Aspirin and Cancer Survival: An Analysis of Molecular Mechanisms
Abstract
Simple Summary
Abstract
1. Introduction
2. Material and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| COX | cyclooxigenase |
| PTGS | prostaglandin-endoperoxide synthase |
| PTGES | prostaglandin E synthase |
| EGFR | epidermal growth factor receptor |
| JAK | Janus kinase |
| BRCA | BReast CAncer gene |
| PARP | Poly (ADP-ribose) polymerases |
| DNA | Deoxyribonucleic acid |
| RCTs | randomized controlled trials |
| HR | hazard ratio |
| MMR | mismatch repair |
| MeSH | Medical Subject heading |
| ASAMET | Aspirin and Metformin in Stage I–III Colorectal Cancer Patients |
References
- Creagan, E.T.; Twito, D.I.; Johansson, S.L.; Schaid, D.J.; Johnson, P.S.; Flaum, M.A.; Buroker, T.R.; Geeraerts, L.H.; Veeder, M.H.; Gesme, D.H. A randomized prospective assessment of recombinant leukocyte A human interferon with or without aspirin in advanced renal adenocarcinoma. J. Clin. Oncol. 1991, 9, 2104–2109. [Google Scholar] [CrossRef] [PubMed]
- Lebeau, B.; Chastang, C.; Muir, J.F.; Vincent, J.; Massin, F.; Fabre, C.; The “Petites Cellules” Group. No effect of an antiaggregant treatment with aspirin in small cell lung cancer treated with CCAVP16 chemotherapy results from a randomized clinical trial of 303 patients. Cancer 1993, 71, 1741–1745. [Google Scholar] [CrossRef] [PubMed]
- Lipton, A.; Scialla, S.; Harvey, H.; Dixon, R.; Gordon, R.; Hamilton, R.; Ramsey, H.; Weltz, M.; Heckard, R.; White, D. Adjuvant antiplatelet therapy with aspirin in colo-rectal cancer. J. Med. 1982, 13, 419–429. [Google Scholar] [PubMed]
- Liu, J.-F.; Jamieson, G.G.; Wu, T.-C.; Zhu, G.-J.; Drew, P.A. A preliminary study on the postoperative survival of patients given aspirin after resection for squamous cell carcinoma of the esophagus or adenocarcinoma of the cardia. Ann. Surg. Oncol. 2009, 16, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- Elwood, P.C.; Morgan, G.; Delon, C.; Protty, M.; Galante, J.; Pickering, J.; Watkins, J.; Weightman, A.; Morris, D. Aspirin and cancer survival: A systematic review and meta-analyses of 118 observational studies of aspirin and 18 cancers. ecancermedicalscience 2021, 15, 1258. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, L.; Nist, A.; Mernberger, M.; Stiewe, T.; Moll, R.; Stabla, K.; Klinge, U.; Mack, E.; Brendel, C.; Neubauer, A. Superior Overall Survival in Patients with Colorectal Cancer, Regular Aspirin Use, and Combined Wild-Type PIK3CA and KRAS-Mutated Tumors. Cancers 2021, 13, 4959. [Google Scholar] [CrossRef]
- Liao, S.; Koshiol, J.; Huang, Y.; Jackson, S.S.; Huang, Y.; Chan, C.; Huang, C.; Liu, P.; Chen, Y.; Hsieh, R.J.; et al. Postdiagnosis Aspirin Use Associated with Decreased Biliary Tract Cancer–Specific Mortality in a Large Nationwide Cohort. Hepatology 2021, 74, 1994–2006. [Google Scholar] [CrossRef]
- Jackson, S.S.; Pfeiffer, R.M.; Liu, Z.; Anderson, L.A.; Tsai, H.-T.; Gadalla, S.M.; Koshiol, J. Association Between Aspirin Use and Biliary Tract Cancer Survival. JAMA Oncol. 2019, 5, 1802–1804. [Google Scholar] [CrossRef]
- Gray, R.T.; Cantwell, M.M.; Coleman, H.G.; Loughrey, M.B.; Bankhead, P.; McQuaid, S.; O’Neill, R.F.; Arthur, K.; Bingham, V.; McGready, C.; et al. Evaluation of PTGS2 Expression, PIK3CA Mutation, Aspirin Use and Colon Cancer Survival in a Population-Based Cohort Study. Clin. Transl. Gastroenterol. 2017, 8, e91. [Google Scholar] [CrossRef]
- Choe, K.S.; Cowan, J.E.; Chan, J.M.; Carroll, P.R.; D’Amico, A.V.; Liauw, S.L. Aspirin use and the risk of prostate cancer mortality in men treated with prostatectomy or radiotherapy. J. Clin. Oncol. 2012, 30, 3540–3544. [Google Scholar] [CrossRef]
- Matsuo, K.; Cahoon, S.S.; Yoshihara, K.; Shida, M.; Kakuda, M.; Adachi, S.; Moeini, A.; Machida, H.; Garcia-Sayre, J.; Ueda, Y.; et al. Association of Low-Dose Aspirin and Survival of Women with Endometrial Cancer. Obstet. Gynecol. 2016, 128, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Haemmerle, M.; Stone, R.L.; Menter, D.G.; Afshar-Kharghan, V.; Sood, A.K. The Platelet Lifeline to Cancer: Challenges and Opportunities. Cancer Cell 2018, 33, 965–983. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.R.; Yousef, G.M.; Ni, H. Cancer and platelet crosstalk: Opportunities and challenges for aspirin and other antiplatelet agents. Blood 2018, 131, 1777–1789. [Google Scholar] [CrossRef] [PubMed]
- Ai, G.; Dachineni, R.; Muley, P.; Tummala, H.; Bhat, G.J. Aspirin and salicylic acid decrease c-Myc expression in cancer cells: A potential role in chemoprevention. Tumor Biol. 2016, 37, 1727–1738. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.-H.; Yuan, L.-Y.; Huang, R.-B.; Chen, G.-A. Aspirin inhibits proliferation and induces apoptosis of multiple myeloma cells through regulation of Bcl-2 and Bax and suppression of VEGF. Eur. J. Haematol. 2014, 93, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Bashir, A.I.; Kankipati, C.S.; Jones, S.; Newman, R.M.; Safrany, S.T.; Perry, C.J.; Nicholl, I.D. A novel mechanism for the anticancer activity of aspirin and salicylates. Int. J. Oncol. 2019, 54, 1256–1270. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, K.; Valanciute, A.; Lima, A.C.S.; Vinuela, P.F.; Jamieson, T.; Rajasekaran, V.; Blackmur, J.; Ochocka-Fox, A.-M.; Guazzelli, A.; Cammareri, P.; et al. Aspirin Rescues Wnt-Driven Stem-like Phenotype in Human Intestinal Organoids and Increases the Wnt Antagonist Dickkopf-1. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 465–489. [Google Scholar] [CrossRef]
- Fernandez, H.R.; Lindén, S.K. The aspirin metabolite salicylate inhibits lysine acetyltransferases and MUC1 induced epithelial to mesenchymal transition. Sci. Rep. 2017, 7, 5626. [Google Scholar] [CrossRef]
- Frouws, M.A.; Reimers, M.S.; Swets, M.; Bastiaannet, E.; Prinse, B.; Van Eijk, R.; Lemmens, V.E.P.P.; Van Herk-Sukel, M.P.P.; Van Wezel, T.; Kuppen, P.J.K.; et al. The Influence of BRAF and KRAS Mutation Status on the Association between Aspirin Use and Survival after Colon Cancer Diagnosis. PLoS ONE 2017, 12, e0170775. [Google Scholar] [CrossRef]
- Gan, H.; Lin, L.; Hu, N.; Yang, Y.; Gao, Y.; Pei, Y.; Chen, K.; Sun, B. Aspirin ameliorates lung cancer by targeting the miR-98/WNT1 axis. Thorac. Cancer 2019, 10, 744–750. [Google Scholar] [CrossRef]
- Guo, H.; Liu, J.; Ben, Q.; Qu, Y.; Li, M.; Wang, Y.; Chen, W.; Zhang, J. The aspirin-induced long non-coding RNA OLA1P2 blocks phosphorylated STAT3 homodimer formation. Genome Biol. 2016, 17, 24. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.C.N.; Benndorf, R.A. Aspirin sensitivity of PIK3CA-mutated Colorectal Cancer: Potential mechanisms revisited. Cell. Mol. Life Sci. 2022, 79, 393. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-R.; Bae, M.-K.; Kim, J.-Y.; Wee, H.-J.; Yoo, M.-A.; Bae, S.-K. Aspirin induces apoptosis through the blockade of IL-6-STAT3 signaling pathway in human glioblastoma A172 cells. Biochem. Biophys. Res. Commun. 2009, 387, 342–347. [Google Scholar] [CrossRef]
- Liao, X.; Lochhead, P.; Nishihara, R.; Morikawa, T.; Kuchiba, A.; Yamauchi, M.; Imamura, Y.; Qian, Z.R.; Baba, Y.; Shima, K.; et al. Aspirin Use, TumorPIK3CAMutation, and Colorectal-Cancer Survival. N Engl. J. Med. 2012, 367, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Mädge, J.C.; Stallmach, A.; Kleebusch, L.; Schlattmann, P. Meta-analysis of aspirin-guided therapy of colorectal cancer. J. Cancer Res. Clin. Oncol. 2022, 148, 1407–1417. [Google Scholar] [CrossRef]
- Ou, Y.-Q.; Zhu, W.-B.; Li, Y.; Qiu, P.-X.; Huang, Y.-J.; Xie, J.; He, S.-M.; Zheng, X.-K.; Leng, T.-D.; Xu, D.; et al. Aspirin inhibits proliferation of gemcitabine-resistant human pancreatic cancer cells and augments gemcitabine-induced cytotoxicity. Acta Pharmacol. Sin. 2010, 31, 73–80. [Google Scholar] [CrossRef][Green Version]
- Park, I.-S.; Jo, J.-R.; Hong, H.; Nam, K.-Y.; Kim, J.-B.; Hwang, S.-H.; Choi, M.-S.; Ryu, N.-H.; Jang, H.-J.; Lee, S.-H.; et al. Aspirin induces apoptosis in YD-8 human oral squamous carcinoma cells through activation of caspases, down-regulation of Mcl-1, and inactivation of ERK-1/2 and AKT. Toxicol. Vitr. 2010, 24, 713–720. [Google Scholar] [CrossRef]
- Reimers, M.S.; Bastiaannet, E.; Langley, R.E.; van Eijk, R.; Van Vlierberghe, R.L.P.; Lemmens, V.E.P.; van Herk-Sukel, M.P.P.; van Wezel, T.; Fodde, R.; Kuppen, P.J.K.; et al. Expression of HLA Class I Antigen, Aspirin Use, and Survival after a Diagnosis of Colon Cancer. JAMA Intern. Med. 2014, 174, 732–739. [Google Scholar] [CrossRef]
- Slattery, M.L.; Lundgreen, A.; Hines, L.M.; Torres-Mejia, G.; Wolff, R.K.; Stern, M.C.; John, E.M. Genetic variation in the JAK/STAT/SOCS signaling pathway influences breast cancer-specific mortality through interaction with cigarette smoking and use of aspirin/NSAIDs: The Breast Cancer Health Disparities Study. Breast Cancer Res. Treat. 2014, 147, 145–158. [Google Scholar] [CrossRef]
- Slattery, M.L.; Lundgreen, A.; Bondurant, K.L.; Wolff, R.K. Interferon-signaling pathway: Associations with colon and rectal cancer risk and subsequent survival. Carcinogenesis 2011, 32, 1660–1667. [Google Scholar] [CrossRef]
- Uddin, S.; Ahmed, M.; Hussain, A.; Assad, L.; Al-Dayel, F.; Bavi, P.; Al-Kuraya, K.S.; Munkarah, A. Cyclooxygenase-2 inhibition inhibits PI3K/AKT kinase activity in epithelial ovarian cancer. Int. J. Cancer 2010, 126, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X. SOX7 is involved in aspirin-mediated growth inhibition of human colorectal cancer cells. World J. Gastroenterol. 2011, 17, 4922–4927. [Google Scholar] [CrossRef] [PubMed]
- Antunes, D.M.; Rodrigues, M.F.S.D.; Guimarães, D.M.; Duarte, C.M.E.; Miguita, L.; Corrêa, L.; DE Oliveira, A.P.L.; Fernandes, K.P.S.; Nunes, F.D. Nonsteroidal Anti-inflammatory Drugs Modulate Gene Expression of Inflammatory Mediators in Oral Squamous Cell Carcinoma. Anticancer. Res. 2019, 39, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Bondurant, K.L.; Lundgreen, A.; Herrick, J.S.; Kadlubar, S.; Wolff, R.K.; Slattery, M.L. Interleukin genes and associations with colon and rectal cancer risk and overall survival. Int. J. Cancer 2013, 132, 905–915. [Google Scholar] [CrossRef] [PubMed]
- Farrugia, M.K.; Long, M.D.; Mattson, D.M.; Flaherty, L.T.; Dong, B.; Gomez, E.C.; Wei, L.; Witkiewicz, A.K.; Yao, S.; Kalinski, P.; et al. Concurrent Aspirin Use Is Associated with Improved Outcome in Rectal Cancer Patients Who Undergo Chemoradiation Therapy. Cancers 2021, 13, 205. [Google Scholar] [CrossRef]
- Feng, Y.; Tao, L.; Wang, G.; Li, Z.; Yang, M.; He, W.; Zhong, X.; Zhang, Y.; Yang, J.; Cheung, S.; et al. Aspirin inhibits prostaglandins to prevents colon tumor formation via down-regulating Wnt production. Eur. J. Pharmacol. 2021, 906, 174173. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Serret, D.; Piqué, M.; Barragán, M.; Cosialls, A.M.; Santidrián, A.F.; González-Gironès, D.M.; Coll-Mulet, L.; de Frias, M.; Pons, G.; Gil, J. Aspirin induces apoptosis in human leukemia cells independently of NF-κB and MAPKs through alteration of the Mcl-1/Noxa balance. Apoptosis 2010, 15, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Vishvakarma, N.K.; Tyagi, A.; Bharti, A.C.; Singh, S.M. Anti-neoplastic action of aspirin against a T-cell lymphoma involves an alteration in the tumour microenvironment and regulation of tumour cell survival. Biosci. Rep. 2012, 32, 91–104. [Google Scholar] [CrossRef][Green Version]
- Kutuk, O.; Basaga, H. Aspirin inhibits TNFα- and IL-1-induced NF-κB activation and sensitizes HeLa cells to apoptosis. Cytokine 2004, 25, 229–237. [Google Scholar] [CrossRef]
- Li, P.; Wu, H.; Zhang, H.; Shi, Y.; Xu, J.; Ye, Y.; Xia, D.; Yang, J.; Cai, J.; Wu, Y. Aspirin use after diagnosis but not prediagnosis improves established colorectal cancer survival: A meta-analysis. Gut 2015, 64, 1419–1425. [Google Scholar] [CrossRef]
- Sakoda, L.C.; Gao, Y.-T.; Chen, B.E.; Chen, J.; Rosenberg, P.S.; Rashid, A.; Deng, J.; Shen, M.-C.; Wang, B.-S.; Han, T.-Q.; et al. Prostaglandin-endoperoxide synthase 2 (PTGS2) gene polymorphisms and risk of biliary tract cancer and gallstones: A population-based study in Shanghai, China. Carcinogenesis 2006, 27, 1251–1256. [Google Scholar] [CrossRef] [PubMed]
- Tougeron, D.; Sha, D.; Manthravadi, S.; Sinicrope, F.A. Aspirin and Colorectal Cancer: Back to the Future. Clin. Cancer Res. 2014, 20, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Galipeau, P.C.; Oman, K.M.; Paulson, T.G.; Sanchez, C.A.; Zhang, Q.; Marty, J.A.; Delrow, J.J.; Kuhner, M.K.; Vaughan, T.L.; Reid, B.J.; et al. NSAID use and somatic exomic mutations in Barrett’s esophagus. Genome Med. 2018, 10, 17. [Google Scholar] [CrossRef] [PubMed]
- Bui, Q.M.; Lin, D.; Ho, W. Approach to Lynch Syndrome for the Gastroenterologist. Dig. Dis. Sci. 2017, 62, 299–304. [Google Scholar] [CrossRef]
- Nicholl; Dibra, H.K.; Brown, J.E.; Hooley, P.; Nicholl, I.D. Aspirin and alterations in DNA repair proteins in the SW480 colorectal cancer cell line. Oncol. Rep. 2010, 24, 37–46. [Google Scholar] [CrossRef]
- Petrera, M.; Paleari, L.; Clavarezza, M.; Puntoni, M.; Caviglia, S.; Briata, I.M.; Oppezzi, M.; Mislej, E.M.; Stabuc, B.; Gnant, M.; et al. The ASAMET trial: A randomized, phase II, double-blind, placebo-controlled, multicenter, 2 × 2 factorial biomarker study of tertiary prevention with low-dose aspirin and metformin in stage I-III colorectal cancer patients. BMC Cancer 2018, 18, 1210. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, W.; Jiang, J. Prevention and treatment of cancer targeting chronic inflammation: Research progress, potential agents, clinical studies and mechanisms. Sci. China Life Sci. 2017, 60, 601–616. [Google Scholar] [CrossRef]
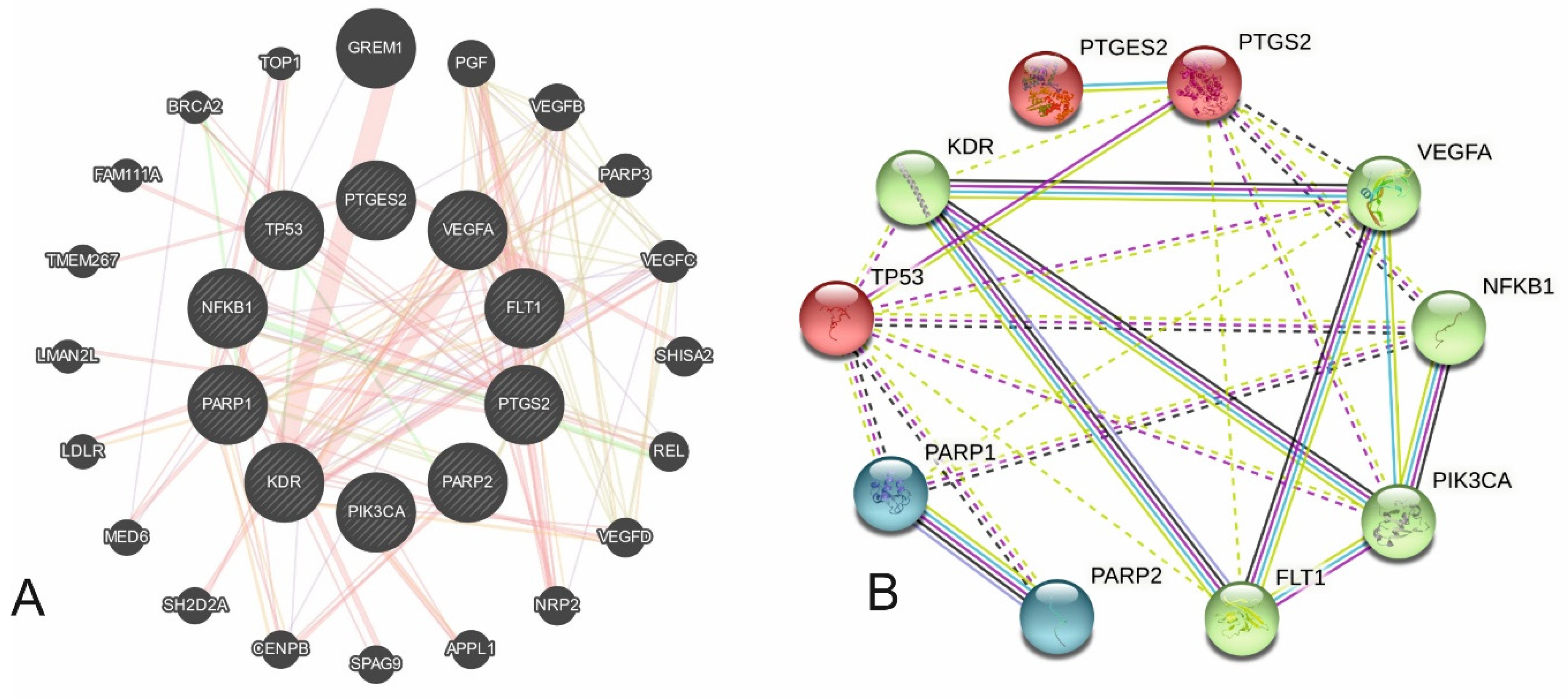
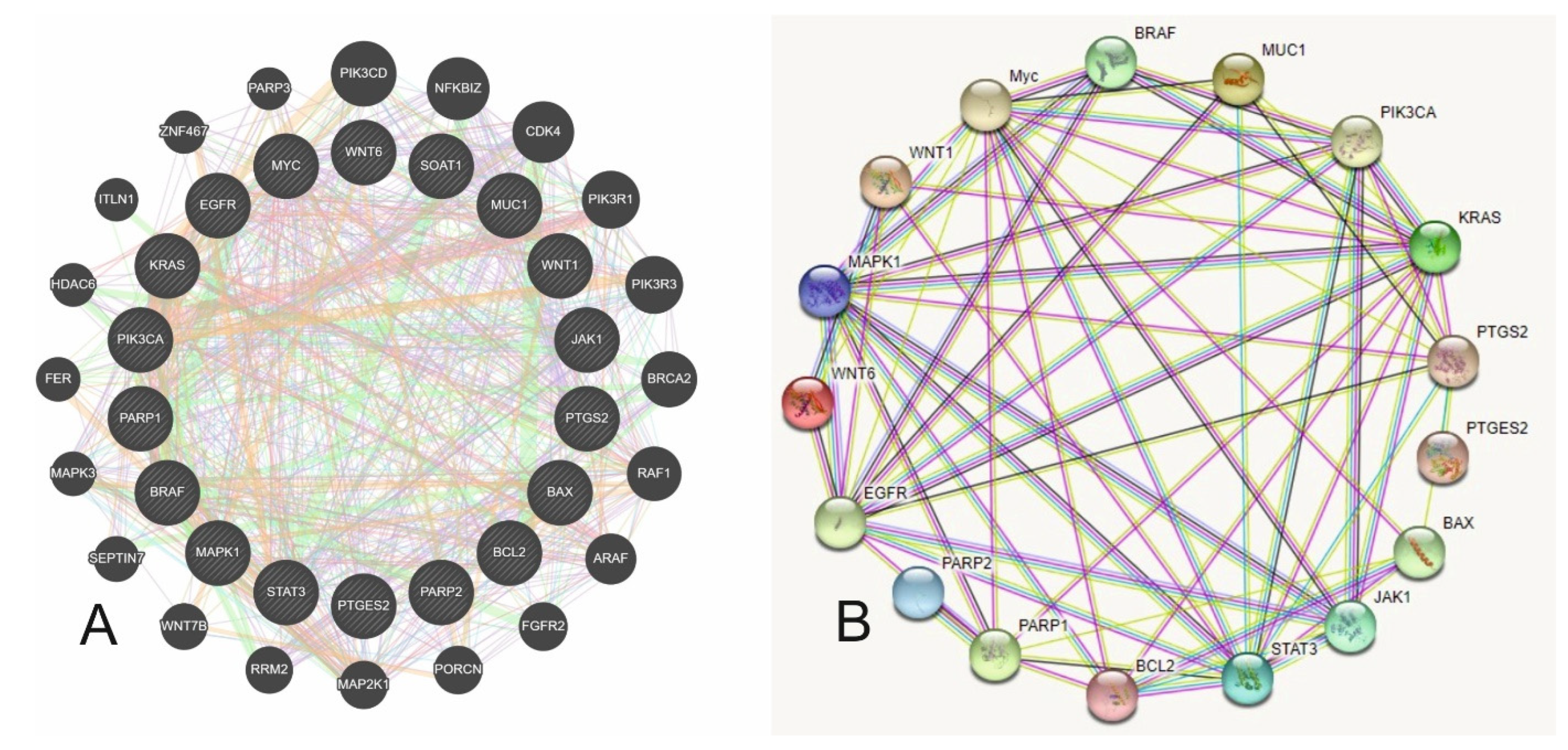
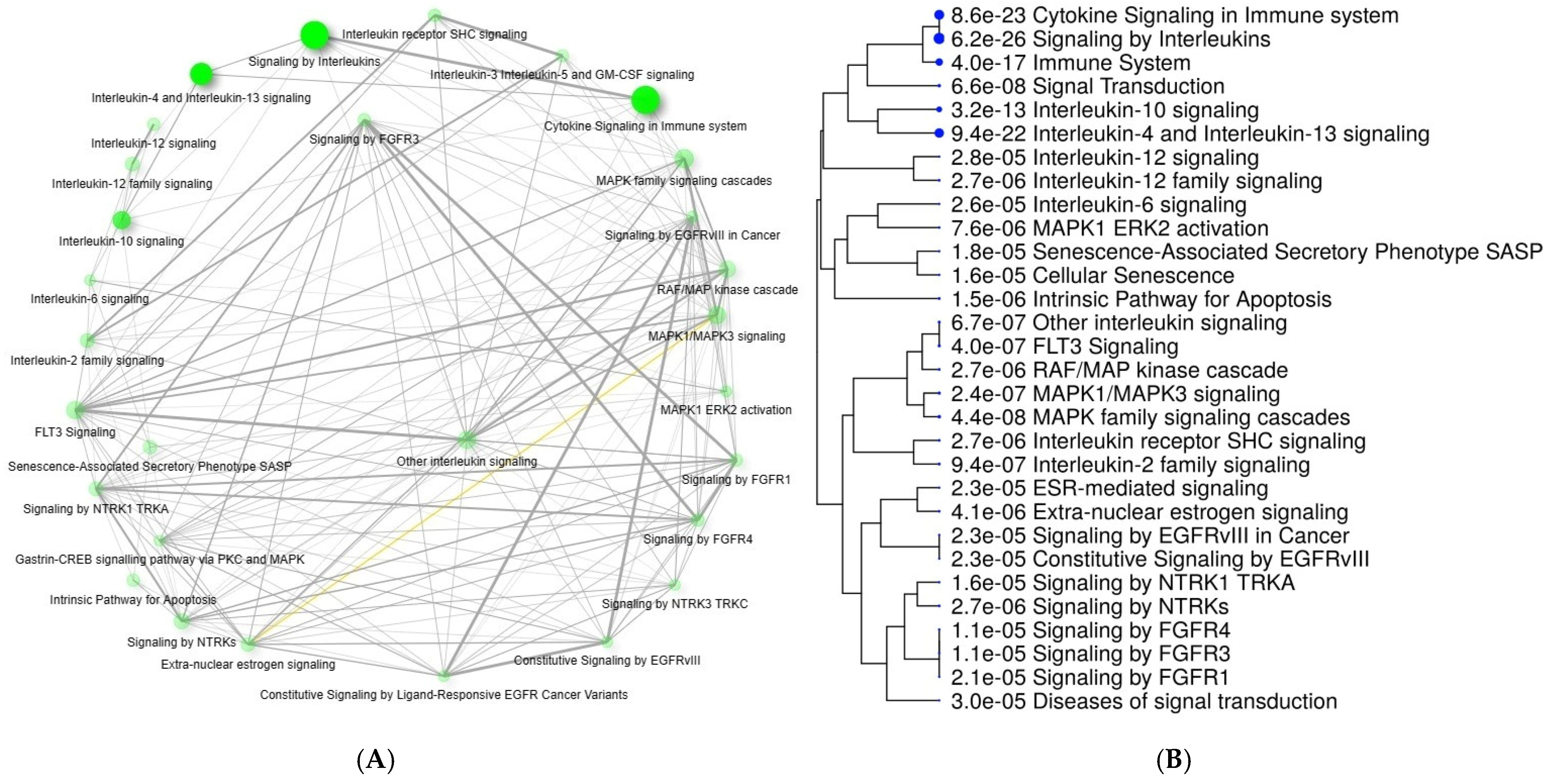
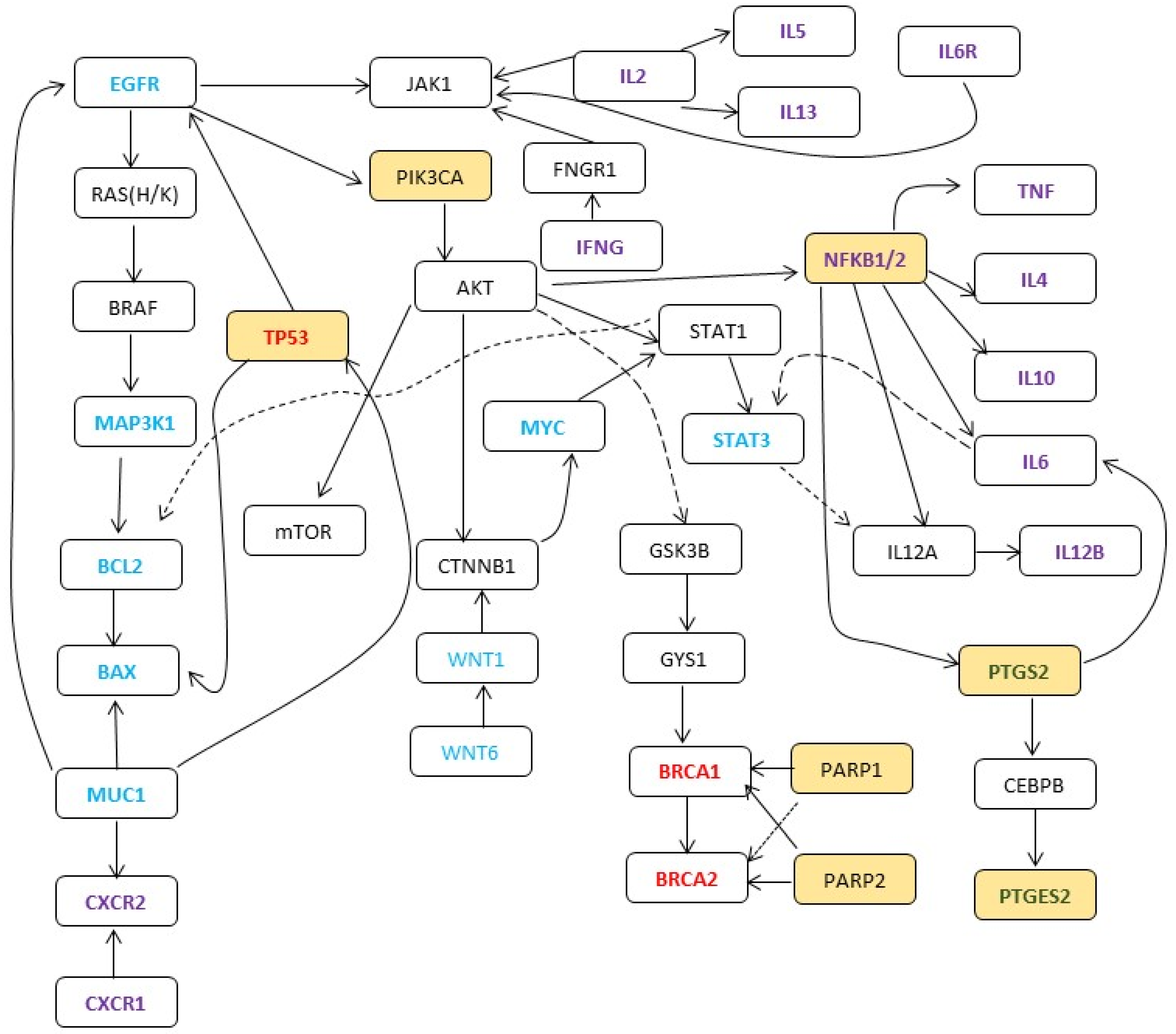
| Group | Gene | Function |
|---|---|---|
| 1 Primary genes | Prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) PTGS2/COX2 | Synthesis of enzyme cyclooxygenase 2 (COX 2) that converts arachidonic acid to prostaglandin endoperoxide H2 |
| PTGES2—prostaglandin E synthase 2 | Encodes membrane-associated prostaglandin E synthase, which catalyzes the conversion of prostaglandin H2 to prostaglandin E2; also activates gamma-interferon-activated transcription element (GATE) | |
| 2 Oncogenes and cell cycle regulators | MYC—MYC proto-oncogene, bHLH transcription factor | Encodes a nuclear phosphoprotein that plays a role in cell cycle progression, apoptosis, and cellular transformation |
| EGFR—epidermal growth factor receptor | Encodes for epidermal growth factor receptor protein that regulates epithelial tissue development and homeostasis | |
| BCL2—B cell lymphoma 2 | Encodes protein that regulates apoptosis. Increases cellular survival by inhibiting proapoptotic proteins | |
| WNT1—WNT family member 1; Proto-oncogene Int-1 homolog | Encodes secreted signaling proteins that increase cell growth and division | |
| KRAS—Kirsten rat sarcoma viral oncogene homolog | Encodes tumor oncogene Kras protein part of RAS/MAPK pathway among other signal transduction pathways | |
| WNT6—Wingless-type MMTV integration site family, member 6 | Highly conserved gene, encodes wnt6 protein that regulates cell growth and differentiation through wnt pathway | |
| BRAF—raf murine sarcoma viral oncogene homolog B | Member of raf kinase family, controls cell proliferation by regulating signal transduction protein kinases | |
| MUC1—Mucin 1, cell surface-associated; polymorphic epithelial mucin (PEM); epithelial membrane antigen (EMA) | Protective function as it binds to foreign pathogens, also regulates cell signaling | |
| PIK3CA—Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha | Encodes oncogenic protein that catalyzes ATP to phosphorylate PtdIns, PtdIns4P, and PtdIns(4,5)P2 | |
| PARP1—Poly[ADP-ribose] polymerase 1 (PARP-1); NAD+ ADP-ribosyltransferase 1 or poly[ADP-ribose] synthase 1 | ADP-ribosylation, repair of single-strand breaks, and with BRCA, double-stranded breaks | |
| PARP2—Poly [ADP-ribose] polymerase 2 | Encodes poly(ADP-ribosyl)transferase-like 2 protein, a catalytic domain capable of catalyzing a poly(ADP-ribosyl)ation reaction | |
| STAT3—Signal transducer and activator of transcription 3 | Transcription activator after being phosphorylated by receptor-associated Janus kinases (JAK) | |
| JAK1—Janus kinase 1 | Tyrosine kinase protein essential for signaling type I and type II cytokines. Promotes cell division | |
| MAPK1—Mitogen-activated protein kinase 1, also known as ERK2 | Member of the MAP kinase family regulating extracellular signal-regulated kinases | |
| STAT—Signal transducer and activator of transcription | Intracellular transcription factors that mediate cellular immunity, proliferation, apoptosis, and differentiation | |
| BAX—bcl-2-like protein 4 | Forms heterodimers with BCL2 and regulates apoptosis. It is a proapoptotic factor that stimulates release of caspases | |
| Group 3—cytokines and Interleukins | IFNG—Interferon gamma | Class II interferon, activator of macrophages and inducer of major histocompatibility complex class II molecule expression |
| IL1B—Interleukin 1Beta, also known as leukocytic pyrogen, leukocytic endogenous mediator, mononuclear cell factor, lymphocyte activating factor | Interleukin 1 family of cytokines, released from macrophages, activated by caspase; it mediates inflammatory response and other cellular activities, including cell proliferation, differentiation, and apoptosis | |
| IL2—Interleukin 2 | Improves tolerance and immunity, primarily via its direct effects on T cells. Improves cell killing by NK cells and activated T cells | |
| IL4—Interleukin 4 | Induces differentiation of naive helper T cells (Th0 cells) to Th2 cells | |
| IL5—Interleukin 5 | Stimulates B cell growth and increases immunoglobulin (IgA) secretion | |
| IL6—Interleukin 6 | Proinflammatory cytokine secreted by macrophage and an anti-inflammatory myokine. It binds to pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) | |
| IL7—Interleukin 7 | Hematopoietic growth factor secreted by stromal cells in the bone marrow and thymus. Participates in proliferation during certain stages of B-cell maturation, T and NK cell survival, development, and homeostasis | |
| IL8—Interleukin 8 | Chemokine produced by macrophages and epithelial cells. Induces chemotaxis and angiogenesis | |
| IL10—Interleukin10; human cytokine synthesis inhibitory factor (CSIF) | Primarily produced by monocytes, participates in immunoregulation and inflammation. Blocks NF-κB, regulates JAK-STAT pathway | |
| IL12B—Interleukin 12 subunit beta; (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor p40, or interleukin-12 subunit p40) | Secreted by activated macrophages, inducer of Th1 cell development. | |
| IL13—Interleukin 13 | Functions similar to IL3. Induces a class of protein-degrading enzymes, known as matrix metalloproteinases (MMPs) | |
| IL17—interleukin 17 | Immune regulatory cytokine, induces and mediates proinflammatory response. Induces expression of keratinocytes | |
| CXCR1—C-X-C motif chemokine receptor 1; Interleukin 8 receptor, alpha or CD181 | G-protein-coupled receptor family that binds to IL8 and increases cellular proliferation | |
| CXCR2—C-X-C motif chemokine receptor 2; Interleukin 8 receptor, beta | Binds with IL8 and transduces the signal through a G-protein-activated second messenger system. Mediates angiogenic effect of IL8 on endothelial cells | |
| NFκB—Nuclear factor kappa-light-chain-enhancer of activated B cells | A primary transcription factor and regulator of signaling response, suppresses TNF, induces cytotoxicity, regulates TRAF1 and TRAF2 | |
| TNF—Tumor necrosis factor (cachexin, or cachectin; also called as tumor necrosis factor alpha or TNF-α) | Adipokine and a cytokine, binds to two receptors, TNFR1 and 2. Regulates NFκB and TRAF1 and 2, activates MAP kinase pathway, cell differentiation, and proliferation | |
| 4—Tumor suppressor genes | TP53—tumor protein 53; transformation-related protein 53 (TRP53) | Tumor suppressor gene, DNA repair, cell cycle arrest in G1/S phase, initiator of apoptosis, senescence response to short telomeres |
| BRCA1—Breast cancer type 1 susceptibility gene | Repair of double-stranded DNA breaks; mismatch repair | |
| BRCA2—breast cancer type 2 susceptibility gene | Encodes BRCA2 protein with functions similar to that of BRCA1. Forms BRCA1-PALB2-BRCA2 complex | |
| MLH1—mutL homolog 1 | DNA mismatch repair of genes | |
| PMS2—PMS1 homolog 2 | DNA mismatch repair of genes | |
| MSH2—mutS homolog 2 | Microsatellite instability-associated gene altered in microsatellite sequences (RER+ phenotype) in HNPCC | |
| MSH6—mutS homolog 6 | DNA mismatch repair; forms MSH recognition complex with MSH2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, M.; Rajput, M.; Singh, P.; Shukla, M.; Zhu, B.; Koshiol, J. Aspirin and Cancer Survival: An Analysis of Molecular Mechanisms. Cancers 2024, 16, 223. https://doi.org/10.3390/cancers16010223
Pandey M, Rajput M, Singh P, Shukla M, Zhu B, Koshiol J. Aspirin and Cancer Survival: An Analysis of Molecular Mechanisms. Cancers. 2024; 16(1):223. https://doi.org/10.3390/cancers16010223
Chicago/Turabian StylePandey, Manoj, Monika Rajput, Pooja Singh, Mridula Shukla, Bin Zhu, and Jill Koshiol. 2024. "Aspirin and Cancer Survival: An Analysis of Molecular Mechanisms" Cancers 16, no. 1: 223. https://doi.org/10.3390/cancers16010223
APA StylePandey, M., Rajput, M., Singh, P., Shukla, M., Zhu, B., & Koshiol, J. (2024). Aspirin and Cancer Survival: An Analysis of Molecular Mechanisms. Cancers, 16(1), 223. https://doi.org/10.3390/cancers16010223








