Neuropilin2 in Mesenchymal Stromal Cells as a Potential Novel Therapeutic Target in Myelofibrosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort
2.2. Immunohistochemistry and Scoring
2.3. Cell Culture and In Vitro Osteoblast Differentiation
2.4. scRNASeq Analysis
2.5. CRISPR/Cas9-Mediated Gene Editing of MC3T3-E1 Cells
2.6. RNAseq Analysis of MC3T3-E1 Clones
2.7. Cell Morphology
2.8. Cell Proliferation
2.9. Analysis of Nrp2 Knockout Mouse Femurs
3. Results
3.1. NRP2 and NCAM1 Expression Is Increased in Myeloid Neoplasms Associated with Myelofibrosis In Situ
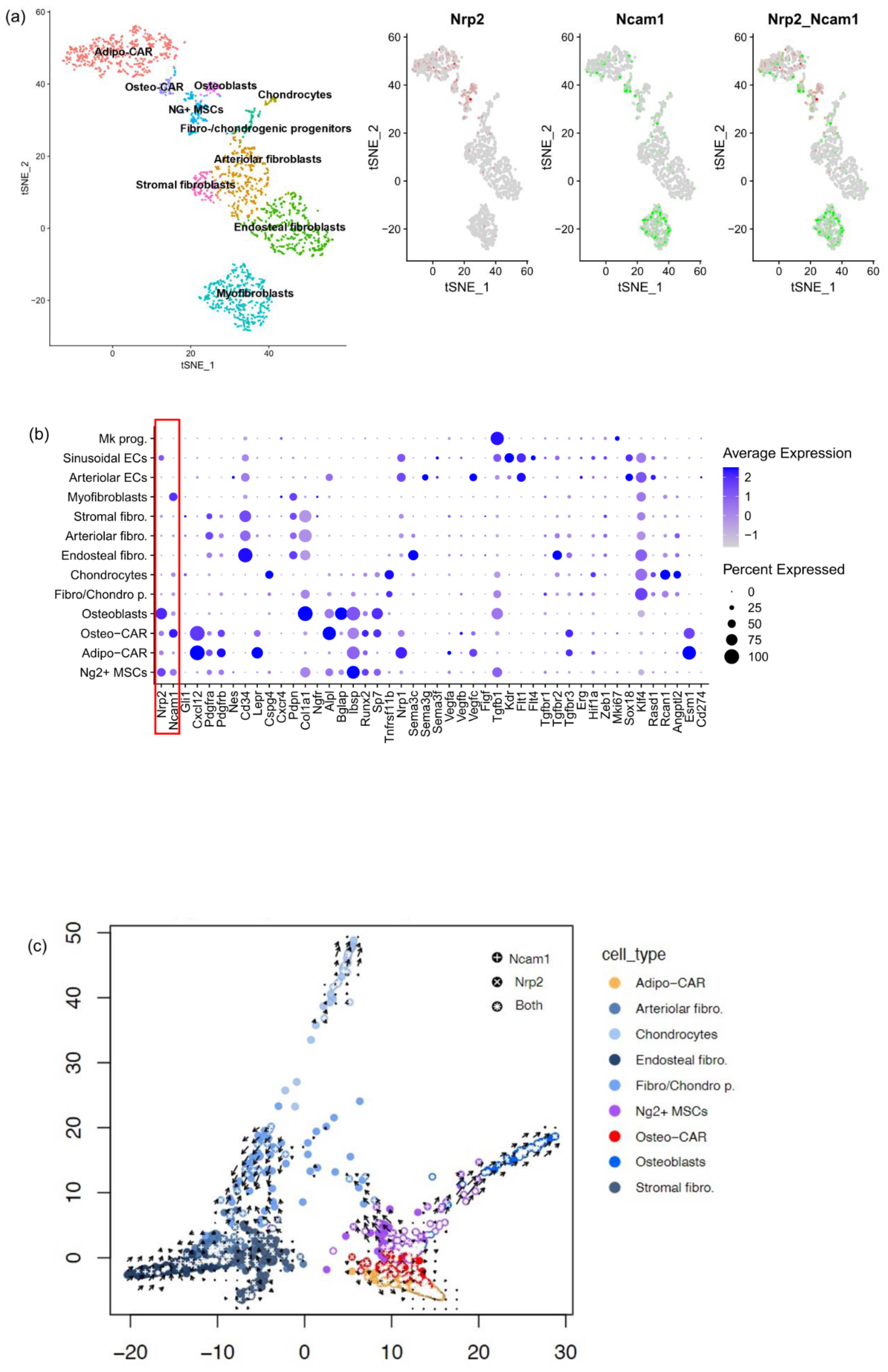
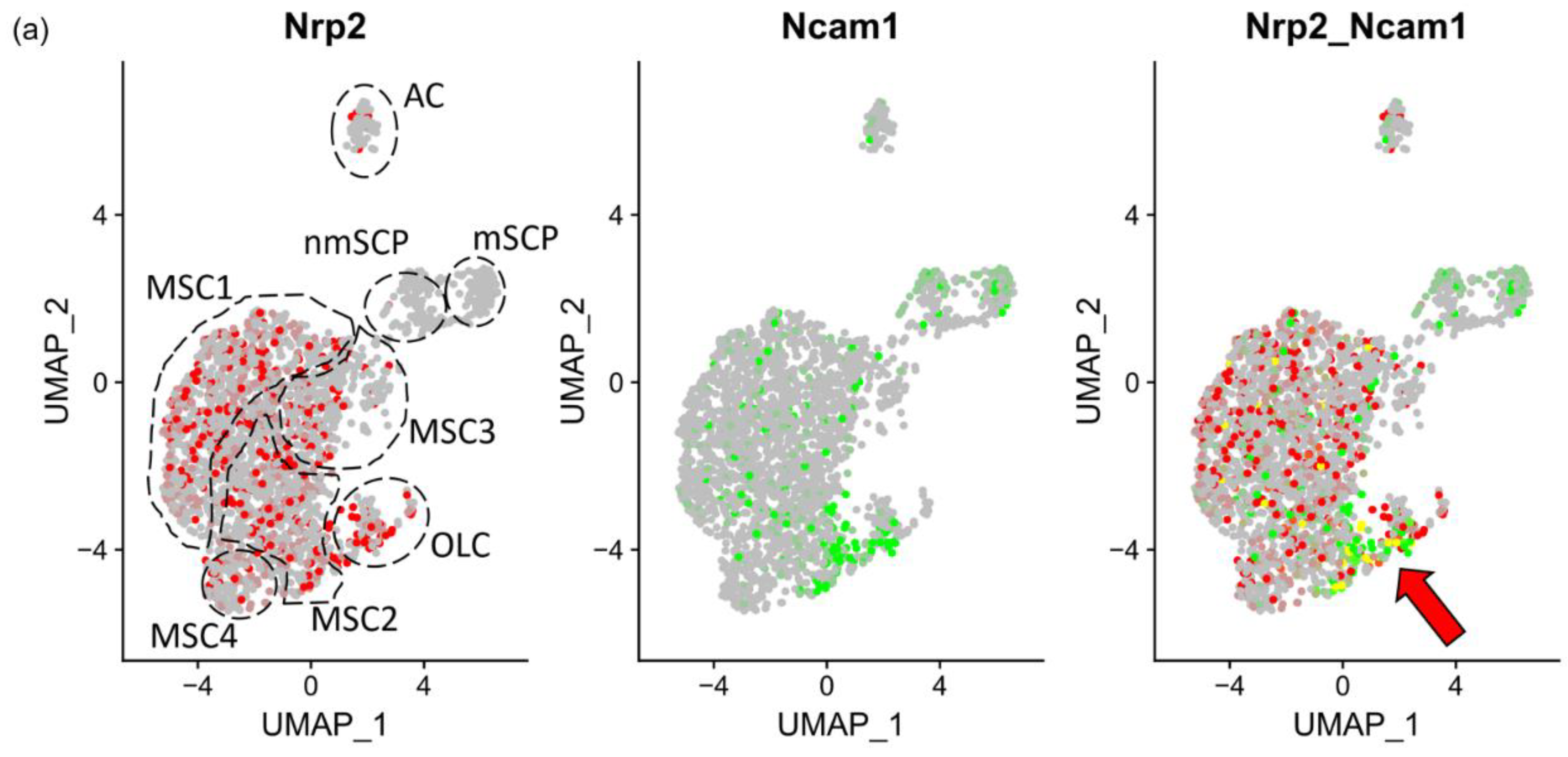
3.2. Phenotypic Identification of NRP2 and NCAM1 Co-Expressing MSC in Normal BM
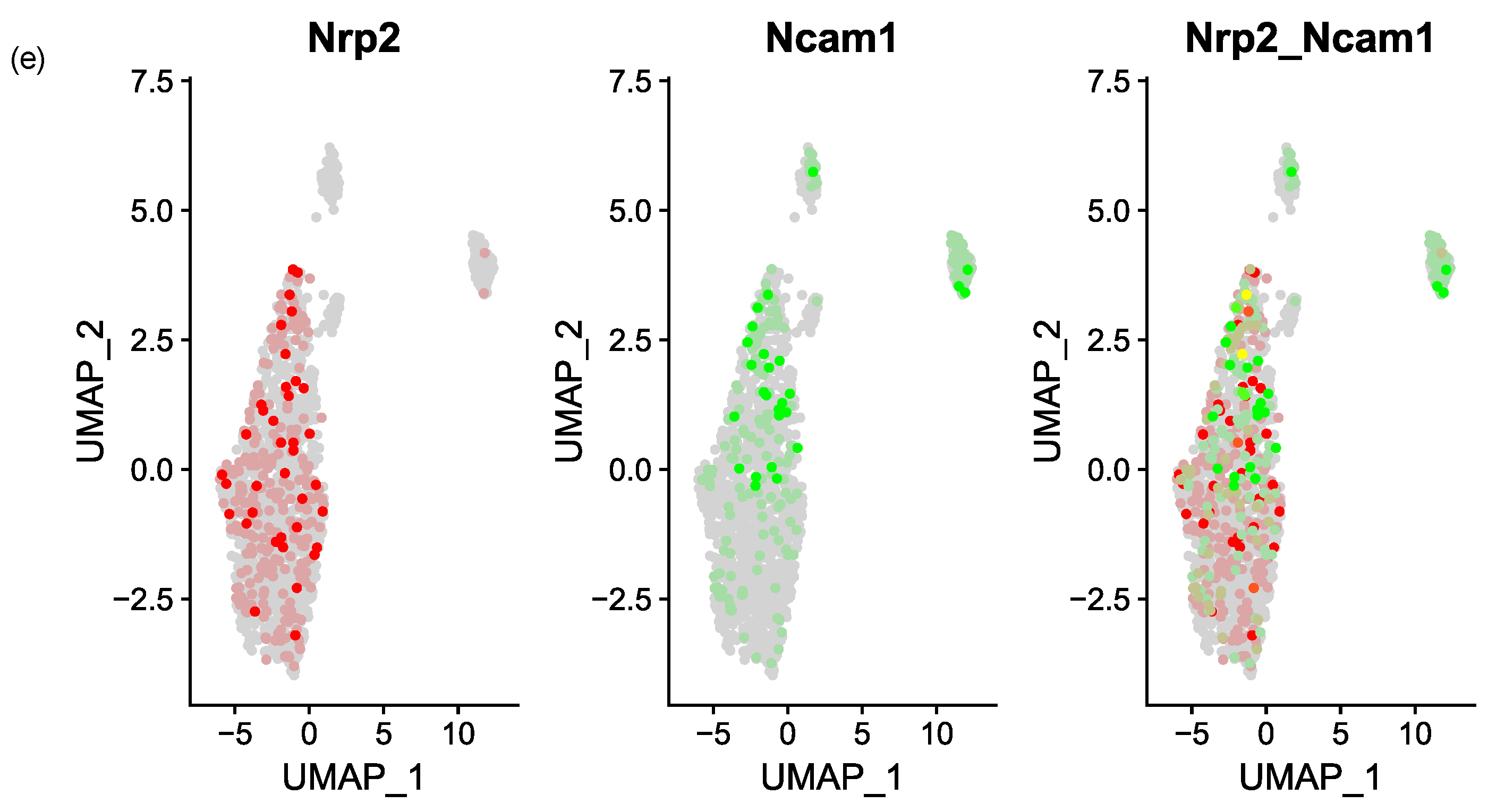
3.3. NRP2 and Ligand Expression in Murine Models of Myelofibrosis
3.4. Nrp2 Is Associated with Increased Osteogenic Differentiation In Vitro
3.5. CRISPR/Cas9-Mediated Nrp2 Ablation in MC3T3-E1 Cells Generates an Osteogenic Differentiation Defect In Vitro
3.6. Osteopenia by Nrp2 Loss In Vivo Is Accompanied by Altered Osteoblast Morphology
4. Discussion
5. Limitations of the Study
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AML | acute myeloid leukemia |
| BM | bone marrow |
| CAR | Cxcl12-abundant reticular |
| EC | endothelial cells |
| ET | essential thrombocythemia |
| IHC | immunohistochemistry |
| KO | knockout |
| MDS | myelodysplastic syndromes |
| MF | myelofibrosis |
| MPN | myeloproliferative neoplasm |
| MSC | mesenchymal stromal cells |
| NCAM1 | neural cell adhesion molecule 1 |
| NRP2 | neuropilin 2 |
| OLC | osteolineage cells |
| PMF | primary myelofibrosis |
| PV | polycythemia vera |
| SCP | Schwann cell progenitors |
| ScRNAseq | single cell RNA sequencing |
| TGFβ1 | transforming growth factor β1 |
| VEGFs | vascular endothelial growth factors |
| WT | wild type |
References
- Rampal, R.; Al-Shahrour, F.; Abdel-Wahab, O.; Patel, J.P.; Brunel, J.-P.; Mermel, C.H.; Bass, A.J.; Pretz, J.; Ahn, J.; Hricik, T.; et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 2014, 123, e123–e133. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A. Primary myelofibrosis: 2023 update on diagnosis, risk-stratification, and management. Am. J. Hematol. 2023, 98, 801–821. [Google Scholar] [CrossRef] [PubMed]
- Harrison, C.N.; Vannucchi, A.M.; Kiladjian, J.-J.; Al-Ali, H.K.; Gisslinger, H.; Knoops, L.; Cervantes, F.; Jones, M.M.; Sun, K.; McQuitty, M.; et al. Long-term findings from COMFORT-II, a phase 3 study of ruxolitinib vs best available therapy for myelofibrosis. Leukemia 2016, 30, 1701–1707. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, D.; Mascarenhas, J. Next Generation Therapeutics for the Treatment of Myelofibrosis. Cells 2021, 10, 1034. [Google Scholar] [CrossRef] [PubMed]
- Wernig, G.; Kharas, M.G.; Okabe, R.; Moore, S.A.; Leeman, D.S.; Cullen, D.E.; Gozo, M.; McDowell, E.P.; Levine, R.L.; Doukas, J.; et al. Efficacy of TG101348, a Selective JAK2 Inhibitor, in Treatment of a Murine Model of JAK2V617F-Induced Polycythemia Vera. Cancer Cell 2008, 13, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Villeval, J.L.; Cohen-Solal, K.; Tulliez, M.; Giraudier, S.; Guichard, J.; Burstein, S.A.; Cramer, E.M.; Vainchenker, W.; Wendling, F. High thrombopoietin production by hematopoietic cells induces a fatal myeloproliferative syndrome in mice. Blood 1997, 90, 4369–4383. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.K.; Mullally, A.; Dugourd, A.; Peisker, F.; Hoogenboezem, R.; Van Strien, P.M.; Bindels, E.M.; Heckl, D.; Büsche, G.; Fleck, D.; et al. Gli1 + Mesenchymal Stromal Cells Are a Key Driver of Bone Marrow Fibrosis and an Important Cellular Therapeutic Target. Cell Stem Cell 2017, 20, 785–800.e8. [Google Scholar] [CrossRef] [PubMed]
- Leimkühler, N.B.; Gleitz, H.F.; Ronghui, L.; Snoeren, I.A.; Fuchs, S.N.; Nagai, J.S.; Banjanin, B.; Lam, K.H.; Vogl, T.; Kuppe, C.; et al. Heterogeneous bone-marrow stromal progenitors drive myelofibrosis via a druggable alarmin axis. Cell Stem Cell 2021, 28, 637–652.e8. [Google Scholar] [CrossRef] [PubMed]
- Harman, J.L.; Sayers, J.; Chapman, C.; Pellet-Many, C. Emerging Roles for Neuropilin-2 in Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 5154. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, A.M. VEGF/Neuropilin Signaling in Cancer Stem Cells. Int. J. Mol. Sci. 2019, 20, 490. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Goel, H.L.; Burkart, C.; Burman, L.; Chong, Y.E.; Barber, A.G.; Geng, Y.; Zhai, L.; Wang, M.; Kumar, A.; et al. Inhibition of VEGF binding to neuropilin-2 enhances chemosensitivity and inhibits metastasis in triple-negative breast cancer. Sci. Transl. Med. 2023, 15, eadf1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wisniewski, C.A.; Xiong, C.; Chhoy, P.; Goel, H.L.; Kumar, A.; Zhu, L.J.; Li, R.; St Louis, P.A.; Ferreira, L.M.; et al. Therapeutic blocking of VEGF binding to neuropilin-2 diminishes PD-L1 expression to activate antitumor immunity in prostate cancer. Sci. Transl. Med. 2023, 15, eade5855. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Polavaram, N.S.; Islam, R.; Bhattacharya, S.; Bodas, S.; Mayr, T.; Roy, S.; Albala, S.A.Y.; Toma, M.I.; Darehshouri, A.; et al. Neuropilin-2 regulates androgen-receptor transcriptional activity in advanced prostate cancer. Oncogene 2022, 41, 3747–3760. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Mishra, J.; Bodas, S.; Bhattacharya, S.; Batra, S.K.; Dutta, S.; Datta, K. Role of Neuropilin-2-mediated signaling axis in cancer progression and therapy resistance. Cancer Metastasis Rev. 2022, 41, 771–787. [Google Scholar] [CrossRef] [PubMed]
- Bühring, H.; Treml, S.; Cerabona, F.; de Zwart, P.; Kanz, L.; Sobiesiak, M. Phenotypic Characterization of Distinct Human Bone Marrow-Derived MSC Subsets. Ann. New York Acad. Sci. 2009, 1176, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Ilas, D.C.; Baboolal, T.G.; Churchman, S.M.; Jones, W.G.; Giannoudis, P.V.; Bühring, H.-J.; McGonagle, D.; Jones, E. The osteogenic commitment of CD271+CD56+ bone marrow stromal cells (BMSCs) in osteoarthritic femoral head bone. Sci. Rep. 2020, 10, 11145. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.-F.; Lian, J.-J.; Yang, H.-J.; Wang, L.; Yu, H.-H.; Bi, J.-J.; Gao, Y.-X.; Chen, S.-J.; Wang, M.; Feng, Z.-W. Neural cell adhesion molecule regulates chondrocyte hypertrophy in chondrogenic differentiation and experimental osteoarthritis. Stem Cells Transl. Med. 2020, 9, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Hwang, P.W.; Horton, J.A. Variable osteogenic performance of MC3T3-E1 subclones impacts their utility as models of osteoblast biology. Sci. Rep. 2019, 9, 8299. [Google Scholar] [CrossRef] [PubMed]
- Walz, A.; Rodriguez, I.; Mombaerts, P. Aberrant Sensory Innervation of the Olfactory Bulb in Neuropilin-2 Mutant Mice. J. Neurosci. 2002, 22, 4025–4035. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Stein, H.; Thiele, J. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; ISBN 9789283244943. [Google Scholar]
- Koerber, R.; Schneider, R.K.; Pritchard, J.E.; Teichmann, L.L.; Schumacher, U.; Brossart, P.; Gütgemann, I. Nestin expression in osteocytes following myeloablation and during bone marrow metastasis. Br. J. Haematol. 2023, 200, 643–651. [Google Scholar] [CrossRef]
- Förster, S.; Chong, Y.E.; Siefker, D.; Becker, Y.; Bao, R.; Escobedo, E.; Qing, Y.; Rauch, K.; Burman, L.; Burkart, C.; et al. Development and Characterization of a Novel Neuropilin-2 Antibody for Immunohistochemical Staining of Cancer and Sarcoidosis Tissue Samples. Monoclon. Antibodies Immunodiagn. Immunother. 2023, 42, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Haddouti, E.-M.; Randau, T.M.; Hilgers, C.; Masson, W.; Pflugmacher, R.; Burger, C.; Gravius, S.; Schildberg, F.A. Vertebral Bone Marrow-Derived Mesenchymal Stromal Cells from Osteoporotic and Healthy Patients Possess Similar Differentiation Properties In Vitro. Int. J. Mol. Sci. 2020, 21, 8309. [Google Scholar] [CrossRef] [PubMed]
- Tolkach, Y.; Kremer, A.; Lotz, G.; Schmid, M.; Mayr, T.; Förster, S.; Garbe, S.; Hosni, S.; Cronauer, M.V.; Kocsmár, I.; et al. Androgen Receptor Splice Variants Contribute to the Upregulation of DNA Repair in Prostate Cancer. Cancers 2022, 14, 4441. [Google Scholar] [CrossRef] [PubMed]
- Baccin, C.; Al-Sabah, J.; Velten, L.; Helbling, P.M.; Grünschläger, F.; Hernández-Malmierca, P.; Nombela-Arrieta, C.; Steinmetz, L.M.; Trumpp, A.; Haas, S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020, 22, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Bräunig, S.; Dhapolar, P.; Karlsson, G.; Lang, S.; Scheding, S. Identification of phenotypically, functionally, and anatomically distinct stromal niche populations in human bone marrow based on single-cell RNA sequencing. eLife 2023, 12, e81656. [Google Scholar] [CrossRef] [PubMed]
- Wolf, F.A.; Angerer, P.; Theis, F.J. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. [Google Scholar] [CrossRef] [PubMed]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef] [PubMed]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegrant Graphics for Data Analysis, 2nd ed.; Springer: Cham, Switzerland, 2016; ISBN 978-3-319-24275-0. [Google Scholar]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Chen, S.; Nurmi, H.; Leppanen, V.-M.; Jeltsch, M.; Scadden, D.; Silberstein, L.; Mikkola, H.; Alitalo, K. VEGF-C protects the integrity of the bone marrow perivascular niche in mice. Blood 2020, 136, 1871–1883. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, L.; Kriebitzsch, C.; Beullens, I.; Tan, B.K.; Carmeliet, G.; Verstuyf, A. Nrp2 deficiency leads to trabecular bone loss and is accompanied by enhanced osteoclast and reduced osteoblast numbers. Bone 2013, 55, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Verlinden, L.; Doms, S.; Janssens, I.; Meyer, M.B.; Pike, J.W.; Carmeliet, G.; Verstuyf, A. Neuropilin 2 in osteoblasts regulates trabecular bone mass in male mice. Front. Endocrinol. 2023, 14, 1223021. [Google Scholar] [CrossRef]
- Dhupar, R.; Powers, A.A.; Eisenberg, S.H.; Gemmill, R.M.; Bardawil, C.E.; Udoh, H.M.; Cubitt, A.; Nangle, L.A.; Soloff, A.C. Orchestrating Resilience: How Neuropilin-2 and Macrophages Contribute to Cardiothoracic Disease. J. Clin. Med. 2024, 13, 1446. [Google Scholar] [CrossRef] [PubMed]
- Chitteti, B.R.; Cheng, Y.-H.; Poteat, B.; Rodriguez-Rodriguez, S.; Goebel, W.S.; Carlesso, N.; Kacena, M.A.; Srour, E.F. Impact of interactions of cellular components of the bone marrow microenvironment on hematopoietic stem and progenitor cell function. Blood 2010, 115, 3239–3248. [Google Scholar] [CrossRef] [PubMed]
- Arranz, L.; Sánchez-Aguilera, A.; Martin-Peérez, D.; Isern, J.; Langa, X.; Tzankov, A.; Lundberg, P.; Muntión, S.; Tzeng, Y.-S.; Lai, D.-M.; et al. Neuropathy of haematopoietic stem cell niche is essential for myeloproliferative neoplasms. Nature 2014, 512, 78–81. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, A.; Lorusso, P.M.; Messersmith, W.A.; Papadopoulos, K.P.; Gore, L.; Beeram, M.; Ramakrishnan, V.; Kim, A.H.; Beyer, J.C.; Mason Shih, L.; et al. A Phase Ib study evaluating MNRP1685A, a fully human anti-NRP1 monoclonal antibody, in combination with bevacizumab and paclitaxel in patients with advanced solid tumors. Cancer Chemother. Pharmacol. 2014, 73, 951–960. [Google Scholar] [CrossRef]
- Culver, D.A.; Aryal, S.; Barney, J.; Hsia, C.C.; James, W.E.; Maier, L.A.; Marts, L.T.; Obi, O.N.; Sporn, P.H.; Sweiss, N.J.; et al. Efzofitimod for the Treatment of Pulmonary Sarcoidosis. Chest 2023, 163, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Martinaud, C.; Desterke, C.; Konopacki, J.; Pieri, L.; Torossian, F.; Golub, R.; Schmutz, S.; Anginot, A.; Guerton, B.; Rochet, N.; et al. Osteogenic Potential of Mesenchymal Stromal Cells Contributes to Primary Myelofibrosis. Cancer Res 2015, 75, 4753–4765. [Google Scholar] [CrossRef] [PubMed]
- El Agha, E.; Kramann, R.; Schneider, R.K.; Li, X.; Seeger, W.; Humphreys, B.D.; Bellusci, S. Mesenchymal Stem Cells in Fibrotic Disease. Cell Stem Cell 2017, 21, 166–177. [Google Scholar] [CrossRef] [PubMed]
- Grandclement, C.; Pallandre, J.R.; Valmary Degano, S.V.; Viel, E.; Bouard, A.; Balland, J.; Rémy-Martin, J.-P.; Simon, B.; Rouleau, A.; Boireau, W.; et al. Neuropilin-2 Expression Promotes TGF-β1-Mediated Epithelial to Mesenchymal Transition in Colorectal Cancer Cells. PLoS ONE 2011, 6, e20444. [Google Scholar] [CrossRef] [PubMed]
- Prud’homme, G.J.; Glinka, Y. Neuropilins are multifunctional coreceptors involved in tumor initiation, growth, metastasis and immunity. Oncotarget 2012, 3, 921–939. [Google Scholar] [CrossRef] [PubMed]
- Polavaram, N.S.; Dutta, S.; Islam, R.; Bag, A.K.; Roy, S.; Poitz, D.; Karnes, J.; Hofbauer, L.C.; Kohli, M.; Costello, B.A.; et al. Tumor- and osteoclast-derived NRP2 in prostate cancer bone metastases. Bone Res. 2021, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.-F.; Feng, X.; Gao, Y.-X.; Jian, S.-Q.; Liu, S.-R.; Wang, M.; Xie, Y.-F.; Wang, L.; Feng, Z.-W.; Yang, H.-J. Neural Cell Adhesion Molecule Regulates Osteoblastic Differentiation Through Wnt/β-Catenin and PI3K-Akt Signaling Pathways in MC3T3-E1 Cells. Front. Endocrinol. 2021, 12, 657953. [Google Scholar] [CrossRef]
- Shi, Y.; Xia, Y.-Y.; Wang, L.; Liu, R.; Khoo, K.-S.; Feng, Z.-W. Neural cell adhesion molecule modulates mesenchymal stromal cell migration via activation of MAPK/ERK signaling. Exp. Cell Res. 2012, 318, 2257–2267. [Google Scholar] [CrossRef] [PubMed]
- Hecht, J.; Seitz, V.; Urban, M.; Wagner, F.; Robinson, P.; Stiege, A.; Dieterich, C.; Kornak, U.; Wilkening, U.; Brieske, N.; et al. Detection of novel skeletogenesis target genes by comprehensive analysis of a Runx2(-/-) mouse model. Gene Expr. Patterns 2007, 7, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, J.; Migliaccio, A.R.; Kosiorek, H.; Bhave, R.; Palmer, J.; Kuykendall, A.; Mesa, R.; Rampal, R.K.; Gerds, A.T.; Yacoub, A.; et al. A Phase Ib Trial of AVID200, a TGFβ 1/3 Trap, in Patients with Myelofibrosis. Clin. Cancer Res. 2023, 29, 3622–3632. [Google Scholar] [CrossRef] [PubMed]
- Leimkühler, N.B.; Costa, I.G.; Schneider, R.K. From cell to cell: Identification of actionable targets in bone marrow fibrosis using single-cell technologies. Exp. Hematol. 2021, 104, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-Y.; Chung, C.-L.; Hu, T.-H.; Chen, J.-J.; Liu, P.-F.; Chen, C.-L. Recent progress in TGF-β inhibitors for cancer therapy. Biomed. Pharmacother. 2021, 134, 111046. [Google Scholar] [CrossRef] [PubMed]
- Baughman, R.P.; Niranjan, V.; Walker, G.; Burkart, C.; Paz, S.; Chong, Y.; Siefker, D.; Sun, E.; Nangle, L.; Forster, S.; et al. Efzofitimod: A novel anti-inflammatory agent for sarcoidosis. Sarcoidosis Vasc. Diffuse Lung Dis. 2023, 40, e2023011. [Google Scholar] [CrossRef] [PubMed]
- Schepers, K.; Pietras, E.M.; Reynaud, D.; Flach, J.; Binnewies, M.; Garg, T.; Wagers, A.J.; Hsiao, E.C.; Passegué, E. Myeloproliferative Neoplasia Remodels the Endosteal Bone Marrow Niche into a Self-Reinforcing Leukemic Niche. Cell Stem Cell 2013, 13, 285–299. [Google Scholar] [CrossRef] [PubMed]
- Marchand, T.; Pinho, S. Leukemic Stem Cells: From Leukemic Niche Biology to Treatment Opportunities. Front. Immunol. 2021, 12, 775128. [Google Scholar] [CrossRef] [PubMed]
- Mosialou, I.; Galan-Diez, M.; Vandenberg, A.; Ali, A.M.; Raza, A.; Kousteni, S. Pharmacological Targeting of Osteoblast-Induced MDS and AML. Blood 2018, 132, 5235. [Google Scholar] [CrossRef]







| Diagnosis | PMF | ET | PV | MDS | MPN/MDS/ CMML | AML | Control | Total |
|---|---|---|---|---|---|---|---|---|
| Number of Cases. N (%) | 25 (20) | 12 (10) | 17 (14) | 16 (13) | 16 (13) | 13 (11) | 23 (19) | 122 (100) |
| Median Age (range) | 63.6 (21–85) | 64.3 (31–81) | 57.8 (41–79) | 75.4 (52–90) | 66.8 (42–84) | 58 (36–73) | 58 (31–79) | 63.4 |
| Gender | ||||||||
| Male | 16 | 7 | 8 | 12 | 14 | 8 | 12 | 77 |
| Female | 9 | 5 | 9 | 4 | 2 | 5 | 11 | 46 |
| MF grade (%) | ||||||||
| 0 | 1 (2) | 10 (20) | 5 (10) | 4 (8) | 4 (8) | 3 (6) | 23 (46) | 50 (100) |
| 1 | 5 (18) | 2 (7) | 5 (18) | 4 (14) | 6 (21) | 6 (21) | 0 (0) | 28 (100) |
| 2 | 11 (35) | 0 (0) | 6 (19) | 7 (23) | 3 (10) | 4 (13) | 0 (0) | 31 (100) |
| 3 | 8 (62) | 0 (0) | 1 (8) | 1 (8) | 3 (23) | 0 (0) | 0 (0) | 13 (100) |
| Mutation | ||||||||
| JAK2V617F | 13 | 6 | 12 | 0 | 1 | 0 | 0 | 32 |
| CALR | 5 | 1 | 0 | 1 | 2 | 0 | 0 | 9 |
| NPM1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 4 |
| MPL | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 4 |
| SRSF2 | 3 | 0 | 0 | 0 | 1 | 1 | 0 | 5 |
| Mean haemoglobin [g/dL] (range) | 9.9 (4.2–15.5) | 14.1 (9.1–16.4) | 14.2 (8.4–19.8) | 9.4 (6.3–13.9) | 9.2 (7–11.8) | 10.2 (6.7–13.4) | 11 (8.5–15.4) | 11.1 (4.2–19.8) |
| Mean platelet [103/µL] (range) | 294.5 (8–993) | 787.2 (23–1143) | 380.6 (31–969) | 83.8 (16–205) | 222.1 (10–1462) | 59.6 (13–130) | 252.1 (82–618) | 297.1 (8–1462) |
| Mean WBC [103/µL] (range) | 13.8 (3.8–36.21) | 12.1 (2.09–14.42) | 18.5 (1.57–32.49) | 2.8 (0.3–6.83) | 15.5 (1.78–56.04) | 26.4 (1.2–25.62) | 8 (5.17–11.53) | 13.9 (1.2–56.04) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vosbeck, K.; Förster, S.; Mayr, T.; Sahu, A.; Haddouti, E.-M.; Al-Adilee, O.; Körber, R.-M.; Bisht, S.; Muders, M.H.; Nesic, S.; et al. Neuropilin2 in Mesenchymal Stromal Cells as a Potential Novel Therapeutic Target in Myelofibrosis. Cancers 2024, 16, 1924. https://doi.org/10.3390/cancers16101924
Vosbeck K, Förster S, Mayr T, Sahu A, Haddouti E-M, Al-Adilee O, Körber R-M, Bisht S, Muders MH, Nesic S, et al. Neuropilin2 in Mesenchymal Stromal Cells as a Potential Novel Therapeutic Target in Myelofibrosis. Cancers. 2024; 16(10):1924. https://doi.org/10.3390/cancers16101924
Chicago/Turabian StyleVosbeck, Karla, Sarah Förster, Thomas Mayr, Anshupa Sahu, El-Mustapha Haddouti, Osamah Al-Adilee, Ruth-Miriam Körber, Savita Bisht, Michael H. Muders, Svetozar Nesic, and et al. 2024. "Neuropilin2 in Mesenchymal Stromal Cells as a Potential Novel Therapeutic Target in Myelofibrosis" Cancers 16, no. 10: 1924. https://doi.org/10.3390/cancers16101924
APA StyleVosbeck, K., Förster, S., Mayr, T., Sahu, A., Haddouti, E.-M., Al-Adilee, O., Körber, R.-M., Bisht, S., Muders, M. H., Nesic, S., Buness, A., Kristiansen, G., Schildberg, F. A., & Gütgemann, I. (2024). Neuropilin2 in Mesenchymal Stromal Cells as a Potential Novel Therapeutic Target in Myelofibrosis. Cancers, 16(10), 1924. https://doi.org/10.3390/cancers16101924







