Metabolic Biomarkers Affecting Cell Proliferation and Prognosis in Polycythemia Vera
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Inclusion and Sample Collection
2.2. Prognostic Risk Stratification
2.3. Detection Method of JAK2 Mutations
2.4. Materials and Reagents
2.5. Experimental Instruments
2.6. Serum Sample Preparation
2.7. LC-MS Analysis
2.8. Statistical Analysis
3. Results
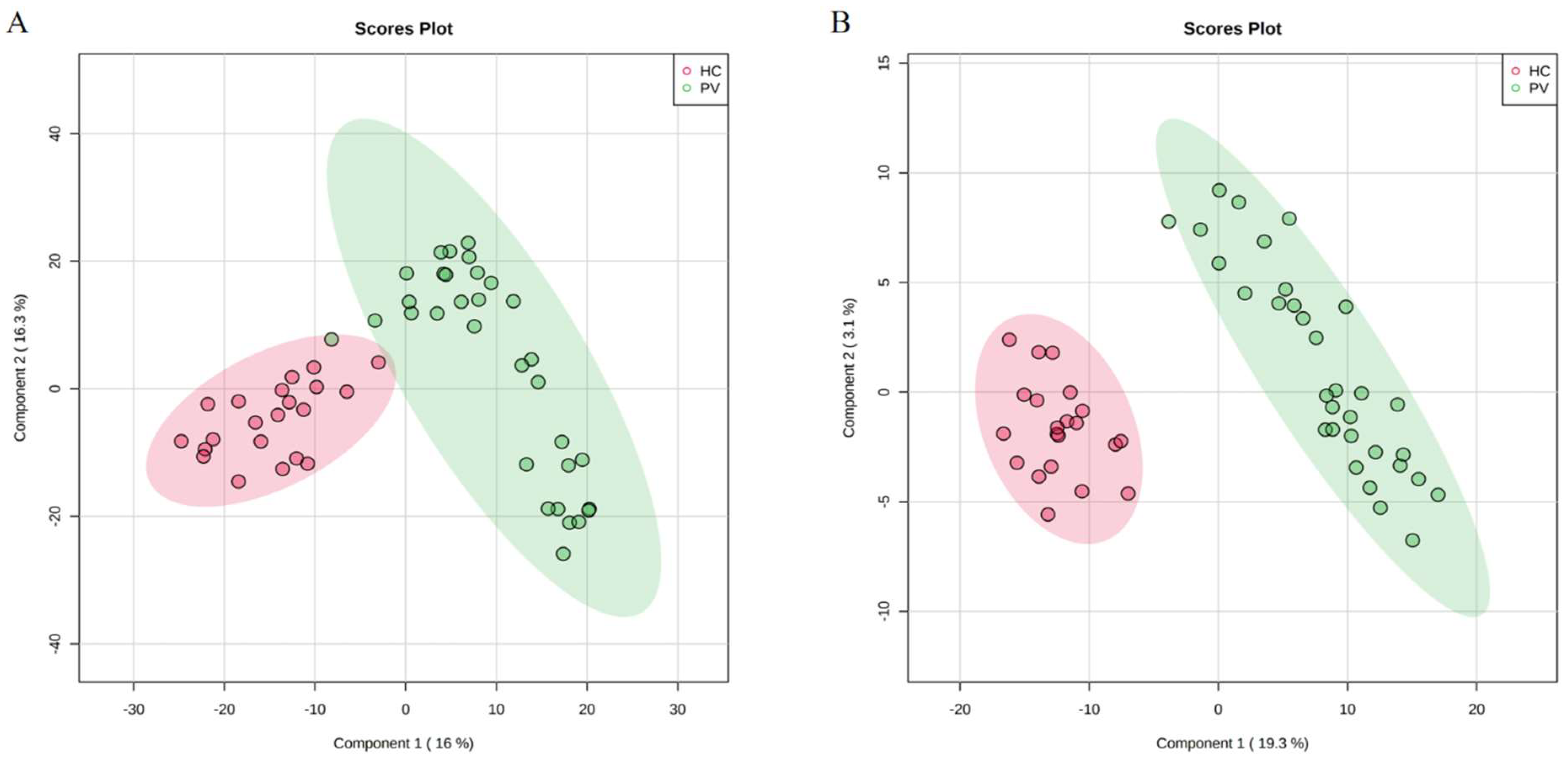
3.1. Analysis of the Metabolic Profiles of PV Patients and Healthy Controls
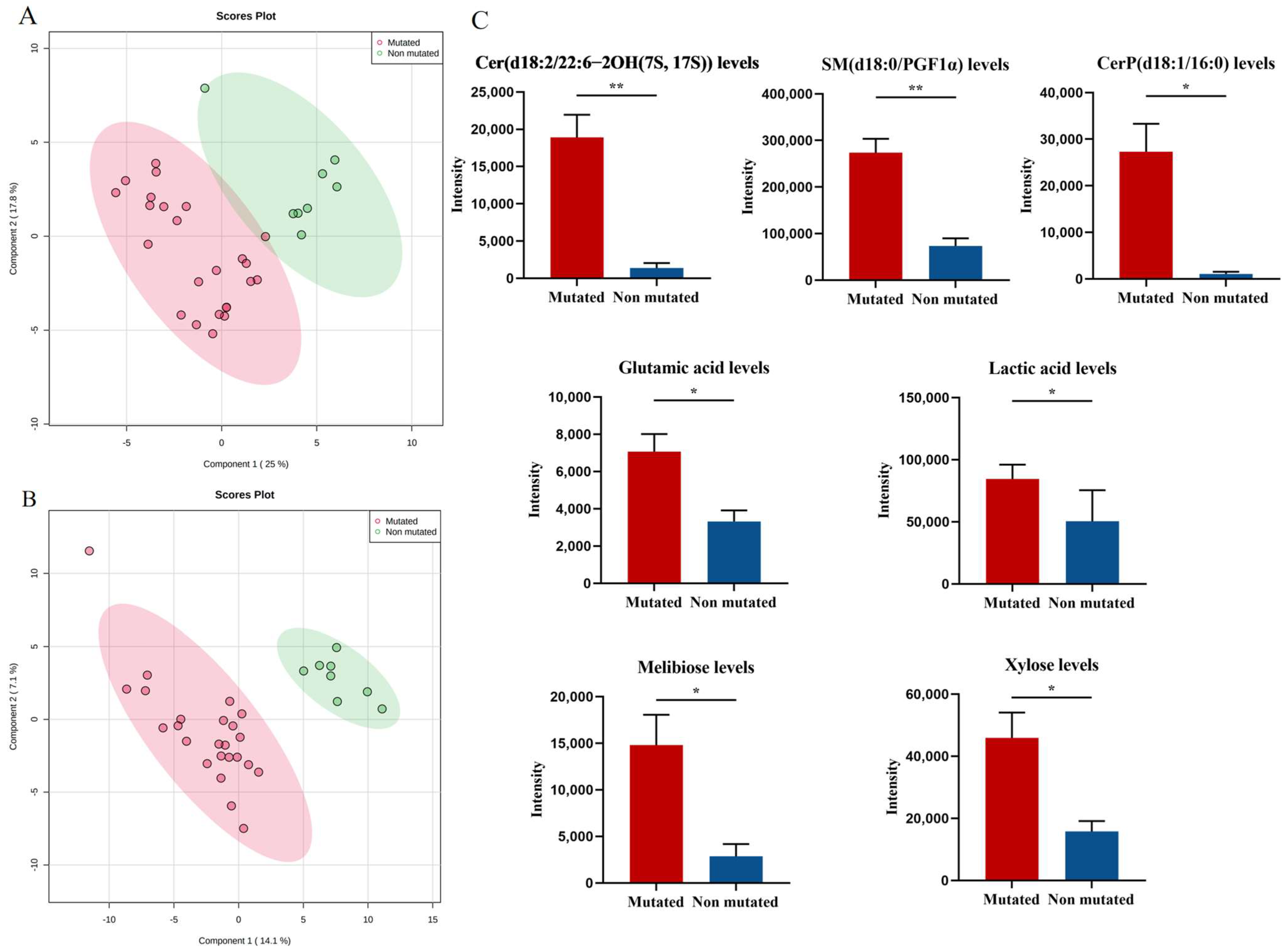
3.2. Effect of JAK2-Associated Metabolic Abnormalities on Cell Proliferation in PV Patients
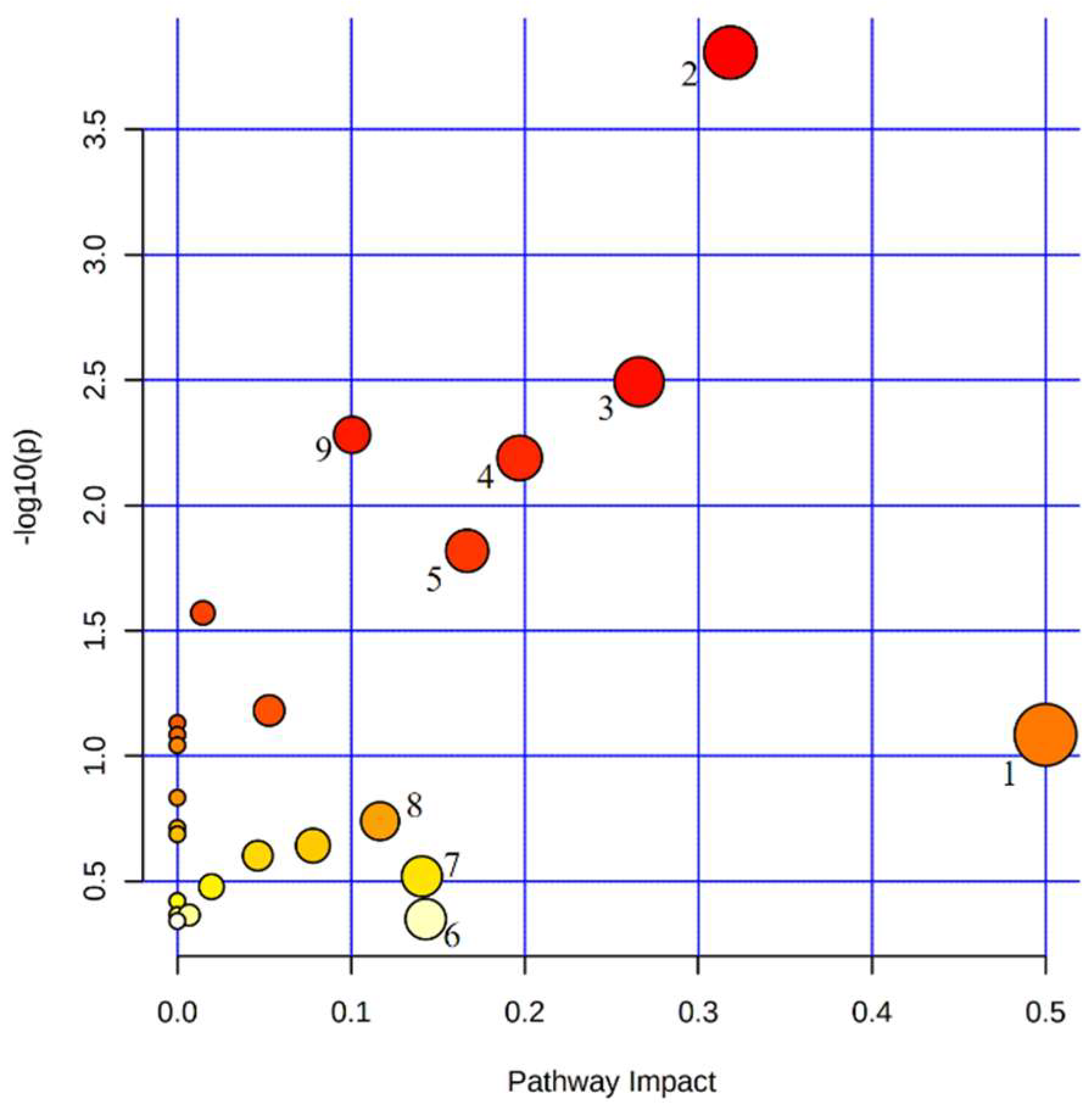
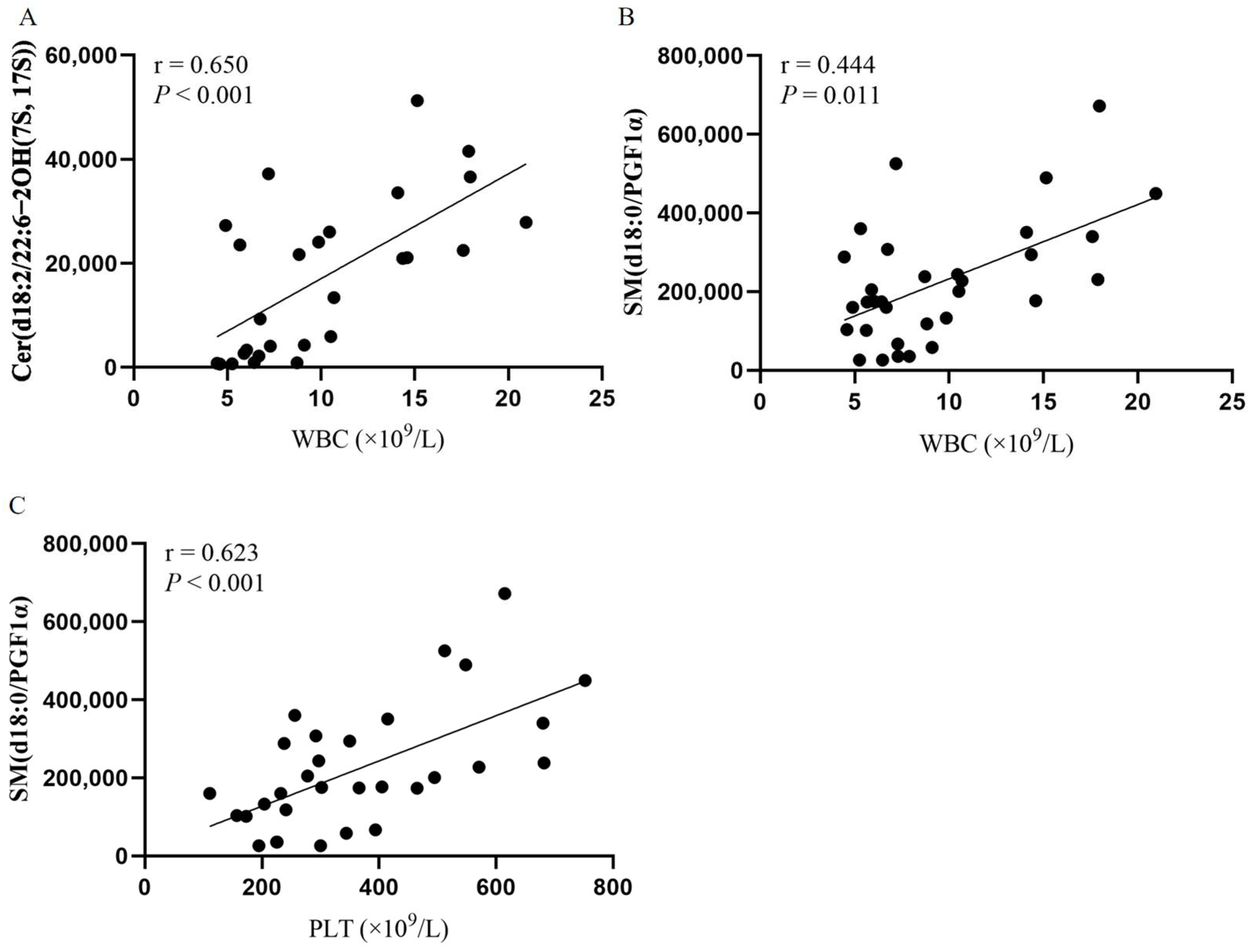
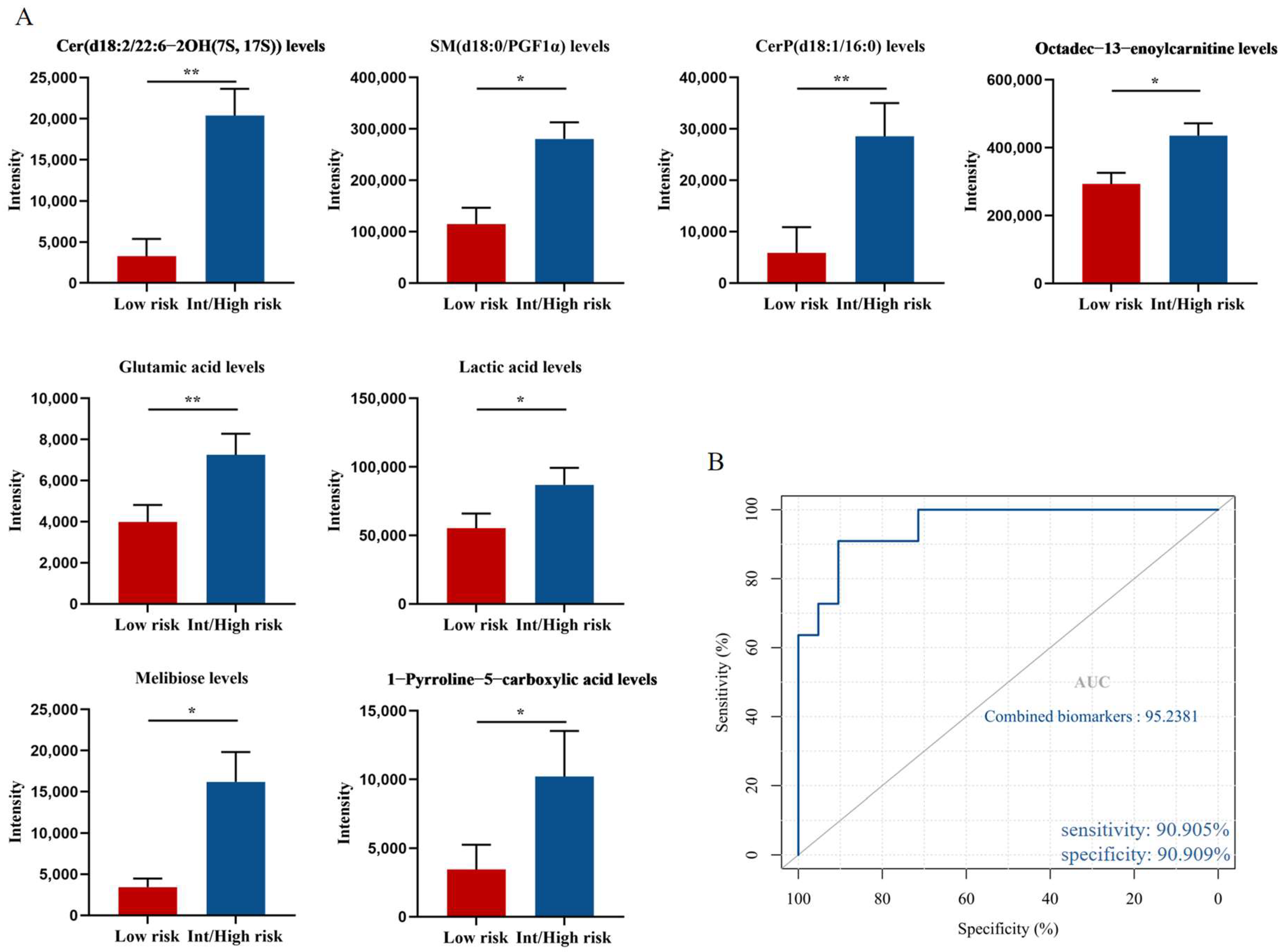
3.3. Potential Metabolic Biomarkers Relevant to the Prognosis of PV Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tefferi, A.; Barbui, T. Polycythemia vera and essential thrombocythemia: 2021 update on diagnosis, risk-stratification and management. Am. J. Hematol. 2020, 95, 1599–1613. [Google Scholar] [CrossRef] [PubMed]
- Srour, S.A.; Devesa, S.S.; Morton, L.M.; Check, D.P.; Curtis, R.E.; Linet, M.S.; Dores, G. Incidence and patient survival of myeloproliferative neoplasms and myelodysplastic/myeloproliferative neoplasms in the United States, 2001–2012. Br. J. Haematol. 2016, 174, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Baxter, E.J.; Scott, L.M.; Campbell, P.J.; East, C.; Fourouclas, N.; Swanton, S.; Vassiliou, G.S.; Bench, A.J.; Boyd, E.M.; Curtin, N.; et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005, 365, 1054–1061. [Google Scholar] [CrossRef]
- Vainchenker, W.; Kralovics, R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood 2017, 129, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- El Hassouni, B.; Granchi, C.; Vallés-Martí, A.; Supadmanaba, I.G.P.; Bononi, G.; Tuccinardi, T.; Funel, N.; Jimenez, C.R.; Peters, G.J.; Giovannetti, E.; et al. The dichotomous role of the glycolytic metabolism pathway in cancer metastasis: Interplay with the complex tumor microenvironment and novel therapeutic strategies. Semin. Cancer Biol. 2019, 60, 238–248. [Google Scholar] [CrossRef]
- Hoy, A.J.; Nagarajan, S.R.; Butler, L.M. Tumour fatty acid metabolism in the context of therapy resistance and obesity. Nat. Cancer 2021, 21, 753–766. [Google Scholar] [CrossRef]
- Stepka, P.; Vsiansky, V.; Raudenska, M.; Gumulec, J.; Adam, V.; Masarik, M. Metabolic and Amino Acid Alterations of the Tumor Microenvironment. Curr. Med. Chem. 2021, 28, 1270–1289. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef]
- Ma, Y.; Temkin, S.M.; Hawkridge, A.M.; Guo, C.; Wang, W.; Wang, X.-Y.; Fang, X. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018, 435, 92–100. [Google Scholar] [CrossRef]
- Rao, T.N.; Hansen, N.; Hilfiker, J.; Rai, S.; Majewska, J.-M.; Leković, D.; Gezer, D.; Andina, N.; Galli, S.; Cassel, T.; et al. JAK2-mutant hematopoietic cells display metabolic alterations that can be targeted to treat myeloproliferative neoplasms. Blood 2019, 134, 1832–1846. [Google Scholar] [CrossRef]
- Zhan, H.; Ciano, K.; Dong, K.; Zucker, S. Targeting glutamine metabolism in myeloproliferative neoplasms. Blood Cells Mol. Dis. 2015, 55, 241–247. [Google Scholar] [CrossRef]
- Forte, D.; Fanelli, F.; Mezzullo, M.; Barone, M.; Corradi, G.; Auteri, G.; Bartoletti, D.; Martello, M.; Ottaviani, E.; Terragna, C.; et al. Disease-Specific Derangement of Circulating Endocannabinoids and N-Acylethanolamines in Myeloproliferative Neoplasms. Int. J. Mol. Sci. 2020, 21, 3399. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Liu, J.; Wang, H.; Lu, H. Metabolomics identified new biomarkers for the precise diagnosis of pancreatic cancer and associated tissue metastasis. Pharmacol. Res. 2020, 156, 104805. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Xu, T.; Huang, J.; Zhang, L.; Xu, S.; Xiong, B.; Wang, Y.; Tang, H. Tissue Metabonomic Phenotyping for Diagnosis and Prognosis of Human Colorectal Cancer. Sci. Rep. 2016, 6, 20790. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef]
- Tefferi, A.; Rumi, E.; Finazzi, G.; Gisslinger, H.; Vannucchi, A.M.; Rodeghiero, F.; Randi, M.L.; Vaidya, R.; Cazzola, M.; Rambaldi, A.; et al. Survival and prognosis among 1545 patients with contemporary polycythemia vera: An international study. Leukemia 2013, 27, 1874–1881. [Google Scholar] [CrossRef] [PubMed]
- Tarasenko, T.N.; Cusmano-Ozog, K.; McGuire, P.J. Tissue acylcarnitine status in a mouse model of mitochondrial β-oxidation deficiency during metabolic decompensation due to influenza virus infection. Mol. Genet. Metab. 2018, 125, 144–152. [Google Scholar] [CrossRef]
- Samudio, I.; Harmancey, R.; Fiegl, M.; Kantarjian, H.; Konopleva, M.; Korchin, B.; Kaluarachchi, K.; Bornmann, W.; Duvvuri, S.; Taegtmeyer, H.; et al. Pharmacologic inhibition of fatty acid oxidation sensitizes human leukemia cells to apoptosis induction. J. Clin. Investig. 2010, 120, 142–156. [Google Scholar] [CrossRef]
- Batchuluun, B.; Al Rijjal, D.; Prentice, K.J.; Eversley, J.A.; Burdett, E.; Mohan, H.; Bhattacharjee, A.; Gunderson, E.P.; Liu, Y.; Wheeler, M.B. Elevated Medium-Chain Acylcarnitines Are Associated With Gestational Diabetes Mellitus and Early Progression to Type 2 Diabetes and Induce Pancreatic β-Cell Dysfunction. Diabetes 2018, 67, 885–897. [Google Scholar] [CrossRef]
- Abbaszadeh, Z.; Çeşmeli, S.; Avcı, .B. Crucial players in glycolysis: Cancer progress. Gene 2019, 726, 144158. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Jiang, Y.; Meisenhelder, J.; Yang, W.; Hawke, D.H.; Zheng, Y.; Xia, Y.; Aldape, K.; He, J.; Hunter, T.; et al. Mitochondria-Translocated PGK1 Functions as a Protein Kinase to Coordinate Glycolysis and the TCA Cycle in Tumorigenesis. Mol. Cell 2016, 61, 705–719. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Sun, N.; Chen, X.; Li, X.; Zhao, J.; Cheng, W.; Hua, H.; Fukatsu, M.; Mori, H.; Takahashi, H.; et al. JAK2V617F Mutation Promoted IL-6 Production and Glycolysis via Mediating PKM1 Stabilization in Macrophages. Front. Immunol. 2021, 11, 3853. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Wright, K.L.; Epling-Burnette, P.K.; Reuther, G.W. Metabolic Vulnerabilities and Epigenetic Dysregulation in Myeloproliferative Neoplasms. Front. Immunol. 2020, 11, 604142. [Google Scholar] [CrossRef] [PubMed]
- Hannun, Y.A.; Obeid, L.M. Sphingolipids and their metabolism in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017, 19, 175–191. [Google Scholar] [CrossRef]
- Pherez-Farah, A.; López-Sánchez, R.D.C.; Villela-Martínez, L.M.; Ortiz-López, R.; Beltrán, B.E.; Hernández-Hernández, J.A. Sphingolipids and Lymphomas: A Double-Edged Sword. Cancers 2022, 14, 2051. [Google Scholar] [CrossRef]
- Kitatani, K.; Taniguchi, M.; Okazaki, A.T. Role of Sphingolipids and Metabolizing Enzymes in Hematological Malignancies. Mol. Cells 2015, 38, 482–495. [Google Scholar] [CrossRef]
- Poulaki, A.; Katsila, T.; Stergiou, I.E.; Giannouli, S.; Gόmez-Tamayo, J.C.; Piperaki, E.-T.; Kambas, K.; Dimitrakopoulou, A.; Patrinos, G.P.; Tzioufas, A.G.; et al. Bioenergetic Profiling of the Differentiating Human MDS Myeloid Lineage with Low and High Bone Marrow Blast Counts. Cancers 2020, 12, 3520. [Google Scholar] [CrossRef]
- Wątek, M.; Durnaś, B.; Wollny, T.; Pasiarski, M.; Góźdź, S.; Marzec, M.; Chabowska, A.; Wolak, P.; Żendzian-Piotrowska, M.; Bucki, R. Unexpected profile of sphingolipid contents in blood and bone marrow plasma collected from patients diagnosed with acute myeloid leukemia. Lipids Health Dis. 2017, 16, 235. [Google Scholar] [CrossRef]
- Lukey, M.J.; Katt, W.P.; Cerione, R.A. Targeting amino acid metabolism for cancer therapy. Drug Discov. Today 2017, 22, 796–804. [Google Scholar] [CrossRef]
- Wang, D.; Tan, G.; Wang, H.; Chen, P.; Hao, J.; Wang, Y. Identification of novel serum biomarker for the detection of acute myeloid leukemia based on liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2019, 166, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Geck, R.; Toker, A. Nonessential amino acid metabolism in breast cancer. Adv. Biol. Regul. 2016, 62, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhang, Y.; Shi, J.; Li, W.; Gan, B. The glutamate/cystine antiporter SLC7A11/xCT enhances cancer cell dependency on glucose by exporting glutamate. J. Biol. Chem. 2017, 292, 14240–14249. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.-H.; Zhai, X.-Y. Role of tryptophan metabolism in cancers and therapeutic implications. Biochimie 2021, 182, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Crodel, C.C.; Jentsch-Ullrich, K.; Reiser, M.; Jacobasch, L.; Sauer, A.; Tesch, H.; Ulshöfer, T.; Wunschel, R.; Palandri, F.; Heidel, F.H. Cytoreductive treatment in real life: A chart review analysis on 1440 patients with polycythemia vera. J. Cancer Res. Clin. Oncol. 2021, 148, 2693–2705. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Lasho, T.L.; Schwager, S.M.; Strand, J.S.; Elliott, M.; Mesa, R.; Li, C.-Y.; Wadleigh, M.; Lee, S.J.; Gilliland, D.G. The clinical phenotype of wild-type, heterozygous, and homozygous JAK2V617F in polycythemia vera. Cancer 2005, 106, 631–635. [Google Scholar] [CrossRef]
- Gou, P.; Zhang, W.; Giraudier, S. Insights into the Potential Mechanisms of JAK2V617F Somatic Mutation Contributing Distinct Phenotypes in Myeloproliferative Neoplasms. Int. J. Mol. Sci. 2022, 23, 1013. [Google Scholar] [CrossRef]
- Lu, S.; Natarajan, S.K.; Mott, J.L.; Kharbanda, K.K.; Harrison-Findik, D.D. Ceramide Induces Human Hepcidin Gene Transcription through JAK/STAT3 Pathway. PLoS ONE 2016, 11, e0147474. [Google Scholar] [CrossRef]
- Jung, J.-S.; Shin, K.-O.; Lee, Y.-M.; Shin, J.A.; Park, E.-M.; Jeong, J.; Kim, D.-H.; Choi, J.W.; Kim, H.-S. Anti-inflammatory mechanism of exogenous C2 ceramide in lipopolysaccharide-stimulated microglia. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2013, 1831, 1016–1026. [Google Scholar] [CrossRef]
- Mazière, C.; Conte, M.-A.; Mazière, J.-C. Activation of the JAK/STAT pathway by ceramide in cultured human fibroblasts. FEBS Lett. 2001, 507, 163–168. [Google Scholar] [CrossRef]
- Tsurumaki, H.; Katano, H.; Sato, K.; Imai, R.; Niino, S.; Hirabayashi, Y.; Ichikawa, S. WP1066, a small molecule inhibitor of the JAK/STAT3 pathway, inhibits ceramide glucosyltransferase activity. Biochem. Biophys. Res. Commun. 2017, 491, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Komuro, M.; Nagane, M.; Fukuyama, T.; Luo, X.; Hiraki, S.; Miyanabe, M.; Ishikawa, M.; Niwa, C.; Murakami, H.; Okamoto, M.; et al. Sphingomyelin maintains the cutaneous barrier via regulation of the STAT3 pathway. FASEB J. 2022, 36, e22111. [Google Scholar] [CrossRef] [PubMed]
- Presti, C.L.; Fauvelle, F.; Jacob, M.-C.; Mondet, J.; Mossuz, P. The metabolic reprogramming in acute myeloid leukemia patients depends on their genotype and is a prognostic marker. Blood Adv. 2021, 5, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Passamonti, F.; Rumi, E.; Pietra, D.; Elena, C.; Boveri, E.; Arcaini, L.; Roncoroni, E.; Astori, C.; Merli, M.; Boggi, S.; et al. A prospective study of 338 patients with polycythemia vera: The impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia 2010, 24, 1574–1579. [Google Scholar] [CrossRef]
- Alvarez-Larrán, A.; Bellosillo, B.; Pereira, A.; Kerguelen, A.; Hernández-Boluda, J.C.; Martínez-Avilés, L.; Fernández-Rodríguez, C.; Gómez, M.; Lombardía, L.; Angona, A.; et al. JAK2V617F monitoring in polycythemia vera and essential thrombocythemia: Clinical usefulness for predicting myelofibrotic transformation and thrombotic events. Am. J. Hematol. 2014, 89, 517–523. [Google Scholar] [CrossRef]
- Ronner, L.; Podoltsev, N.; Gotlib, J.; Heaney, M.L.; Kuykendall, A.T.; O’Connell, C.; Shammo, J.; Fleischman, A.G.; Scherber, R.M.; Mesa, R.; et al. Persistent leukocytosis in polycythemia vera is associated with disease evolution but not thrombosis. Blood 2020, 135, 1696–1703. [Google Scholar] [CrossRef]


| No. | Metabolite a | Mass | Retention Time/Min | Tendency b | Ion Mode | Pathway |
|---|---|---|---|---|---|---|
| 1 | Arachidic acid | 312.3028 | 0.9840 | Up | Positive | Fatty acid metabolism |
| 2 | Dodecanoic acid | 200.1776 | 1.4151 | Up | Positive | Fatty acid metabolism |
| 3 | Myristic acid | 228.2089 | 1.3200 | Up | Positive | Fatty acid metabolism |
| 4 | Palmitic acid | 256.2402 | 1.4951 | Up | Positive | Fatty acid metabolism |
| 5 | Octadec-13-enoylcarnitine | 425.3505 | 0.9802 | Up | Positive | Fatty acid oxidation |
| 6 | Dodec-9-enedioylcarnitine | 371.2308 | 1.3390 | Up | Positive | Fatty acid oxidation |
| 7 | Dodecanoylcarnitine | 344.2795 | 1.3580 | Up | Positive | Fatty acid oxidation |
| 8 | Hexacosanoyl carnitine | 539.4914 | 0.9802 | Up | Positive | Fatty acid oxidation |
| 9 | Octanoylcarnitine | 288.2175 | 1.4684 | Up | Positive | Fatty acid oxidation |
| 10 | Glucose | 180.0634 | 5.0102 | Up | Negative | Glycolysis/Gluconeogenesis |
| 11 | Lactic acid | 90.0317 | 4.2093 | Up | Negative | Glycolysis/Gluconeogenesis |
| 12 | Pyruvic acid | 88.0160 | 3.4841 | Down | Negative | Glycolysis/Gluconeogenesis |
| 13 | Melibiose | 342.1162 | 9.6866 | Up | Positive | Galactose metabolism |
| 14 | Xylose | 150.0528 | 3.0263 | Up | Negative | Pentose and glucuronate interconversions |
| 15 | Cer(d18:1/24:1) | 647.6216 | 0.8812 | Up | Positive | Sphingolipid metabolism |
| 16 | Cer(d18:2/6 keto-PGF1α) | 649.4918 | 0.8051 | Up | Positive | Sphingolipid metabolism |
| 17 | CerP(d18:1/16:0) | 617.4784 | 4.1358 | Up | Positive | Sphingolipid metabolism |
| 18 | Phytosphingosine | 317.2930 | 0.9840 | Up | Positive | Sphingolipid metabolism |
| 19 | SM(d18:0/12:0) | 650.5363 | 4.7082 | Up | Positive | Sphingolipid metabolism |
| 20 | SM(d18:0/22:0) | 788.6771 | 4.4175 | Down | Positive | Sphingolipid metabolism |
| 21 | SM(d18:0/PGF1α) | 804.5993 | 0.9878 | Up | Positive | Sphingolipid metabolism |
| 22 | Sphingosine | 299.2824 | 1.3542 | Up | Positive | Sphingolipid metabolism |
| 23 | Cer(d18:2/22:6-2OH(7S, 17S)) | 639.4863 | 4.1180 | Up | Negative | Sphingolipid metabolism |
| 24 | SM(d20:1/PGE2) | 826.5836 | 2.8587 | Up | Negative | Sphingolipid metabolism |
| 25 | SM(d20:1/PGF2α) | 828.5993 | 2.6873 | Up | Negative | Sphingolipid metabolism |
| 26 | Glutamic acid | 147.0532 | 2.5652 | Up | Negative | Glutamate metabolism |
| 27 | 1-Pyrroline-5-carboxylic acid | 113.0477 | 3.2366 | Up | Negative | Glutamate Metabolism |
| 28 | Lactoylglutathione | 379.1049 | 1.1106 | Down | Positive | Pyruvate metabolism |
| 29 | Aminoadipic acid | 161.0688 | 0.8012 | Up | Positive | Lysine biosynthesis |
| 30 | Homo-L-arginine | 188.1273 | 10.8460 | Down | Positive | Arginine metabolism |
| 31 | Tryptophan | 204.0899 | 1.6445 | Up | Positive | Tryptophan metabolism |
| 32 | Cytidine | 243.0855 | 3.2290 | Up | Negative | Pyrimidine metabolism |
| 33 | Bilirubin | 584.2635 | 0.8703 | Up | Negative | Porphyrin metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Lv, Y.; Yang, E.; Li, Y.; Wang, D.; Hu, G.; Li, Y.; Wang, M.; Liu, W.; Sun, M.; et al. Metabolic Biomarkers Affecting Cell Proliferation and Prognosis in Polycythemia Vera. Cancers 2022, 14, 4913. https://doi.org/10.3390/cancers14194913
Wang Z, Lv Y, Yang E, Li Y, Wang D, Hu G, Li Y, Wang M, Liu W, Sun M, et al. Metabolic Biomarkers Affecting Cell Proliferation and Prognosis in Polycythemia Vera. Cancers. 2022; 14(19):4913. https://doi.org/10.3390/cancers14194913
Chicago/Turabian StyleWang, Ziqing, Yan Lv, Erpeng Yang, Yujin Li, Dehao Wang, Guang Hu, Yumeng Li, Mingjing Wang, Weiyi Liu, Mingqian Sun, and et al. 2022. "Metabolic Biomarkers Affecting Cell Proliferation and Prognosis in Polycythemia Vera" Cancers 14, no. 19: 4913. https://doi.org/10.3390/cancers14194913
APA StyleWang, Z., Lv, Y., Yang, E., Li, Y., Wang, D., Hu, G., Li, Y., Wang, M., Liu, W., Sun, M., & Hu, X. (2022). Metabolic Biomarkers Affecting Cell Proliferation and Prognosis in Polycythemia Vera. Cancers, 14(19), 4913. https://doi.org/10.3390/cancers14194913





