kieser, A.; Sterz, K.R. The Latent Membrane Protein 1 (LMP1). Curr. Top. Microbiol. Immunol. 2015, 391, 119–149. [Google Scholar] [CrossRef]
- Münz, C. Latency and Lytic Replication in Epstein–Barr Virus-Associated Oncogenesis. Nat. Rev. Microbiol. 2019, 17, 691–700. [Google Scholar] [CrossRef]
- Liu, J.; Martin, H.J.; Liao, G.; Hayward, S.D. The Kaposi’s Sarcoma-Associated Herpesvirus LANA Protein Stabilizes and Activates c-Myc. J. Virol. 2007, 81, 10451–10459. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Robertson, E.S. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded Latency-Associated Nuclear Antigen Induces Chromosomal Instability through Inhibition of P53 Function. J. Virol. 2006, 80, 697–709. [Google Scholar] [CrossRef]
- Sun, Z.; Xiao, B.; Jha, H.C.; Lu, J.; Banerjee, S.; Robertson, E.S. Kaposi’s Sarcoma-Associated Herpesvirus-Encoded LANA Can Induce Chromosomal Instability through Targeted Degradation of the Mitotic Checkpoint Kinase Bub1. J. Virol. 2014, 88, 7367–7378. [Google Scholar] [CrossRef]
- Godden-Kent, D.; Talbot, S.J.; Boshoff, C.; Chang, Y.; Moore, P.; Weiss, R.A.; Mittnacht, S. The Cyclin Encoded by Kaposi’s Sarcoma-Associated Herpesvirus Stimulates Cdk6 to Phosphorylate the Retinoblastoma Protein and Histone H1. J. Virol. 1997, 71, 4193–4198. [Google Scholar] [CrossRef]
- Laman, H.; Coverley, D.; Krude, T.; Laskey, R.; Jones, N. Viral Cyclin–Cyclin-Dependent Kinase 6 Complexes Initiate Nuclear DNA Replication. Mol. Cell. Biol. 2001, 21, 624–635. [Google Scholar] [CrossRef]
- Li, T.; Gao, S.-J. Metabolic Reprogramming and Metabolic Sensors in KSHV-Induced Cancers and KSHV Infection. Cell Biosci. 2021, 11, 176. [Google Scholar] [CrossRef]
- Kumar, A.; Tripathy, M.K.; Pasquereau, S.; Al Moussawi, F.; Abbas, W.; Coquard, L.; Khan, K.A.; Russo, L.; Algros, M.-P.; Valmary-Degano, S.; et al. The Human Cytomegalovirus Strain DB Activates Oncogenic Pathways in Mammary Epithelial Cells. EBioMedicine 2018, 30, 167–183. [Google Scholar] [CrossRef]
- Moussawi, F.A.; Kumar, A.; Pasquereau, S.; Tripathy, M.K.; Karam, W.; Diab-Assaf, M.; Herbein, G. The Transcriptome of Human Mammary Epithelial Cells Infected with the HCMV-DB Strain Displays Oncogenic Traits. Sci. Rep. 2018, 8, 12574. [Google Scholar] [CrossRef] [PubMed]
- Nehme, Z.; Pasquereau, S.; Haidar Ahmad, S.; Coaquette, A.; Molimard, C.; Monnien, F.; Algros, M.-P.; Adotevi, O.; Diab Assaf, M.; Feugeas, J.-P.; et al. Polyploid Giant Cancer Cells, Stemness and Epithelial-Mesenchymal Plasticity Elicited by Human Cytomegalovirus. Oncogene 2021, 40, 3030–3046. [Google Scholar] [CrossRef] [PubMed]
- Haidar Ahmad, S.; Pasquereau, S.; El Baba, R.; Nehme, Z.; Lewandowski, C.; Herbein, G. Distinct Oncogenic Transcriptomes in Human Mammary Epithelial Cells Infected With Cytomegalovirus. Front. Immunol. 2021, 12, 772160. [Google Scholar] [CrossRef] [PubMed]
- Nehme, Z.; Pasquereau, S.; Haidar Ahmad, S.; El Baba, R.; Herbein, G. Polyploid Giant Cancer Cells, EZH2 and Myc Upregulation in Mammary Epithelial Cells Infected with High-Risk Human Cytomegalovirus. eBioMedicine 2022, 80, 104056. [Google Scholar] [CrossRef] [PubMed]
- Haidar Ahmad, S.; El Baba, R.; Herbein, G. Polyploid Giant Cancer Cells, Cytokines and Cytomegalovirus in Breast Cancer Progression. Cancer Cell Int. 2023, 23, 119. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Pasquereau, S.; Haidar Ahmad, S.; Monnien, F.; Abad, M.; Bibeau, F.; Herbein, G. EZH2-Myc Driven Glioblastoma Elicited by Cytomegalovirus Infection of Human Astrocytes. Oncogene 2023, 42, 2031–2045. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Haidar Ahmad, S.; Monnien, F.; Mansar, R.; Bibeau, F.; Herbein, G. Polyploidy, EZH2 Upregulation, and Transformation in Cytomegalovirus-Infected Human Ovarian Epithelial Cells. Oncogene 2023, 42, 3047–3061. [Google Scholar] [CrossRef] [PubMed]
- El Baba, R.; Pasquereau, S.; Haidar Ahmad, S.; Diab-Assaf, M.; Herbein, G. Oncogenic and Stemness Signatures of the High-Risk HCMV Strains in Breast Cancer Progression. Cancers 2022, 14, 4271. [Google Scholar] [CrossRef]
- Bouezzedine, F.; El Baba, R.; Haidar Ahmad, S.; Herbein, G. Polyploid Giant Cancer Cells Generated from Human Cytomegalovirus-Infected Prostate Epithelial Cells. Cancers 2023, 15, 4994. [Google Scholar] [CrossRef]
- Müller-Coan, B.G.; Caetano, B.F.R.; Pagano, J.S.; Elgui De Oliveira, D. Cancer Progression Goes Viral: The Role of Oncoviruses in Aggressiveness of Malignancies. Trends Cancer 2018, 4, 485–498. [Google Scholar] [CrossRef]
- Compton, T.; Kurt-Jones, E.A.; Boehme, K.W.; Belko, J.; Latz, E.; Golenbock, D.T.; Finberg, R.W. Human Cytomegalovirus Activates Inflammatory Cytokine Responses via CD14 and Toll-Like Receptor 2. J. Virol. 2003, 77, 4588–4596. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Yao, J.; Bast, R.C.; Sood, A.K.; Liu, J. IL-6 Promotes Drug Resistance through Formation of Polyploid Giant Cancer Cells and Stromal Fibroblast Reprogramming. Oncogenesis 2021, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.E.; Mason, G.M.; Wills, M.R. Human Cytomegalovirus Immunity and Immune Evasion. Virus Res. 2011, 157, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Herbein, G. Epigenetic Regulation of Human Cytomegalovirus Latency: An Update. Epigenomics 2014, 6, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Renzette, N.; Gibson, L.; Bhattacharjee, B.; Fisher, D.; Schleiss, M.R.; Jensen, J.D.; Kowalik, T.F. Rapid Intrahost Evolution of Human Cytomegalovirus Is Shaped by Demography and Positive Selection. PLoS Genet. 2013, 9, e1003735. [Google Scholar] [CrossRef]
- Renzette, N.; Bhattacharjee, B.; Jensen, J.D.; Gibson, L.; Kowalik, T.F. Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants. PLoS Pathog. 2011, 7, e1001344. [Google Scholar] [CrossRef] [PubMed]
- Dhingra, A.; Götting, J.; Varanasi, P.R.; Steinbrueck, L.; Camiolo, S.; Zischke, J.; Heim, A.; Schulz, T.F.; Weissinger, E.M.; Kay-Fedorov, P.C.; et al. Human Cytomegalovirus Multiple-Strain Infections and Viral Population Diversity in Haematopoietic Stem Cell Transplant Recipients Analysed by High-Throughput Sequencing. Med. Microbiol. Immunol. 2021, 210, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Götting, J.; Lazar, K.; Suárez, N.M.; Steinbrück, L.; Rabe, T.; Goelz, R.; Schulz, T.F.; Davison, A.J.; Hamprecht, K.; Ganzenmueller, T. Human Cytomegalovirus Genome Diversity in Longitudinally Collected Breast Milk Samples. Front. Cell. Infect. Microbiol. 2021, 11, 664247. [Google Scholar] [CrossRef] [PubMed]
- Suárez, N.M.; Wilkie, G.S.; Hage, E.; Camiolo, S.; Holton, M.; Hughes, J.; Maabar, M.; Vattipally, S.B.; Dhingra, A.; Gompels, U.A.; et al. Human Cytomegalovirus Genomes Sequenced Directly From Clinical Material: Variation, Multiple-Strain Infection, Recombination, and Gene Loss. J. Infect. Dis. 2019, 220, 781–791. [Google Scholar] [CrossRef]
- Hage, E.; Wilkie, G.S.; Linnenweber-Held, S.; Dhingra, A.; Suárez, N.M.; Schmidt, J.J.; Kay-Fedorov, P.C.; Mischak-Weissinger, E.; Heim, A.; Schwarz, A.; et al. Characterization of Human Cytomegalovirus Genome Diversity in Immunocompromised Hosts by Whole-Genome Sequencing Directly From Clinical Specimens. J. Infect. Dis. 2017, 215, 1673–1683. [Google Scholar] [CrossRef]
- Suárez, N.M.; Blyth, E.; Li, K.; Ganzenmueller, T.; Camiolo, S.; Avdic, S.; Withers, B.; Linnenweber-Held, S.; Gwinner, W.; Dhingra, A.; et al. Whole-Genome Approach to Assessing Human Cytomegalovirus Dynamics in Transplant Patients Undergoing Antiviral Therapy. Front. Cell. Infect. Microbiol. 2020, 10, 267. [Google Scholar] [CrossRef] [PubMed]
- Görzer, I.; Guelly, C.; Trajanoski, S.; Puchhammer-Stöckl, E. Deep Sequencing Reveals Highly Complex Dynamics of Human Cytomegalovirus Genotypes in Transplant Patients over Time. J. Virol. 2010, 84, 7195–7203. [Google Scholar] [CrossRef] [PubMed]
- Gatherer, D.; Seirafian, S.; Cunningham, C.; Holton, M.; Dargan, D.J.; Baluchova, K.; Hector, R.D.; Galbraith, J.; Herzyk, P.; Wilkinson, G.W.G.; et al. High-Resolution Human Cytomegalovirus Transcriptome. Proc. Natl. Acad. Sci. USA 2011, 108, 19755–19760. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.W.G.; Davison, A.J.; Tomasec, P.; Fielding, C.A.; Aicheler, R.; Murrell, I.; Seirafian, S.; Wang, E.C.Y.; Weekes, M.; Lehner, P.J.; et al. Human Cytomegalovirus: Taking the Strain. Med. Microbiol. Immunol. 2015, 204, 273–284. [Google Scholar] [CrossRef] [PubMed]
- Sijmons, S.; Thys, K.; Mbong Ngwese, M.; Van Damme, E.; Dvorak, J.; Van Loock, M.; Li, G.; Tachezy, R.; Busson, L.; Aerssens, J.; et al. High-Throughput Analysis of Human Cytomegalovirus Genome Diversity Highlights the Widespread Occurrence of Gene-Disrupting Mutations and Pervasive Recombination. J. Virol. 2015, 89, 7673–7695. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Schafer, X.; Munger, J. Expression of Oncogenic Alleles Induces Multiple Blocks to Human Cytomegalovirus Infection. J. Virol. 2016, 90, 4346–4356. [Google Scholar] [CrossRef]
- Branch, K.M.; Garcia, E.C.; Chen, Y.M.; McGregor, M.; Min, M.; Prosser, R.; Whitney, N.; Spencer, J.V. Productive Infection of Human Breast Cancer Cell Lines with Human Cytomegalovirus (HCMV). Pathogens 2021, 10, 641. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Neumann, D.M. Persistent HCMV Infection of a Glioblastoma Cell Line Contributes to the Development of Resistance to Temozolomide. Virus Res. 2020, 276, 197829. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Tang, X.; McMullen, T.P.W.; Brindley, D.N.; Hemmings, D.G. PDGFRα Enhanced Infection of Breast Cancer Cells with Human Cytomegalovirus but Infection of Fibroblasts Increased Prometastatic Inflammation Involving Lysophosphatidate Signaling. IJMS 2021, 22, 9817. [Google Scholar] [CrossRef]
- Herbein, G. High-Risk Oncogenic Human Cytomegalovirus. Viruses 2022, 14, 2462. [Google Scholar] [CrossRef]
- Cohen, A.; Brodie, C.; Sarid, R. An Essential Role of ERK Signalling in TPA-Induced Reactivation of Kaposi’s Sarcoma-Associated Herpesvirus. J. Gen. Virol. 2006, 87, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Van Sciver, N.; Ohashi, M.; Pauly, N.P.; Bristol, J.A.; Nelson, S.E.; Johannsen, E.C.; Kenney, S.C. Hippo Signaling Effectors YAP and TAZ Induce Epstein-Barr Virus (EBV) Lytic Reactivation through TEADs in Epithelial Cells. PLoS Pathog. 2021, 17, e1009783. [Google Scholar] [CrossRef] [PubMed]
- Cerasuolo, A.; Annunziata, C.; Tortora, M.; Starita, N.; Stellato, G.; Greggi, S.; Maglione, M.G.; Ionna, F.; Losito, S.; Botti, G.; et al. Comparative Analysis of HPV16 Gene Expression Profiles in Cervical and in Oropharyngeal Squamous Cell Carcinoma. Oncotarget 2017, 8, 34070–34081. [Google Scholar] [CrossRef] [PubMed]
- Guyon, J.; Haidar Ahmad, S.; El Baba, R.; Le Quang, M.; Bikfalvi, A.; Daubon, T.; Herbein, G. Generation of Glioblastoma in Mice Engrafted with Human Cytomegalovirus-Infected Astrocytes. Cancer Gene Ther. 2024. [Google Scholar] [CrossRef] [PubMed]
- Guven-Maiorov, E.; Tsai, C.-J.; Nussinov, R. Oncoviruses Can Drive Cancer by Rewiring Signaling Pathways Through Interface Mimicry. Front. Oncol. 2019, 9, 1236. [Google Scholar] [CrossRef] [PubMed]
- Aghamajidi, A.; Farhangnia, P.; Pashangzadeh, S.; Damavandi, A.R.; Jafari, R. Tumor-Promoting Myeloid Cells in the Pathogenesis of Human Oncoviruses: Potential Targets for Immunotherapy. Cancer Cell Int. 2022, 22, 327. [Google Scholar] [CrossRef] [PubMed]
- Mocarski, E.S. Immunomodulation by Cytomegaloviruses: Manipulative Strategies beyond Evasion. Trends Microbiol. 2002, 10, 332–339. [Google Scholar] [CrossRef]
- Semmes, E.C.; Hurst, J.H.; Walsh, K.M.; Permar, S.R. Cytomegalovirus as an Immunomodulator across the Lifespan. Curr. Opin. Virol. 2020, 44, 112–120. [Google Scholar] [CrossRef]
- El Baba, R.; Herbein, G. Immune Landscape of CMV Infection in Cancer Patients: From “Canonical” Diseases Toward Virus-Elicited Oncomodulation. Front. Immunol. 2021, 12, 730765. [Google Scholar] [CrossRef]
- Cinatl, J.; Scholz, M.; Doerr, H.W. Role of Tumor Cell Immune Escape Mechanisms in Cytomegalovirus-mediated Oncomodulation. Med. Res. Rev. 2005, 25, 167–185. [Google Scholar] [CrossRef]
- Cinatl, J.; Scholz, M.; Kotchetkov, R.; Vogel, J.-U.; Wilhelm Doerr, H. Molecular Mechanisms of the Modulatory Effects of HCMV Infection in Tumor Cell Biology. Trends Mol. Med. 2004, 10, 19–23. [Google Scholar] [CrossRef]
- Margulies, B.J.; Browne, H.; Gibson, W. Identification of the Human Cytomegalovirus G Protein-Coupled Receptor Homologue Encoded by UL33 in Infected Cells and Enveloped Virus Particles. Virology 1996, 225, 111–125. [Google Scholar] [CrossRef]
- Chen, Y.; Song, Y.; Du, W.; Gong, L.; Chang, H.; Zou, Z. Tumor-Associated Macrophages: An Accomplice in Solid Tumor Progression. J. Biomed. Sci. 2019, 26, 78. [Google Scholar] [CrossRef] [PubMed]
- Baasch, S.; Giansanti, P.; Kolter, J.; Riedl, A.; Forde, A.J.; Runge, S.; Zenke, S.; Elling, R.; Halenius, A.; Brabletz, S.; et al. Cytomegalovirus Subverts Macrophage Identity. Cell 2021, 184, 3774–3793.e25. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Coaquette, A.; Davrinche, C.; Herbein, G. Bcl-3-Regulated Transcription from Major Immediate-Early Promoter of Human Cytomegalovirus in Monocyte-Derived Macrophages. J. Immunol. 2009, 182, 7784–7794. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hein, S.M.; Haricharan, S.; Johnston, A.N.; Toneff, M.J.; Reddy, J.P.; Dong, J.; Bu, W.; Li, Y. Luminal Epithelial Cells within the Mammary Gland Can Produce Basal Cells upon Oncogenic Stress. Oncogene 2016, 35, 1461–1467. [Google Scholar] [CrossRef]
- Rodilla, V.; Fre, S. Cellular Plasticity of Mammary Epithelial Cells Underlies Heterogeneity of Breast Cancer. Biomedicines 2018, 6, 103. [Google Scholar] [CrossRef] [PubMed]
- Tata, P.R.; Mou, H.; Pardo-Saganta, A.; Zhao, R.; Prabhu, M.; Law, B.M.; Vinarsky, V.; Cho, J.L.; Breton, S.; Sahay, A.; et al. Dedifferentiation of Committed Epithelial Cells into Stem Cells in Vivo. Nature 2013, 503, 218–223. [Google Scholar] [CrossRef]
- Yu, W.; Ma, Y.; Ochoa, A.C.; Shankar, S.; Srivastava, R.K. Cellular Transformation of Human Mammary Epithelial Cells by SATB2. Stem Cell Res. 2017, 19, 139–147. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, V.; Yang, Y.; Schubert, M.C.; Reyhan, E.; Tetzlaff, S.K.; Wißmann, N.; Botz, M.; Soyka, S.J.; Beretta, C.A.; Pramatarov, R.L.; et al. Glioblastoma Hijacks Neuronal Mechanisms for Brain Invasion. Cell 2022, 185, 2899–2917.e31. [Google Scholar] [CrossRef]
- Merchut-Maya, J.M.; Bartek, J.; Bartkova, J.; Galanos, P.; Pantalone, M.R.; Lee, M.; Cui, H.L.; Shilling, P.J.; Brøchner, C.B.; Broholm, H.; et al. Human Cytomegalovirus Hijacks Host Stress Response Fueling Replication Stress and Genome Instability. Cell Death Differ. 2022, 29, 1639–1653. [Google Scholar] [CrossRef]
- Fortunato, E.A.; Spector, D.H. Viral Induction of Site-specific Chromosome Damage. Rev. Med. Virol. 2003, 13, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Siew, V.-K.; Duh, C.-Y.; Wang, S.-K. Human Cytomegalovirus UL76 Induces Chromosome Aberrations. J. Biomed. Sci. 2009, 16, 107. [Google Scholar] [CrossRef]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Roberts, C.W.M. Targeting EZH2 in Cancer. Nat. Med. 2016, 22, 128–134. [Google Scholar] [CrossRef] [PubMed]
- Veneti, Z.; Gkouskou, K.; Eliopoulos, A. Polycomb Repressor Complex 2 in Genomic Instability and Cancer. IJMS 2017, 18, 1657. [Google Scholar] [CrossRef]
- Guo, S.; Li, X.; Rohr, J.; Wang, Y.; Ma, S.; Chen, P.; Wang, Z. EZH2 Overexpression in Different Immunophenotypes of Breast Carcinoma and Association with Clinicopathologic Features. Diagn. Pathol. 2016, 11, 41. [Google Scholar] [CrossRef]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.A.B.; Otte, A.P.; Hayes, D.F.; et al. EZH2 Is a Marker of Aggressive Breast Cancer and Promotes Neoplastic Transformation of Breast Epithelial Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef]
- Reid, B.M.; Vyas, S.; Chen, Z.; Chen, A.; Kanetsky, P.A.; Permuth, J.B.; Sellers, T.A.; Saglam, O. Morphologic and Molecular Correlates of EZH2 as a Predictor of Platinum Resistance in High-Grade Ovarian Serous Carcinoma. BMC Cancer 2021, 21, 714. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.E.; Moore, H.M.; Li, X.; Toy, K.A.; Huang, W.; Sabel, M.S.; Kidwell, K.M.; Kleer, C.G. EZH2 Expands Breast Stem Cells through Activation of NOTCH1 Signaling. Proc. Natl. Acad. Sci. USA 2014, 111, 3098–3103. [Google Scholar] [CrossRef]
- Wu, J.; Crowe, D.L. The Histone Methyltransferase EZH2 Promotes Mammary Stem and Luminal Progenitor Cell Expansion, Metastasis and Inhibits Estrogen Receptor-Positive Cellular Differentiation in a Model of Basal Breast Cancer. Oncol. Rep. 2015, 34, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Koh, C.M.; Iwata, T.; Zheng, Q.; Bethel, C.; Yegnasubramanian, S.; De Marzo, A.M. Myc Enforces Overexpression of EZH2 in Early Prostatic Neoplasia via Transcriptional and Post-Transcriptional Mechanisms. Oncotarget 2011, 2, 669–683. [Google Scholar] [CrossRef]
- Kuser-Abali, G.; Alptekin, A.; Cinar, B. Overexpression of MYC and EZH2 Cooperates to Epigenetically Silence MST1 Expression. Epigenetics 2014, 9, 634–643. [Google Scholar] [CrossRef] [PubMed]
- Dzutsev, A.; Badger, J.H.; Perez-Chanona, E.; Roy, S.; Salcedo, R.; Smith, C.K.; Trinchieri, G. Microbes and Cancer. Annu. Rev. Immunol. 2017, 35, 199–228. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The Microbiome, Cancer, and Cancer Therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Godoy-Vitorino, F.; Romaguera, J.; Zhao, C.; Vargas-Robles, D.; Ortiz-Morales, G.; Vázquez-Sánchez, F.; Sanchez-Vázquez, M.; De La Garza-Casillas, M.; Martinez-Ferrer, M.; White, J.R.; et al. Cervicovaginal Fungi and Bacteria Associated With Cervical Intraepithelial Neoplasia and High-Risk Human Papillomavirus Infections in a Hispanic Population. Front. Microbiol. 2018, 9, 2533. [Google Scholar] [CrossRef]
- Puchhammer-Stöckl, E.; Görzer, I. Cytomegalovirus and Epstein-Barr Virus Subtypes—The Search for Clinical Significance. J. Clin. Virol. 2006, 36, 239–248. [Google Scholar] [CrossRef]
- Kohda, C.; Ino, S.; Ishikawa, H.; Kuno, Y.; Nagashima, R.; Iyoda, M. The Essential Role of Intestinal Microbiota in Cytomegalovirus Reactivation. Microbiol. Spectr. 2023, 11, e02341-23. [Google Scholar] [CrossRef]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef]
- Kowald, A.; Passos, J.F.; Kirkwood, T.B.L. On the Evolution of Cellular Senescence. Aging Cell 2020, 19, e13270. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Kohli, J.; Demaria, M. Senescent Cells in Cancer Therapy: Friends or Foes? Trends Cancer 2020, 6, 838–857. [Google Scholar] [CrossRef]
- Birch, J.; Gil, J. Senescence and the SASP: Many Therapeutic Avenues. Genes. Dev. 2020, 34, 1565–1576. [Google Scholar] [CrossRef]
- Faget, D.V.; Ren, Q.; Stewart, S.A. Unmasking Senescence: Context-Dependent Effects of SASP in Cancer. Nat. Rev. Cancer 2019, 19, 439–453. [Google Scholar] [CrossRef]
- Gorgoulis, V.; Adams, P.D.; Alimonti, A.; Bennett, D.C.; Bischof, O.; Bishop, C.; Campisi, J.; Collado, M.; Evangelou, K.; Ferbeyre, G.; et al. Cellular Senescence: Defining a Path Forward. Cell 2019, 179, 813–827. [Google Scholar] [CrossRef] [PubMed]
- De Blander, H.; Morel, A.-P.; Senaratne, A.P.; Ouzounova, M.; Puisieux, A. Cellular Plasticity: A Route to Senescence Exit and Tumorigenesis. Cancers 2021, 13, 4561. [Google Scholar] [CrossRef] [PubMed]
- Baker, D.J.; Childs, B.G.; Durik, M.; Wijers, M.E.; Sieben, C.J.; Zhong, J.; Saltness, R.A.; Jeganathan, K.B.; Verzosa, G.C.; Pezeshki, A.; et al. Naturally Occurring p16Ink4a-Positive Cells Shorten Healthy Lifespan. Nature 2016, 530, 184–189. [Google Scholar] [CrossRef]
- Liu, J.; Erenpreisa, J.; Sikora, E. Polyploid Giant Cancer Cells: An Emerging New Field of Cancer Biology. Semin. Cancer Biol. 2022, 81, 1–4. [Google Scholar] [CrossRef]
- Pienta, K.J.; Hammarlund, E.U.; Brown, J.S.; Amend, S.R.; Axelrod, R.M. Cancer Recurrence and Lethality Are Enabled by Enhanced Survival and Reversible Cell Cycle Arrest of Polyaneuploid Cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2020838118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yao, J.; Li, X.; Niu, N.; Liu, Y.; Hajek, R.A.; Peng, G.; Westin, S.; Sood, A.K.; Liu, J. Targeting Polyploid Giant Cancer Cells Potentiates a Therapeutic Response and Overcomes Resistance to PARP Inhibitors in Ovarian Cancer. Sci. Adv. 2023, 9, eadf7195. [Google Scholar] [CrossRef] [PubMed]
- Seoane, R.; Vidal, S.; Bouzaher, Y.H.; El Motiam, A.; Rivas, C. The Interaction of Viruses with the Cellular Senescence Response. Biology 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Bouezzedine, F.; El Baba, R.; Morot-Bizot, S.; Diab-Assaf, M.; Herbein, G. Cytomegalovirus at the Crossroads of Immunosenescence and Oncogenesis. Explor. Immunol. 2023, 3, 17–27. [Google Scholar] [CrossRef]
- Hewavisenti, R.V.; Arena, J.; Ahlenstiel, C.L.; Sasson, S.C. Human Papillomavirus in the Setting of Immunodeficiency: Pathogenesis and the Emergence of next-Generation Therapies to Reduce the High Associated Cancer Risk. Front. Immunol. 2023, 14, 1112513. [Google Scholar] [CrossRef]
| Viral Agent | Oncoproteins | Cancer Cell Lines/Tumor Types | Associated/Described Outcomes | References |
|---|---|---|---|---|
| HPV | E6, E7, E5 |
|
| [27,28,29,30,31] |
| HBV | HBx, LHBs |
|
| [32,33,34] |
| HCV | None (HCV core, NS3, NS5A, NS5B *) | PGCCs | [35,36,37] | |
| HTLV-1 | Tax |
|
| [38,39,40,41,42] |
| MCPyV | LT-Ag | PGCCs | [43,44,45,46] | |
| EBV | LMP1, EBNA2, EBNA3C, BNRF1 |
|
| [47,48,49,50,51,52,53] |
| KSHV | LANA Cyclin K |
|
| [54,55,56,57,58,59] |
| HCMV | IE1, IE2, US28 ** |
|
| [60,61,62,63,64,65,66,67,68,69] |
| HCMV-Transformed Primary Human Cell | Oncogenic High-Risk HCMV Strains | Phenotypic Features of HCMV-Transformed Cells | Molecular Characteristics of HCMV-Transformed Cells | References |
|---|---|---|---|---|
| CTH cell (CMV-transformed human mammary epithelial cells) | DB, BL HCMV strains isolated from TNBC tumors |
|
| [60,61,62,63,64,65,68] |
| CTO cells (CMV-transformed ovarian epithelial cells) | DB, BL HCMV strains isolated from HGSOC tumors |
|
| [67] |
| CTP cell (CMV-transformed prostate epithelial cells) | DB, BL |
|
| [69] |
| CEGBC (CMV-elicited glioblastoma Cells) | DB, BL HCMV strains isolated from glioblastoma tumors |
|
| [66] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herbein, G. Cellular Transformation by Human Cytomegalovirus. Cancers 2024, 16, 1970. https://doi.org/10.3390/cancers16111970
Herbein G. Cellular Transformation by Human Cytomegalovirus. Cancers. 2024; 16(11):1970. https://doi.org/10.3390/cancers16111970
Chicago/Turabian StyleHerbein, Georges. 2024. "Cellular Transformation by Human Cytomegalovirus" Cancers 16, no. 11: 1970. https://doi.org/10.3390/cancers16111970
APA StyleHerbein, G. (2024). Cellular Transformation by Human Cytomegalovirus. Cancers, 16(11), 1970. https://doi.org/10.3390/cancers16111970




