Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Cell Lines
2.3. CRISPR Knock-Out
2.4. qPCR
2.5. Lentivirus Production and Cell Infection
2.6. Western Blot Assays
2.7. Flow Cytometry
2.8. Melanosphere Culture Assay
2.9. In Vivo Studies
3. Results
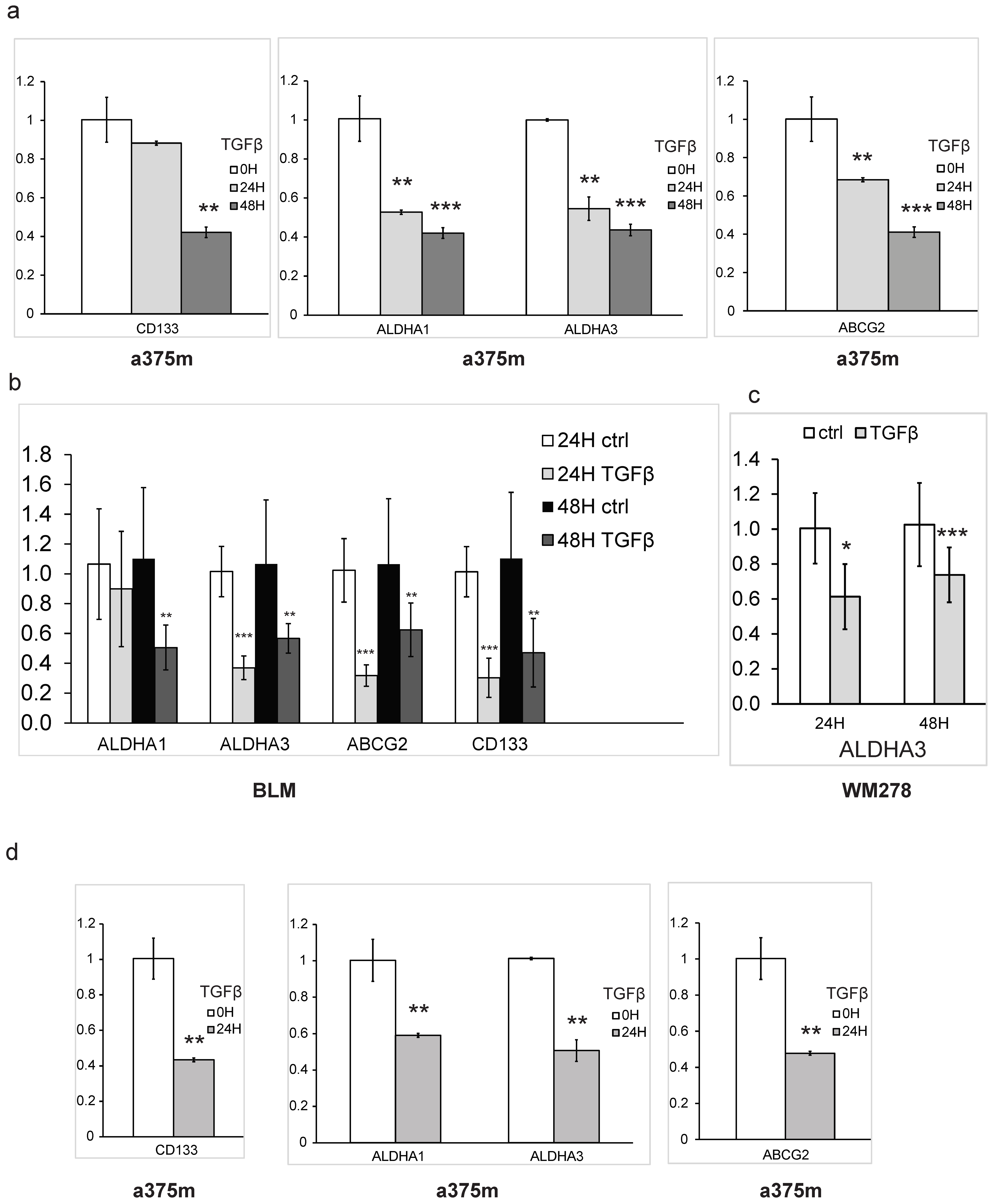
3.1. TGFβ Inhibits Stem-Cell Maintenance and CSC Self-Renewal Capacity in Melanoma
3.2. The Smad3/4 Pathway Is Required for TGFβ-Mediated Inhibition of Melanoma Cancer Self-Renewal
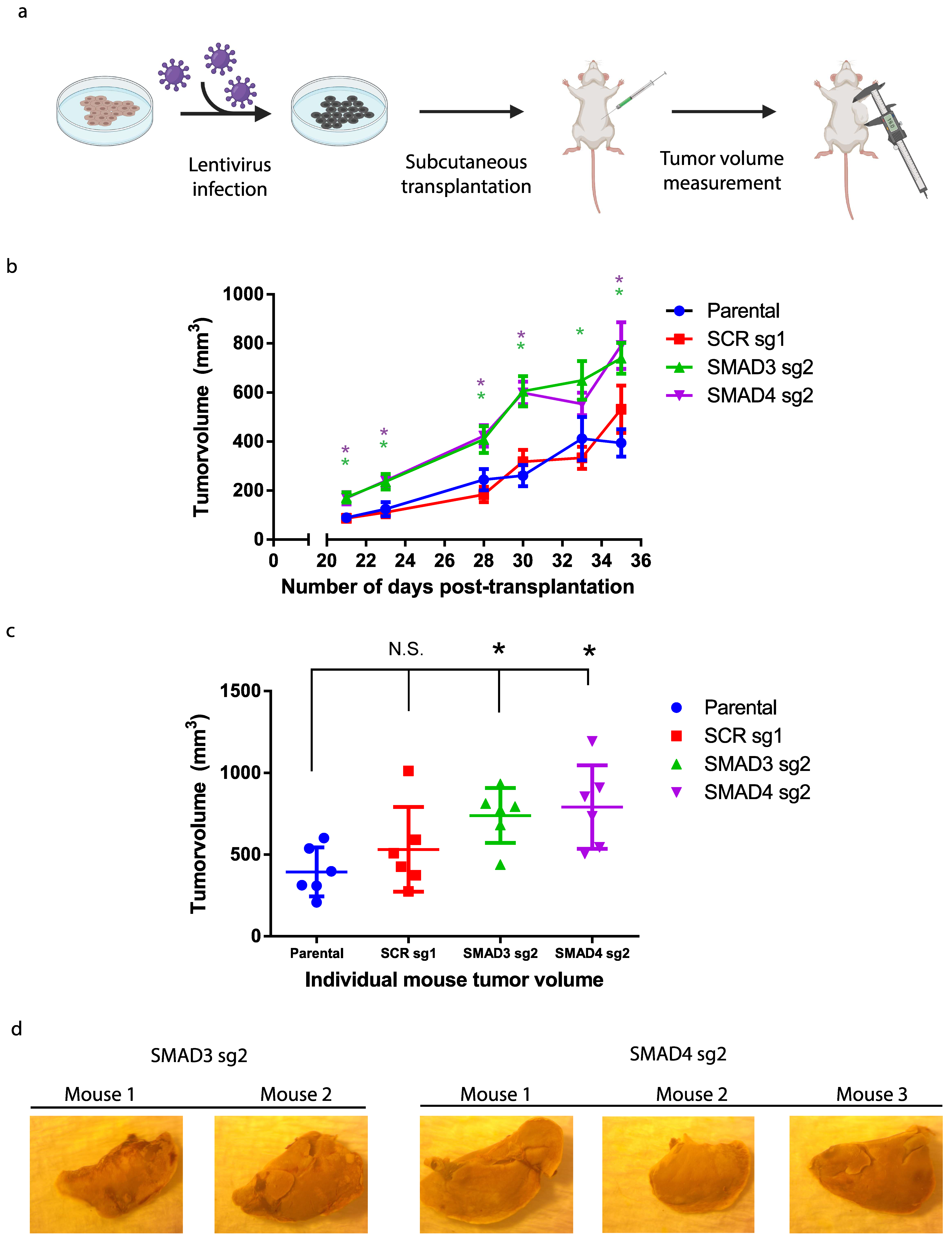
3.3. Blocking TGFβ/Smad Signaling Promotes Melanoma Tumor Growth In Vivo
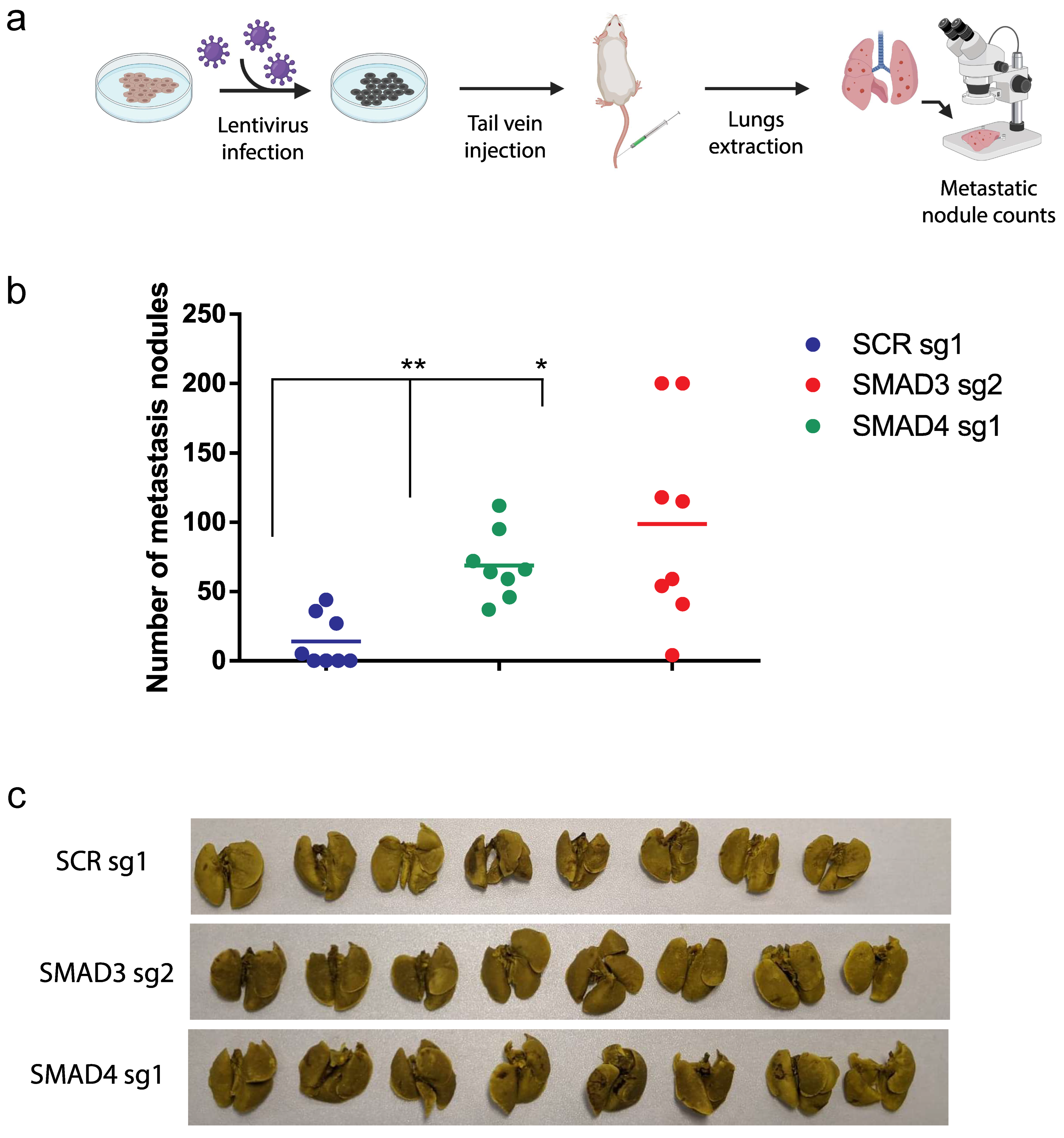
3.4. The TGFβ/Smad Pathway Inhibits Melanoma Lung Metastasis In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Domingues, B.; Lopes, J.; Soares, P.; Populo, H. Melanoma treatment in review. ImmunoTargets Ther. 2018, 7, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Damsky, W.E.; Theodosakis, N.; Bosenberg, M. Melanoma metastasis: New concepts and evolving paradigms. Oncogene 2014, 33, 2413–2422. [Google Scholar] [CrossRef] [PubMed]
- Shannan, B.; Perego, M.; Somasundaram, R.; Herlyn, M. Heterogeneity in melanoma. Cancer Treat. Res. 2016, 167, 1–15. [Google Scholar] [PubMed]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy (Review). Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Kwong, L.N.; Davies, M.A. Navigating the Therapeutic Complexity of PI3K Pathway Inhibition in Melanoma. Clin. Cancer Res. 2013, 19, 5310–5319. [Google Scholar] [CrossRef] [PubMed]
- Xue, G.; Romano, E.; Massi, D.; Mandalà, M. Wnt/β-catenin signaling in melanoma: Preclinical rationale and novel therapeutic insights. Cancer Treat. Rev. 2016, 49, 1–12. [Google Scholar] [CrossRef]
- Ueda, Y.; Richmond, A. NF-κB activation in melanoma. Pigment Cell Res. 2006, 19, 112–124. [Google Scholar] [CrossRef]
- Hammouda, M.B.; Ford, A.E.; Liu, Y.; Zhang, J.Y. The JNK Signaling Pathway in Inflammatory Skin Disorders and Cancer. Cells 2020, 9, 857. [Google Scholar] [CrossRef]
- Thomas, S.J.; Snowden, J.A.; Zeidler, M.P.; Danson, S.J. The role of JAK/STAT signalling in the pathogenesis, prognosis and treatment of solid tumours. Br. J. Cancer 2015, 113, 365–371. [Google Scholar] [CrossRef]
- Javelaud, D.; Alexaki, V.I.; Mauviel, A. Transforming growth factor-β in cutaneous melanoma. Pigment Cell Melanoma Res. 2008, 21, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Humbert, L.; Lebrun, J.J. TGF-beta inhibits human cutaneous melanoma cell migration and invasion through regulation of the plasminogen activator system. Cell Signal. 2013, 25, 490–500. [Google Scholar] [CrossRef] [PubMed]
- Humbert, L.; Ghozlan, M.; Canaff, L.; Tian, J.; Lebrun, J.J. The leukemia inhibitory factor (LIF) and p21 mediate the TGFβ tumor suppressive effects in human cutaneous melanoma. BMC Cancer 2015, 15, 200. [Google Scholar] [CrossRef] [PubMed]
- Ramont, L.; Pasco, S.; Hornebeck, W.; Maquart, F.X.; Monboisse, J.C. Transforming growth factor-β1 inhibits tumor growth in a mouse melanoma model by down-regulating the plasminogen activation system. Exp. Cell Res. 2003, 291, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Weigel, K.J.; Zhou, H.; Wang, X.J. Paradoxical roles of TGF-β signaling in suppressing and promoting squamous cell carcinoma. Acta Biochim. Biophys. Sin. 2018, 50, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Neel, J.-C.; Humbert, L.; Lebrun, J.-J. The Dual Role of TGFβ in Human Cancer: From Tumor Suppression to Cancer Metastasis. ISRN Mol. Biol. 2012, 2012, 381428. [Google Scholar] [CrossRef] [PubMed]
- Stingl, J.; Caldas, C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat. Rev. Cancer 2007, 7, 791–799. [Google Scholar] [CrossRef]
- Visvader, J.E.; Lindeman, G.J. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat. Rev. Cancer 2008, 8, 755–768. [Google Scholar] [CrossRef]
- Bellomo, C.; Caja, L.; Moustakas, A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br. J. Cancer 2016, 115, 761–769. [Google Scholar] [CrossRef]
- Dupin, E.; Sommer, L. Neural crest progenitors and stem cells: From early development to adulthood. Dev. Biol. 2012, 366, 83–95. [Google Scholar] [CrossRef]
- Fang, D.; Nguyen, T.K.; Leishear, K.; Finko, R.; Kulp, A.N.; Hotz, S.; Van Belle, P.A.; Xu, X.; Elder, D.E.; Herlyn, M. Asubpopulation with stem cell properties in melanomas. Cancer Res. 2005, 65, 9328–9337. [Google Scholar]
- Monzani, E.; Facchetti, F.; Galmozzi, E.; Corsini, E.; Benetti, A.; Cavazzin, C.; Gritti, A.; Piccinini, A.; Porro, D.; Santinami, M.; et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur. J. Cancer 2007, 43, 935–946. [Google Scholar] [CrossRef] [PubMed]
- Frank, N.Y.; Margaryan, A.; Huang, Y.; Schatton, T.; Waaga-Gasser, A.M.; Gasser, M.; Sayegh, M.H.; Sadee, W.; Frank, M. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005, 65, 4320–4333. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Nguyen, N.; Fujita, M. Isolation of human melanoma stem cells UNIT 3.8 using ALDH as a marker. Curr. Protoc. Stem Cell Biol. 2013, 1, 3–8. [Google Scholar]
- Boonyaratanakornkit, J.B.; Yue, L.; Strachan, L.R.; Scalapino, K.J.; Leboit, P.E.; Lu, Y.; Leong, S.P.; Smith, J.E.; Ghadially, R. Selection of tumorigenic melanoma cells using ALDH. J. Investig. Dermatol. 2010, 130, 2799–2808. [Google Scholar] [CrossRef] [PubMed]
- Klein, W.M.; Wu, B.P.; Zhao, S.; Wu, H.; Klein-Szanto, A.J.P.; Tahan, S.R. Increased expression of stem cell markers in malignant melanoma. Mod. Pathol. 2007, 20, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Redmer, T.; Walz, I.; Klinger, B.; Khouja, S.; Welte, Y.; Schäfer, R.; Regenbrecht, C. The role of the cancer stem cell marker CD271 in DNA damage response and drug resistance of melanoma cells. Oncogenesis 2017, 6, e291. [Google Scholar] [CrossRef]
- Iwasaki, H.; Suda, T. Cancer stem cells and their niche. Cancer Sci. 2009, 100, 1166–1172. [Google Scholar] [CrossRef]
- Sakaki-Yumoto, M.; Katsuno, Y.; Derynck, R. TGF-β family signaling in stem cells. Biochim. Biophys. Acta—Gen. Subj. 2013, 1830, 2280–2296. [Google Scholar] [CrossRef]
- Tian, J.; Hachim, M.Y.; Hachim, I.Y.; Dai, M.; Lo, C.; Al Raffa, F.; Ali, S.; Lebrun, J.J. Cyclooxygenase-2 regulates TGFβ-induced cancer stemness in triple-negative breast cancer. Sci. Rep. 2017, 7, 40258. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [PubMed]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Concordet, J.-P.; Haeussler, M. CRISPOR: Intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018, 46, W242–W245. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Yoo, N.; Vu, M.; Mamura, M.; Nam, J.S.; Ooshima, A.; Du, Z.; Desprez, P.Y.; Anver, M.R.; Michalowska, A.M.; et al. Transforming growth factor-β can suppress tumorigenesis through effects on the putative cancer stem or early progenitor cell and committed progeny in a breast cancer xenograft model. Cancer Res. 2007, 67, 8643–8652. [Google Scholar] [CrossRef] [PubMed]
- Bhola, N.E.; Balko, J.M.; Dugger, T.C.; Kuba, M.G.; Sánchez, V.; Sanders, M.; Stanford, J.; Cook, R.S.; Arteaga, C.L. TGF-β inhibition enhances chemotherapy action against triple-negative breast cancer. J. Clin. Investig. 2013, 123, 1348–1358. [Google Scholar] [CrossRef]
- Dai, M.; Zhang, C.; Ali, A.; Hong, X.; Tian, J.; Lo, C.; Fils-Aimé, N.; Burgos, S.A.; Ali, S.; Lebrun, J.-J. CDK4 regulates cancer stemness and is a novel therapeutic target for triple-negative breast cancer. Sci. Rep. 2016, 6, 35383. [Google Scholar] [CrossRef]
- Peñuelas, S.; Anido, J.; Prieto-Sánchez, R.M.; Folch, G.; Barba, I.; Cuartas, I.; García-Dorado, D.; Poca, M.A.; Sahuquillo, J.; Baselga, J.; et al. TGF-β Increases Glioma-Initiating Cell Self-Renewal through the Induction of LIF in Human Glioblastoma. Cancer Cell 2009, 15, 315–327. [Google Scholar] [CrossRef]
- Ehata, S.; Johansson, E.; Katayama, R.; Koike, S.; Watanabe, A.; Hoshino, Y.; Katsuno, Y.; Komuro, A.; Koinuma, D.; Kano, M.R.; et al. Transforming growth factor-Β decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene 2011, 30, 1693–1705. [Google Scholar] [CrossRef]
- Oshimori, N.; Oristian, D.; Fuchs, E. TGF-β Promotes Heterogeneity and Drug Resistance in Squamous Cell Carcinoma. Cell 2015, 160, 963–976. [Google Scholar] [CrossRef]
- Samson, J.M.; Ravindran Menon, D.; Smith, D.E.; Baird, E.; Kitano, T.; Gao, D.; Tan, A.C.; Fujita, M. Clinical implications of ALDH1A1 and ALDH1A3 mRNA expression in melanoma subtypes. Chem. Biol. Interact. 2019, 314, 108822. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yu, C.; Wang, B.; Chang, W. Tumorsphere as an effective in vitro platform for screening anti-cancer stem cell drugs. Oncotarget 2016, 7, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Rodeck, U.; Nishiyama, T.; Mauviel, A. Independent regulation of growth and SMAD-mediated transcription by transforming growth factor β in human melanoma cells. Cancer Res. 1999, 59, 547–550. [Google Scholar] [PubMed]
- Shams, A.; Binothman, N.; Boudreault, J.; Wang, N.; Shams, F.; Hamam, D.; Tian, J.; Moamer, A.; Dai, M.; Lebrun, J.J.; et al. Prolactin receptor-driven combined luminal and epithelial differentiation in breast cancer restricts plasticity, stemness, tumorigenesis and metastasis. Oncogenesis 2021, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Boudreault, J.; Wang, N.; Poulet, S.; Daliah, G.; Yan, G.; Moamer, A.; Burgos, S.A.; Sabri, S.; Ali, S.; et al. Differential regulation of cancer progression by CDK4/6 plays a central role in DNA replication and repair pathways. Cancer Res. 2021, 81, 1332–1346. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Dai, M.; Zhang, C.; Poulet, S.; Moamer, A.; Wang, N.; Boudreault, J.; Ali, S.; Lebrun, J.J. TGFβ/cyclin D1/Smad-mediated inhibition of BMP4 promotes breast cancer stem cell self-renewal activity. Oncogenesis 2021, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Anselmi, M.; Fontana, F.; Marzagalli, M.; Gagliano, N.; Sommariva, M.; Limonta, P. Melanoma Stem Cells Educate Neutrophils to Support Cancer Progression. Cancers 2022, 14, 3391. [Google Scholar] [CrossRef] [PubMed]
- Yin, Q.; Shi, X.; Lan, S.; Jin, H.; Wu, D. Effect of melanoma stem cells on melanoma metastasis (Review). Oncol. Lett. 2021, 22, 566. [Google Scholar] [CrossRef]
- Grasso, C.; Anaka, M.; Hofmann, O.; Sompallae, R.; Broadley, K.; Hide, W.; Berridge, M.V.; Cebon, J.; Behren, A.; McConnell, M.J. Iterative sorting reveals CD133+ and CD133- melanoma cells as phenotypically distinct populations. BMC Cancer 2016, 16, 726. [Google Scholar] [CrossRef]
- Simbulan-Rosenthal, C.M.; Dougherty, R.; Vakili, S.; Ferraro, A.M.; Kuo, L.W.; Alobaidi, R.; Aljehane, L.; Gaur, A.; Sykora, P.; Glasgow, E.; et al. CRISPR-Cas9 knockdown and induced expression of CD133 reveal essential roles in melanoma invasion and metastasis. Cancers 2019, 11, 1490. [Google Scholar] [CrossRef]
- Hoshino, Y.; Nishida, J.; Katsuno, Y.; Koinuma, D.; Aoki, T.; Kokudo, N.; Miyazono, K.; Ehata, S. Smad4 decreases the population of pancreatic cancer—Initiating cells through transcriptional repression of ALDH1A1. Am. J. Pathol. 2015, 185, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Kaji, H.; Canaff, L.; Lebrun, J.-J.; Goltzman, D.; Hendy, G.N. Inactivation of menin, a Smad3-interacting protein, blocks transforming growth factor type β signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 3837–3842. [Google Scholar] [CrossRef] [PubMed]
- Lacerte, A.; Korah, J.; Roy, M.; Yang, X.J.; Lemay, S.; Lebrun, J.J. Transforming growth factor-β inhibits telomerase through SMAD3 and E2F transcription factors. Cell Signal. 2008, 20, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Lee, J.K.; Miyake, S.; Kim, S.J. A Novel E1A-like Inhibitor of Differentiation (EID) Family Member, EID-2, Suppresses Transforming Growth Factor (TGF)-β Signaling by Blocking TGF-β-induced Formation of Smad3-Smad4 Complexes. J. Biol. Chem. 2004, 279, 2666–2672. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xu, D.; Li, J.; Berndt, M.C.; Liu, J.P. Transforming growth factor β suppresses human telomerase reverse transcriptase (hTERT) by Smad3 interactions with c-Myc and the hTERT gene. J. Biol. Chem. 2006, 281, 25588–25600. [Google Scholar] [CrossRef] [PubMed]
- Kretschmer, A.; Moepert, K.; Dames, S.; Sternberger, M.; Kaufmann, J.; Klippel, A. Differential regulation of TGF-β signaling through Smad2, Smad3 and Smad4. Oncogene 2003, 22, 6748–6763. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Solal, K.A.; Merrigan, K.T.; Chan, J.L.K.; Goydos, J.S.; Chen, W.; Foran, D.J.; Liu, F.; Lasfar, A.; Reiss, M. Constitutive Smad linker phosphorylation in melanoma: A mechanism of resistance to transforming growth factor-β-mediated growth inhibition. Pigment Cell Melanoma Res. 2011, 24, 512–524. [Google Scholar] [CrossRef]
- Lüönd, F.; Pirkl, M.; Hisano, M.; Prestigiacomo, V.; Kalathur, R.K.R.; Beerenwinkel, N.; Christofori, G. Hierarchy of TGFβ/SMAD, Hippo/YAP/TAZ, and Wnt/ β-catenin signaling in melanoma phenotype switching. Life Sci. Alliance 2022, 5, 1–14. [Google Scholar] [CrossRef]
- Goding, C.R. A picture of Mitf in melanoma immortality. Oncogene 2011, 30, 2304–2306. [Google Scholar] [CrossRef]
- Pierrat, M.J.; Marsaud, V.; Mauviel, A.; Javelaud, D. Expression of microphthalmia-associated transcription factor (MITF), which is critical for melanoma progression, is inhibited by both transcription factor GLI2 and transforming growth factor-β. J. Biol. Chem. 2012, 287, 17996–18004. [Google Scholar] [CrossRef]
- Yeh, H.W.; Lee, S.S.; Chang, C.Y.; Lang, Y.D.; Jou, Y.S. A new switch for TGFβ in cancer. Cancer Res. 2019, 79, 3797–3805. [Google Scholar] [CrossRef] [PubMed]
- Bierie, B.; Moses, H.L. TGF-β and cancer. Cytokine Growth Factor Rev. 2006, 17, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.B.; Wakefield, L.M. The two faces of transforming growth factor β in carcinogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 8621–8623. [Google Scholar] [CrossRef] [PubMed]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Javelaud, D.; Mohammad, K.S.; McKenna, C.R.; Fournier, P.; Luciani, F.; Niewolna, M.; André, J.; Delmas, V.; Larue, L.; Guise, T.A.; et al. Stable overexpression of Smad7 in human melanoma cells impairs bone metastasis. Cancer Res. 2007, 67, 2317–2324. [Google Scholar] [CrossRef] [PubMed]
- Javelaud, D.; Delmas, V.; Möller, M.; Sextius, P.; André, J.; Menashi, S.; Larue, L.; Mauviel, A. Stable overexpression of Smad7 in human melanoma cells inhibits their tumorigenicity in vitro and in vivo. Oncogene 2005, 24, 7624–7629. [Google Scholar] [CrossRef]
- Derakhshani, A.; Silvestris, N.; Hemmat, N.; Asadzadeh, Z.; Shadbad, M.A.; Nourbakhsh, N.S.; Mobasheri, L.; Vahedi, P.; Shahmirzaie, M.; Brunetti, O.; et al. Targeting TGF-β-Mediated SMAD Signaling pathway via novel recombinant cytotoxin II: A potent protein from naja naja oxiana venom in Melanoma. Molecules 2020, 25, 5148. [Google Scholar] [CrossRef]
- Laverty, H.G.; Wakefield, L.M.; Occleston, N.L.; O’Kane, S.; Ferguson, M.W.J. TGF-β3 and cancer: A review. Cytokine Growth Factor Rev. 2009, 20, 305–317. [Google Scholar] [CrossRef]
- Durani, P.; Occleston, N.; O’Kane, S.; Ferguson, M.W.J. Avotermin: A novel antiscarring agent. Int. J. Low. Extrem. Wounds 2008, 7, 160–168. [Google Scholar] [CrossRef]
- Marzagalli, M.; Moretti, R.M.; Messi, E.; Marelli, M.M.; Fontana, F.; Anastasia, A.; Bani, M.R.; Beretta, G.; Limonta, P. Targeting melanoma stem cells with the Vitamin E derivative δ-tocotrienol. Sci. Rep. 2018, 8, 587. [Google Scholar] [CrossRef]


| Primer Name | Single-Guide Primer Sequence |
|---|---|
| scrsg1-F | 5′-CACCGACGGAGGCTAAGCGTCGCAA-3′ |
| scrsg1-R | 5′-AAACTTGCGACGCTTAGCCTCCGTC-3′ |
| scrsg2-F | 5′-CACCGCGCTTCCGCGGCCCGTTCAA-3′ |
| scrsg2-R | 5′-AAACTTGAACGGGCCGCGGAAGCGC-3′ |
| scrsg3-F | 5′-CACCGATCGTTTCCGCTTAACGGCG-3′ |
| scrsg3-R | 5′-AAACCGCCGTTAAGCGGAAACGATC-3′ |
| Smad2sg4-F | 5′-CACCGTGGCGGCGTGAATGGCAAGA-3′ |
| Smad2sg4-R | 5′-AAACTCTTGCCATTCACGCCGCCAC-3′ |
| Smad3sg2-F | 5′-CACCGTTCACGATCGGGGGAGTGAA-3′ |
| Smad3sg2-R | 5′-AAACTTCACTCCCCCGATCGTGAAC-3′ |
| Smad4sg1-F | 5′-CACCGAACTCTGTACAAAGACCGCG-3′ |
| Smad4sg1-R | 5′-AAACCGCGGTCTTTGTACAGAGTTC-3′ |
| Primer Name | Primer Sequence for qPCR |
|---|---|
| CD133-F | TACCAAGGACAAGGGGTTCAC |
| CD133-R | CAGTCGTGGTTTGGCGTTGTA |
| ABCG2-F | GCTCAGGAGGCCTTGGGATA |
| ABCG2-R | GGCTCTATGATCTCTGTGGCTTT |
| ALDH1A1-F | CTGTGTTCCAGGAGCCGAAT |
| ALDH1A1-R | CTGCCTTGTCAACATCCTCCTTA |
| ALDH1A3-F | GGAAGAAGGAGATAAGCCCGAC |
| ALDH1A3-R | AGCCCTCCAGGTCGATGAAA |
| GAPDH-F | GACAGTCAGCCGCATCTTCT |
| GAPDH-R | GCGCCCAATACGACCAAATC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boudreault, J.; Wang, N.; Ghozlan, M.; Lebrun, J.-J. Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis. Cancers 2024, 16, 224. https://doi.org/10.3390/cancers16010224
Boudreault J, Wang N, Ghozlan M, Lebrun J-J. Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis. Cancers. 2024; 16(1):224. https://doi.org/10.3390/cancers16010224
Chicago/Turabian StyleBoudreault, Julien, Ni Wang, Mostafa Ghozlan, and Jean-Jacques Lebrun. 2024. "Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis" Cancers 16, no. 1: 224. https://doi.org/10.3390/cancers16010224
APA StyleBoudreault, J., Wang, N., Ghozlan, M., & Lebrun, J.-J. (2024). Transforming Growth Factor-β/Smad Signaling Inhibits Melanoma Cancer Stem Cell Self-Renewal, Tumor Formation and Metastasis. Cancers, 16(1), 224. https://doi.org/10.3390/cancers16010224




