Identifying Key Regulators of Keratinization in Lung Squamous Cell Cancer Using Integrated TCGA Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
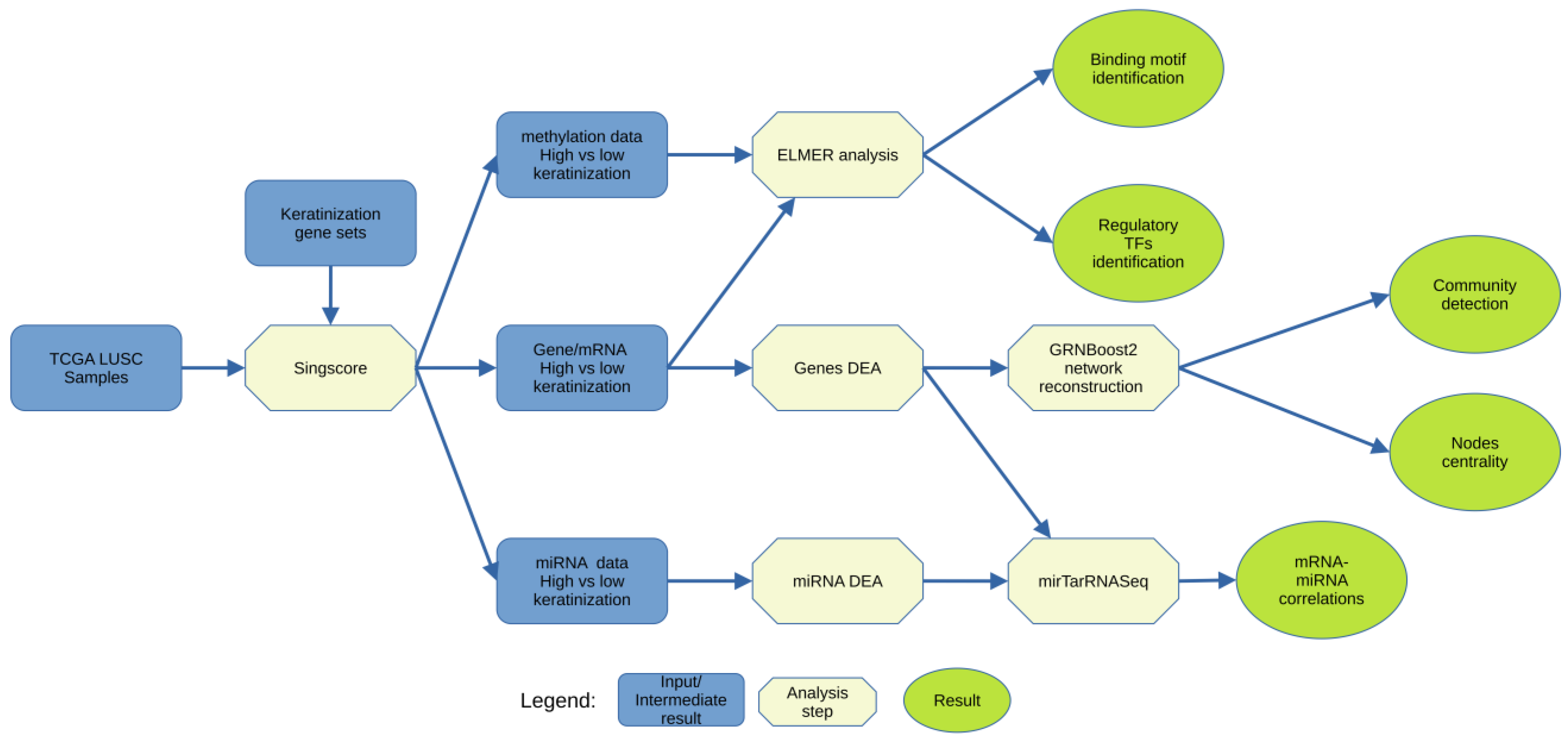
2.1. Overview
2.2. Data Acquisition and Preparation
2.3. Data Single-Sample Scoring and Clustering
2.4. Differential Expression Analysis (DEA) of the Genes
2.5. DEGs Network Reconstruction and Analysis
2.6. Methylation Motif and Regulatory Transcription Factor Identification
2.7. miRNA–mRNA Relationships Analysis
2.8. Source Code
3. Results
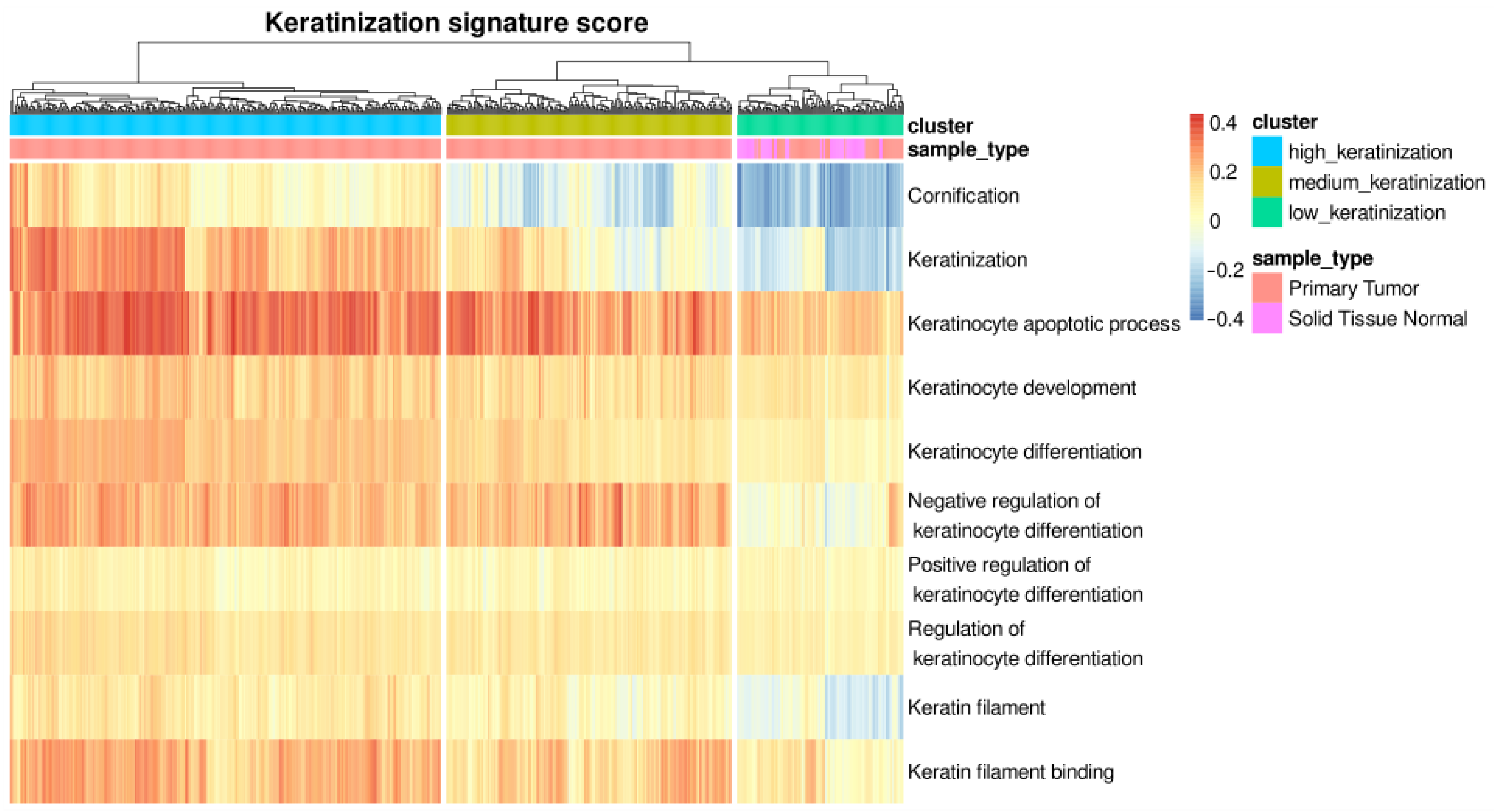
3.1. Hierarchical Clustering Based on Single-Sample Scoring against Keratinization-Related Gene Set Identifies Three LUSC Phenotypes
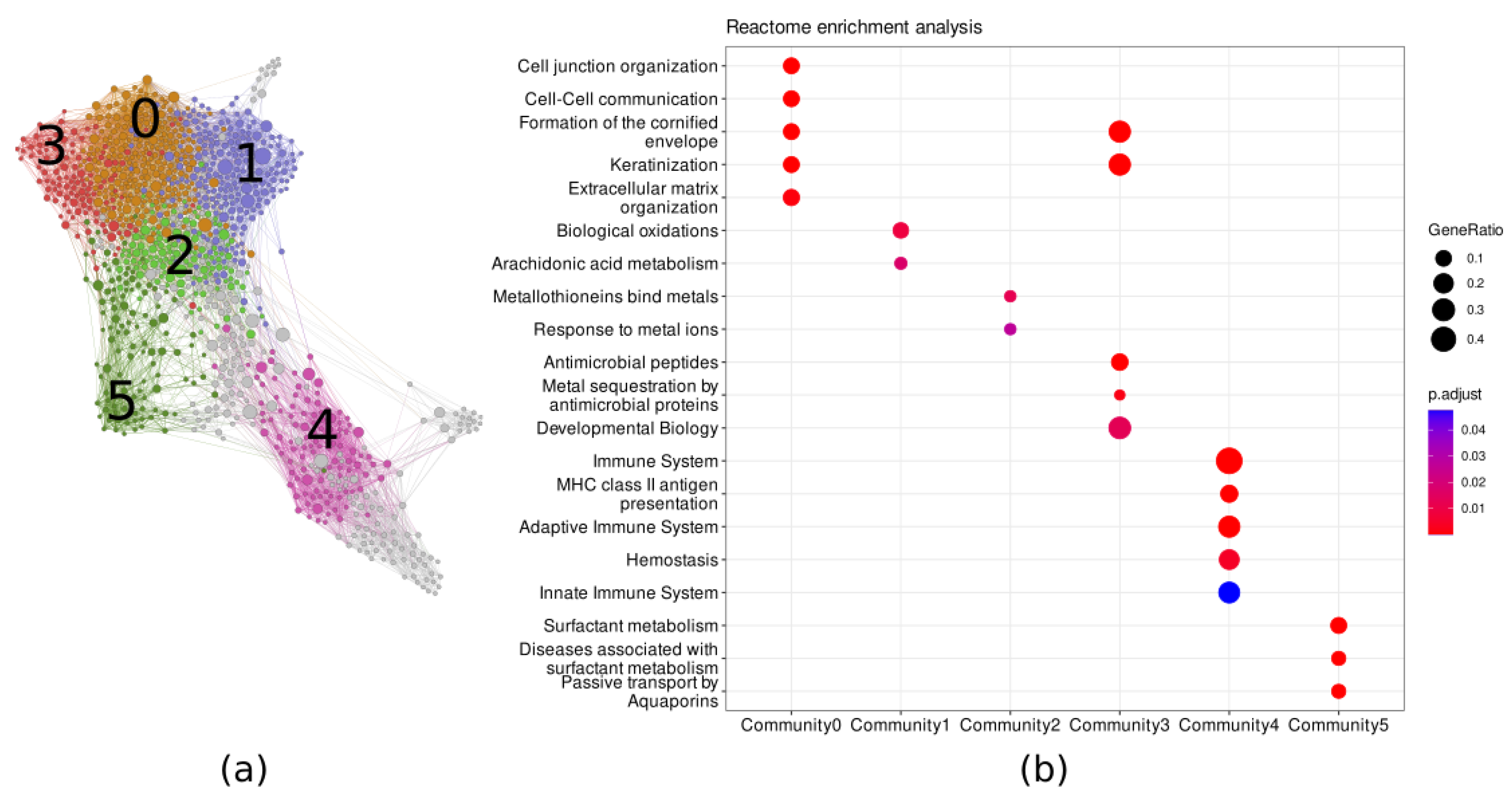
3.2. DEGs Network Reconstruction and Analysis
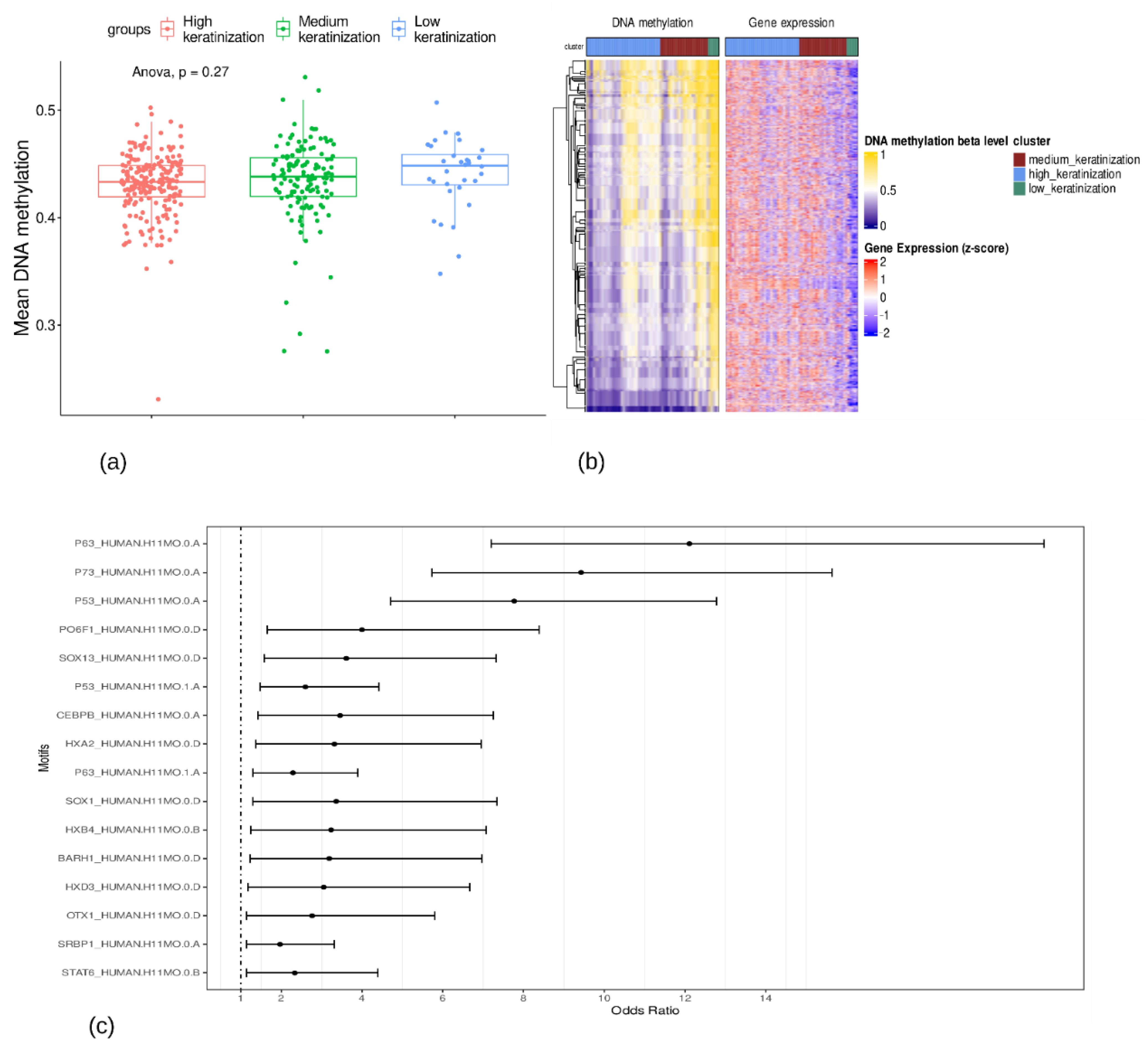
3.3. P63, P73, and P53 Were the Top Three Enriched Motifs for Hypomethylated Probes
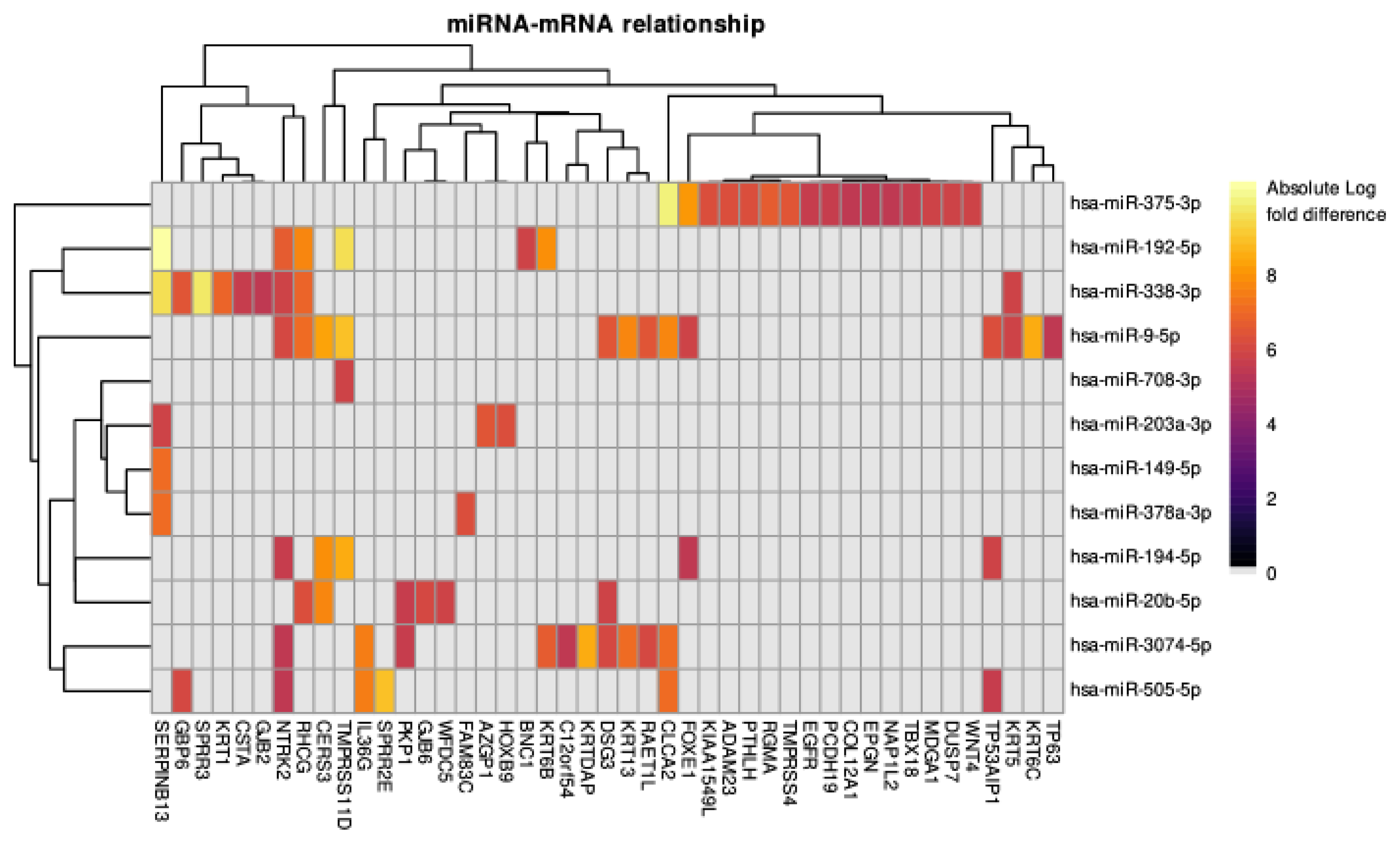
3.4. The miRNA–mRNA Relationships Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lewis, D.R.; Check, D.P.; Caporaso, N.E.; Travis, W.D.; Devesa, S.S. US Lung Cancer Trends by Histologic Type. Cancer 2014, 120, 2883–2892. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Abbas, A.K.; Aster, J.C.; Turner, J.R.; Perkins, J.A.; Robbins, S.L.; Cotran, R.S. (Eds.) Robbins & Cotran Pathologic Basis of Disease, 10th ed.; Elsevier: Philadelphia, PA, USA, 2021; ISBN 978-0-323-53113-9. [Google Scholar]
- Eckhart, L.; Lippens, S.; Tschachler, E.; Declercq, W. Cell Death by Cornification. Biochim. Biophys. Acta 2013, 1833, 3471–3480. [Google Scholar] [CrossRef]
- Deo, P.N.; Deshmukh, R. Pathophysiology of Keratinization. J. Oral Maxillofac. Pathol. 2018, 22, 86. [Google Scholar] [CrossRef] [PubMed]
- Karantza, V. Keratins in Health and Cancer: More than Mere Epithelial Cell Markers. Oncogene 2011, 30, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Werner, S.; Keller, L.; Pantel, K. Epithelial Keratins: Biology and Implications as Diagnostic Markers for Liquid Biopsies. Mol. Asp. Med. 2020, 72, 100817. [Google Scholar] [CrossRef]
- Sharma, P.; Alsharif, S.; Fallatah, A.; Chung, B.M. Intermediate Filaments as Effectors of Cancer Development and Metastasis: A Focus on Keratins, Vimentin, and Nestin. Cells 2019, 8, 497. [Google Scholar] [CrossRef]
- Park, H.J.; Cha, Y.-J.; Kim, S.H.; Kim, A.; Kim, E.Y.; Chang, Y.S. Keratinization of Lung Squamous Cell Carcinoma Is Associated with Poor Clinical Outcome. Tuberc. Respir. Dis. 2017, 80, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Heryanto, Y.D.; Katayama, K.; Imoto, S. Analyzing Integrated Network of Methylation and Gene Expression Profiles in Lung Squamous Cell Carcinoma. Sci. Rep. 2022, 12, 15799. [Google Scholar] [CrossRef]
- Silva, T.C.; Coetzee, S.G.; Gull, N.; Yao, L.; Hazelett, D.J.; Noushmehr, H.; Lin, D.-C.; Berman, B.P. ELMER v.2: An R/Bioconductor Package to Reconstruct Gene Regulatory Networks from DNA Methylation and Transcriptome Profiles. Bioinformatics 2019, 35, 1974–1977. [Google Scholar] [CrossRef]
- Movassagh, M.; Morton, S.U.; Hehnly, C.; Smith, J.; Doan, T.T.; Irizarry, R.; Broach, J.R.; Schiff, S.J.; Bailey, J.A.; Paulson, J.N. MirTarRnaSeq: An R/Bioconductor Statistical Package for MiRNA-MRNA Target Identification and Interaction Analysis. BMC Genom. 2022, 23, 439. [Google Scholar] [CrossRef] [PubMed]
- Foroutan, M.; Bhuva, D.D.; Lyu, R.; Horan, K.; Cursons, J.; Davis, M.J. Single Sample Scoring of Molecular Phenotypes. BMC Bioinform. 2018, 19, 404. [Google Scholar] [CrossRef]
- Narang, V.; Ramli, M.A.; Singhal, A.; Kumar, P.; de Libero, G.; Poidinger, M.; Monterola, C. Automated Identification of Core Regulatory Genes in Human Gene Regulatory Networks. PLOS Comput. Biol. 2015, 11, e1004504. [Google Scholar] [CrossRef]
- Yao, L.; Shen, H.; Laird, P.W.; Farnham, P.J.; Berman, B.P. Inferring Regulatory Element Landscapes and Transcription Factor Networks from Cancer Methylomes. Genome Biol. 2015, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.C.; Colaprico, A.; Olsen, C.; D’Angelo, F.; Bontempi, G.; Ceccarelli, M.; Noushmehr, H. TCGA Workflow: Analyze Cancer Genomics and Epigenomics Data Using Bioconductor Packages. F1000Research 2016, 5, 1542. [Google Scholar] [CrossRef]
- Moerman, T.; Aibar Santos, S.; Bravo González-Blas, C.; Simm, J.; Moreau, Y.; Aerts, J.; Aerts, S. GRNBoost2 and Arboreto: Efficient and Scalable Inference of Gene Regulatory Networks. Bioinformatics 2019, 35, 2159–2161. [Google Scholar] [CrossRef] [PubMed]
- Traag, V.A.; Waltman, L.; van Eck, N.J. From Louvain to Leiden: Guaranteeing Well-Connected Communities. Sci. Rep. 2019, 9, 5233. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Enright, A.J.; John, B.; Gaul, U.; Tuschl, T.; Sander, C.; Marks, D.S. MicroRNA Targets in Drosophila. Genome Biol. 2003, 5, R1. [Google Scholar] [CrossRef]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA Interference Reveals That Oncogenic KRAS-Driven Cancers Require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Yi, M.; Nissley, D.V.; McCormick, F.; Stephens, R.M. SsGSEA Score-Based Ras Dependency Indexes Derived from Gene Expression Data Reveal Potential Ras Addiction Mechanisms with Possible Clinical Implications. Sci. Rep. 2020, 10, 10258. [Google Scholar] [CrossRef]
- He, Y.; Jiang, Z.; Chen, C.; Wang, X. Classification of Triple-Negative Breast Cancers Based on Immunogenomic Profiling. J. Exp. Clin. Cancer Res. 2018, 37, 327. [Google Scholar] [CrossRef]
- Peng, Z.; Chen, Y.; Cao, H.; Zou, H.; Wan, X.; Zeng, W.; Liu, Y.; Hu, J.; Zhang, N.; Xia, Z.; et al. Protein Disulfide Isomerases Are Promising Targets for Predicting the Survival and Tumor Progression in Glioma Patients. Aging 2020, 12, 2347–2372. [Google Scholar] [CrossRef] [PubMed]
- Melino, G.; Memmi, E.M.; Pelicci, P.G.; Bernassola, F. Maintaining Epithelial Stemness with P63. Sci. Signal. 2015, 8, re9. [Google Scholar] [CrossRef] [PubMed]
- Bilodeau, C.; Shojaie, S.; Goltsis, O.; Wang, J.; Luo, D.; Ackerley, C.; Rogers, I.M.; Cox, B.; Post, M. TP63 Basal Cells Are Indispensable during Endoderm Differentiation into Proximal Airway Cells on Acellular Lung Scaffolds. NPJ Regen. Med. 2021, 6, 12. [Google Scholar] [CrossRef]
- Blanpain, C.; Fuchs, E. P63: Revving up Epithelial Stem-Cell Potential. Nat. Cell Biol. 2007, 9, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Ma, Q.; Peng, S.; Adelmant, G.; Swain, D.; Song, W.; Fox, C.; Francis, J.M.; Pedamallu, C.S.; DeLuca, D.S.; et al. SOX2 and P63 Colocalize at Genetic Loci in Squamous Cell Carcinomas. J. Clin. Investig. 2014, 124, 1636–1645. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, R.P.; Lessard, J.C.; Coulombe, P.A. Keratin Intermediate Filament Proteins—Novel Regulators of Inflammation and Immunity in Skin. J. Cell Sci. 2012, 125, 5257–5258. [Google Scholar] [CrossRef] [PubMed]
- DePianto, D.; Kerns, M.L.; Dlugosz, A.A.; Coulombe, P.A. Keratin 17 Promotes Epithelial Proliferation and Tumor Growth by Polarizing the Immune Response in Skin. Nat. Genet. 2010, 42, 910–914. [Google Scholar] [CrossRef]
- Lessard, J.C.; Piña-Paz, S.; Rotty, J.D.; Hickerson, R.P.; Kaspar, R.L.; Balmain, A.; Coulombe, P.A. Keratin 16 Regulates Innate Immunity in Response to Epidermal Barrier Breach. Proc. Natl. Acad. Sci. USA 2013, 110, 19537–19542. [Google Scholar] [CrossRef] [PubMed]
- Sequeira, I.; Neves, J.F.; Carrero, D.; Peng, Q.; Palasz, N.; Liakath-Ali, K.; Lord, G.M.; Morgan, P.R.; Lombardi, G.; Watt, F.M. Immunomodulatory Role of Keratin 76 in Oral and Gastric Cancer. Nat. Commun. 2018, 9, 3437. [Google Scholar] [CrossRef]
- Nathan, N.; Taytard, J.; Duquesnoy, P.; Thouvenin, G.; Corvol, H.; Amselem, S.; Clement, A. Surfactant Protein A: A Key Player in Lung Homeostasis. Int. J. Biochem. Cell Biol. 2016, 81, 151–155. [Google Scholar] [CrossRef] [PubMed]
- King, S.D.; Chen, S.-Y. Recent Progress on Surfactant Protein A: Cellular Function in Lung and Kidney Disease Development. Am. J. Physiol.-Cell Physiol. 2020, 319, C316–C320. [Google Scholar] [CrossRef] [PubMed]
- Dötsch, V.; Bernassola, F.; Coutandin, D.; Candi, E.; Melino, G. P63 and P73, the Ancestors of P53. Cold Spring Harb. Perspect. Biol. 2010, 2, a004887. [Google Scholar] [CrossRef] [PubMed]
- Mistry, D.S.; Chen, Y.; Wang, Y.; Sen, G.L. SNAI2 Controls the Undifferentiated State of Human Epidermal Progenitor Cells. Stem Cells 2014, 32, 3209–3218. [Google Scholar] [CrossRef]
- Chen, X.; Lloyd, S.M.; Kweon, J.; Gamalong, G.M.; Bao, X. Epidermal Progenitors Suppress GRHL3-Mediated Differentiation through Intronic Polyadenylation Promoted by CPSF-HNRNPA3 Collaboration. Nat. Commun. 2021, 12, 448. [Google Scholar] [CrossRef]
- Hegde, G.V.; de la Cruz, C.; Giltnane, J.M.; Crocker, L.; Venkatanarayan, A.; Schaefer, G.; Dunlap, D.; Hoeck, J.D.; Piskol, R.; Gnad, F.; et al. NRG1 Is a Critical Regulator of Differentiation in TP63-Driven Squamous Cell Carcinoma. eLife 2019, 8, e46551. [Google Scholar] [CrossRef] [PubMed]
- Hazawa, M.; Lin, D.-C.; Handral, H.; Xu, L.; Chen, Y.; Jiang, Y.-Y.; Mayakonda, A.; Ding, L.-W.; Meng, X.; Sharma, A.; et al. ZNF750 Is a Lineage-Specific Tumour Suppressor in Squamous Cell Carcinoma. Oncogene 2017, 36, 2243–2254. [Google Scholar] [CrossRef]
- Eichberger, T.; Regl, G.; Ikram, M.S.; Neill, G.W.; Philpott, M.P.; Aberger, F.; Frischauf, A.-M. FOXE1, A New Transcriptional Target of GLI2 Is Expressed in Human Epidermis and Basal Cell Carcinoma. J. Investig. Dermatol. 2004, 122, 1180–1187. [Google Scholar] [CrossRef]
- Suzuki, H.I.; Katsura, A.; Matsuyama, H.; Miyazono, K. MicroRNA Regulons in Tumor Microenvironment. Oncogene 2015, 34, 3085–3094. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- de Koning, P.J.A.; Bovenschen, N.; Leusink, F.K.J.; Broekhuizen, R.; Quadir, R.; van Gemert, J.T.M.; Hordijk, G.J.; Chang, W.-S.W.; van der Tweel, I.; Tilanus, M.G.J.; et al. Downregulation of SERPINB13 Expression in Head and Neck Squamous Cell Carcinomas Associates with Poor Clinical Outcome. Int. J. Cancer 2009, 125, 1542–1550. [Google Scholar] [CrossRef]
- Oomizu, S.; Sahuc, F.; Asahina, K.; Inamatsu, M.; Matsuzaki, T.; Sasaki, M.; Obara, M.; Yoshizato, K. Kdap, a Novel Gene Associated with the Stratification of the Epithelium. Gene 2000, 256, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Crimi, S.; Falzone, L.; Gattuso, G.; Grillo, C.M.; Candido, S.; Bianchi, A.; Libra, M. Droplet Digital PCR Analysis of Liquid Biopsy Samples Unveils the Diagnostic Role of Hsa-MiR-133a-3p and Hsa-MiR-375-3p in Oral Cancer. Biology 2020, 9, 379. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, P.; Zhang, P.; Zhang, X.; Du, H.; Liu, Q.; Huang, B.; Qian, C.; Zhang, S.; Zhu, W.; et al. An Integrative Bioinformatics Analysis Identified MiR-375 as a Candidate Key Regulator of Malignant Breast Cancer. J. Appl. Genet. 2019, 60, 335–346. [Google Scholar] [CrossRef]
- Hudcova, K.; Raudenska, M.; Gumulec, J.; Binkova, H.; Horakova, Z.; Kostrica, R.; Babula, P.; Adam, V.; Masarik, M. Expression Profiles of MiR-29c, MiR-200b and MiR-375 in Tumour and Tumour-Adjacent Tissues of Head and Neck Cancers. Tumor Biol. 2016, 37, 12627–12633. [Google Scholar] [CrossRef]
- Li, Q.; Huyan, T.; Cai, S.; Huang, Q.; Zhang, M.; Peng, H.; Zhang, Y.; Liu, N.; Zhang, W. The Role of Exosomal MiR-375-3p: A Potential Suppressor in Bladder Cancer via the Wnt/β-Catenin Pathway. FASEB J. 2020, 34, 12177–12196. [Google Scholar] [CrossRef]
- Zhan, S.; Ni, B. Hsa-MiR-9-5p Down-Regulates HK2 and Confers Radiosensitivity to Nasopharyngeal Carcinoma. Technol. Cancer Res. Treat. 2021, 20. [Google Scholar] [CrossRef]
- Wei, Y.-Q.; Jiao, X.-L.; Zhang, S.-Y.; Xu, Y.; Li, S.; Kong, B.-H. MiR-9-5p Could Promote Angiogenesis and Radiosensitivity in Cervical Cancer by Targeting SOCS5. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7314–7326. [Google Scholar] [CrossRef]
- Kania, E.E.; Carvajal-Moreno, J.; Hernandez, V.A.; English, A.; Papa, J.L.; Shkolnikov, N.; Ozer, H.G.; Yilmaz, A.S.; Yalowich, J.C.; Elton, T.S. Hsa-MiR-9-3p and Hsa-MiR-9-5p as Post-Transcriptional Modulators of DNA Topoisomerase IIα in Human Leukemia K562 Cells with Acquired Resistance to Etoposide. Mol. Pharmacol. 2020, 97, 159–170. [Google Scholar] [CrossRef] [PubMed]
| GO Subset | Name | GO Description |
|---|---|---|
| Biological Process | KERATINIZATION | The process in which the cytoplasm of the outermost cells of the vertebrate epidermis is replaced by keratin. |
| Biological Process | CORNIFICATION | A unique type of programmed cell death that leads to the formation of keratin layer. |
| Biological Process | KERATINOCYTE APOPTOTIC PROCESS | Any apoptotic process in a keratinocyte. |
| Biological Process | KERATINOCYTE DEVELOPMENT | The process whose specific outcome is the progression of a keratinocyte over time, from its formation to the mature structure. |
| Biological Process | KERATINOCYTE DIFFERENTIATION | The process in which a relatively unspecialized cell acquires specialized features of a keratinocyte. |
| Biological Process | NEGATIVE_REGULATION OF KERATINOCYTE DIFFERENTIATION | Any process that stops, prevents, or reduces the frequency, rate, or extent of keratinocyte differentiation. |
| Biological Process | POSITIVE REGULATION OF KERATINOCYTE DIFFERENTIATION | Any process that activates or increases the frequency, rate, or extent of keratinocyte differentiation. |
| Biological Process | REGULATION OF KERATINOCYTE DIFFERENTIATION | Any process that modulates the frequency, rate, or extent of keratinocyte differentiation. |
| Cellular Component | KERATIN FILAMENT | A filament composed of acidic and basic keratins (types I and II), typically expressed in epithelial cells. |
| Molecular Function | KERATIN FILAMENT BINDING | Binding to a keratin filament. |
| Top 20 genes ranked by betweenness centrality | SOX2, FOXE1, SYNE1, SPIB, JMJD7-PLA2G4B, FER1L4, DLX6, ARNT2, GNG11, VAMP5, ABCC5, ICAM1, ACSL5, TREM1, PKP1, TSPAN18, COL7A1, MICAL1, ANKRD36BP2, CXCL1 |
| Top 20 genes ranked by nodes out-degree index | TP63, CD53, NCKAP1L, KRT6A, PKP1, SOX2, PTPRC, NTRK2, CD3E, GBP6, CD2, HLA-DMB, GIMAP4, SASH3, GJB5, FAT2, CLCA2, ITK, DOCK2, ABCC5 |
| Top 5% TFs related to hypomethylated probes | SNAI2, GRHL3, TP63, ZNF750, FOXE1, IRF6, BNC1, ZNF385A, PITX1, HES2, KLF5, SOX15, FOXN1, HOMEZ, OVOL1, NFE2L2, ZBTB7C, GRHL1, RARG, ZNF488, SOX2, ARNTL2, KLF3, DLX5, IRX4, SOX21, YBX3, BCL11B, ZNF365, RAG1, PPARA, TCF20, TBX18, MAF, EEA1, TSHZ2, HOXA1, FLYWCH1, HOXD11, TEF, ZIC5, DMRT2, NR1D1, ZBTB7A, FEZF1, HOXD10, FOXD1, MXD1, ZNF703, ELF4, KLF4, FOXQ1, TP73, HES1, GLI2, PRRX2, AEBP2, SHOX2, TFAP2C, FOXF2 |
| Significant regulatory miRNA | hsa-miR-20b-5p, hsa-miR-3074-5p, hsa-miR-375-3p, hsa-miR-194-5p, hsa-miR-505-5p, hsa-miR-9-5p, hsa-miR-338-3p, hsa-miR-378a-3p, hsa-miR-192-5p, hsa-miR-708-3p, hsa-miR-203a-3p, hsa-miR-149-5p |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heryanto, Y.D.; Imoto, S. Identifying Key Regulators of Keratinization in Lung Squamous Cell Cancer Using Integrated TCGA Analysis. Cancers 2023, 15, 2066. https://doi.org/10.3390/cancers15072066
Heryanto YD, Imoto S. Identifying Key Regulators of Keratinization in Lung Squamous Cell Cancer Using Integrated TCGA Analysis. Cancers. 2023; 15(7):2066. https://doi.org/10.3390/cancers15072066
Chicago/Turabian StyleHeryanto, Yusri Dwi, and Seiya Imoto. 2023. "Identifying Key Regulators of Keratinization in Lung Squamous Cell Cancer Using Integrated TCGA Analysis" Cancers 15, no. 7: 2066. https://doi.org/10.3390/cancers15072066
APA StyleHeryanto, Y. D., & Imoto, S. (2023). Identifying Key Regulators of Keratinization in Lung Squamous Cell Cancer Using Integrated TCGA Analysis. Cancers, 15(7), 2066. https://doi.org/10.3390/cancers15072066





