Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma
Simple Summary
Abstract
1. Introduction
2. Need for Markers of Malignant SCC
3. Structure and Function of CSPG4
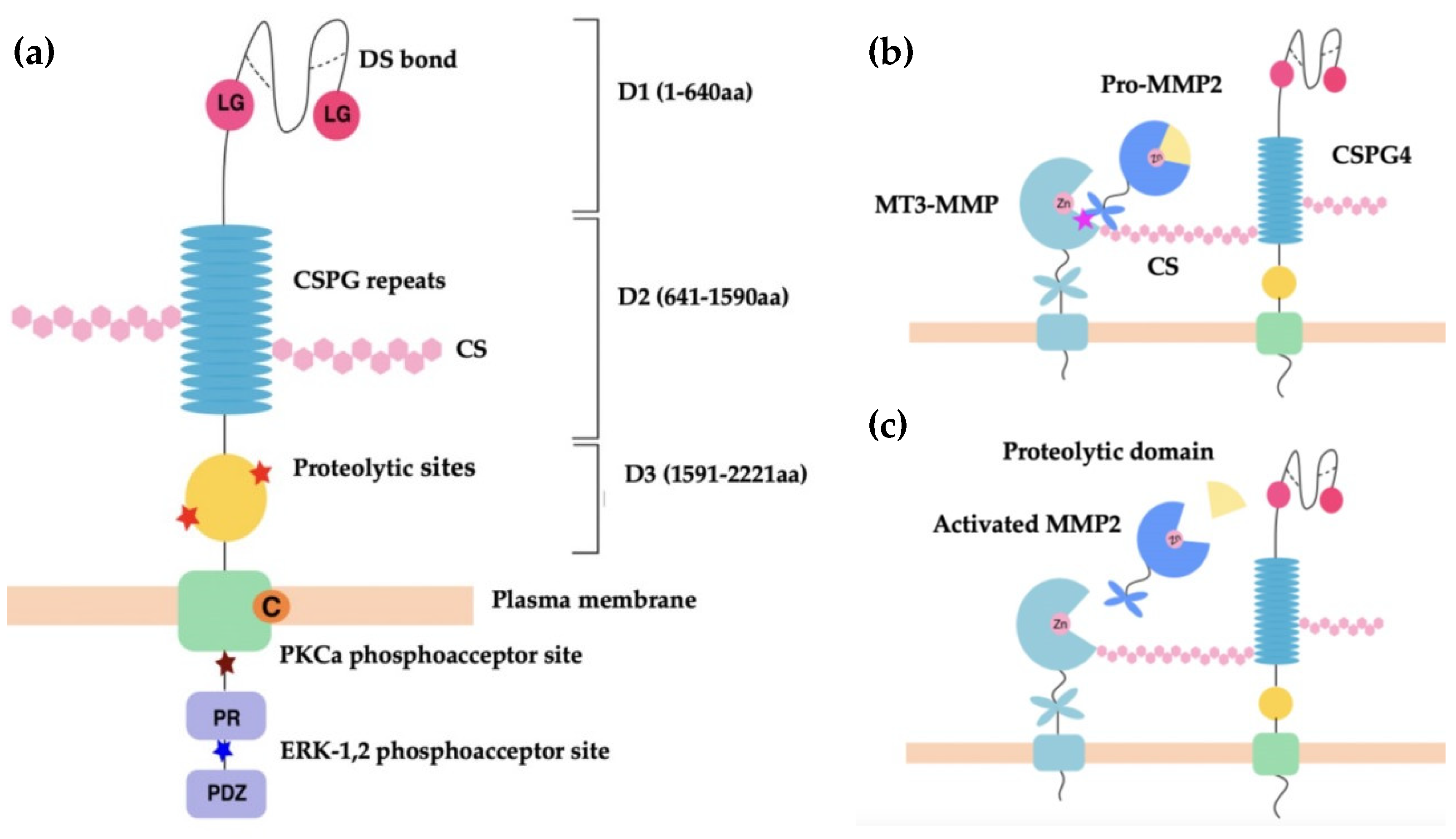
3.1. CSPG4/NG2 Structure
3.2. CSPG4 Function in Malignancy
3.2.1. Regulation of Extracellular Proteases
3.2.2. CSPG4 as a Co-Receptor
3.2.3. CSPG4 & Integrin Signalling
3.2.4. CSPG4 as a Receptor for Structural Components of the ECM
3.2.5. c-Met, CSPG4 and the EMT
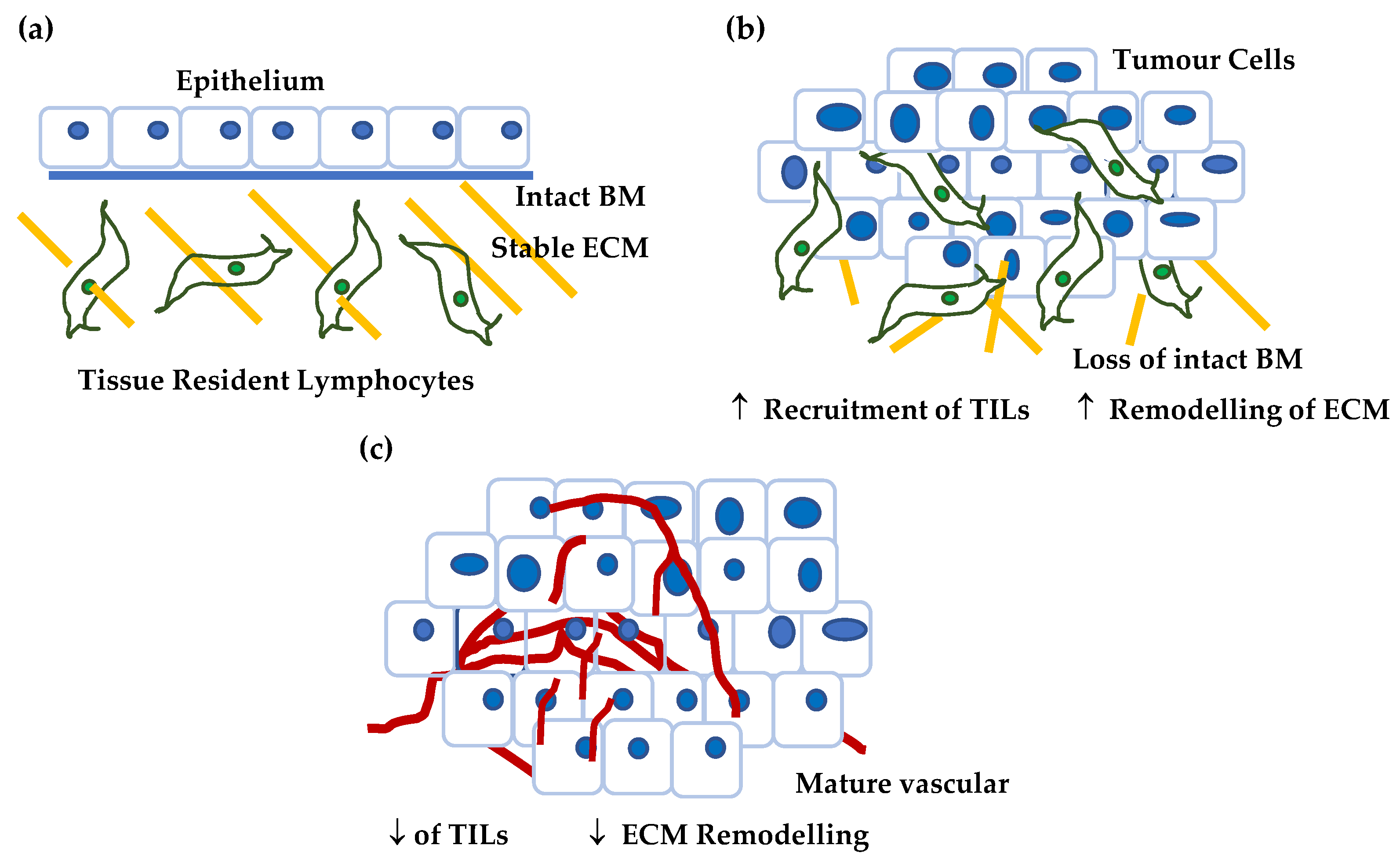
3.2.6. CSPG4 & Angiogenesis
4. CSPG4 Expression in Disease and Development
4.1. An Overview of Methodological Approach
4.2. Tissue Distribution of CSPG4
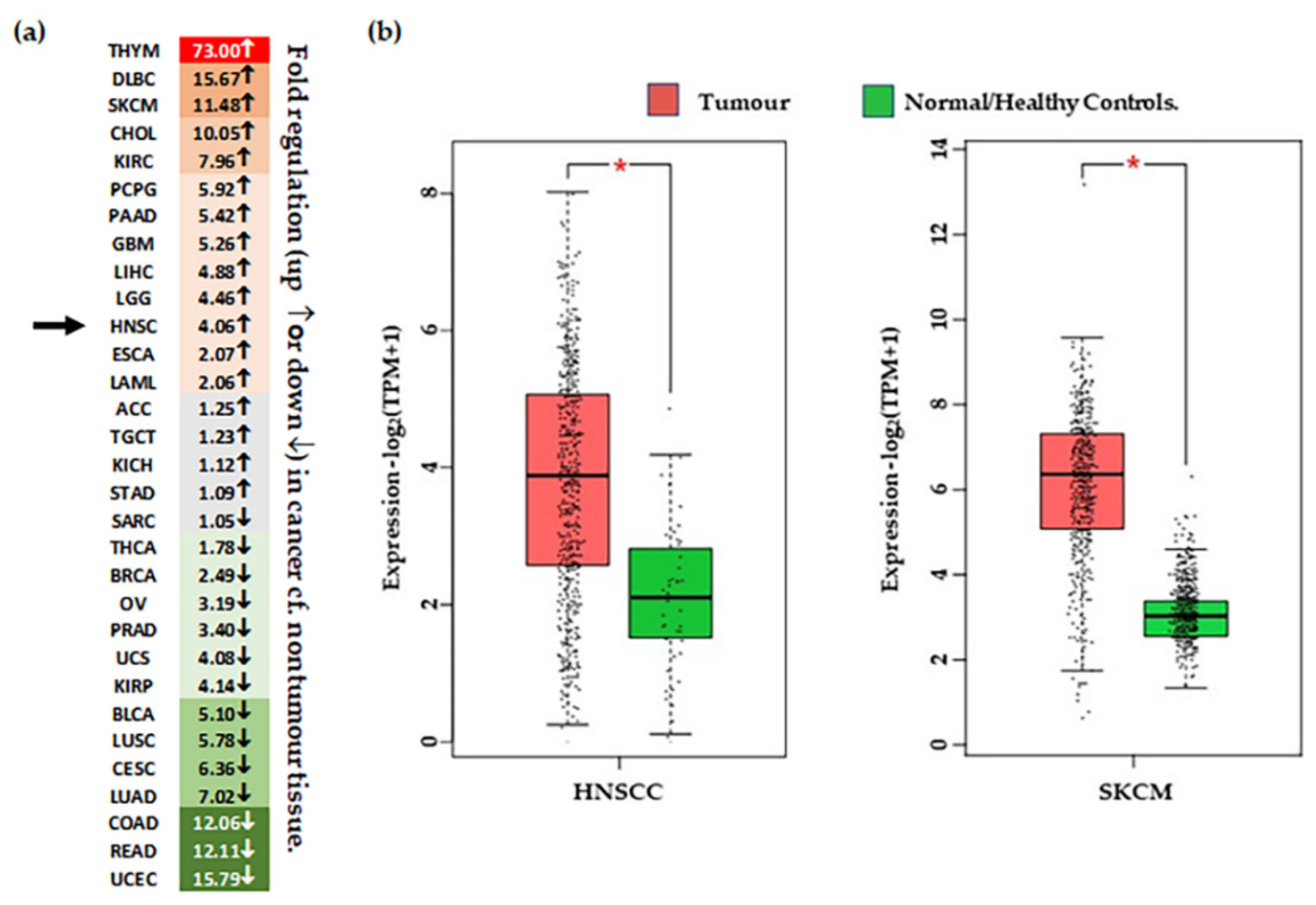
4.3. CSPG4 Expression in Malignancy
4.4. CSPG4 Expression, Patient Survival & Stage
5. Genes Co-Regulated with CSPG4 in SCCs
5.1. CSPG4 Co-Expressed Genes in HNSCC and cSCC
5.2. Comparison with Results of Single Cell Analysis
5.3. CSPG4 Co-Expressed Genes in RDEB SCCs
5.4. Skin Stem Cells, the p-EMT/TSK Programs and CSPG4
6. CSPG4 as a Diagnostic Marker in Cancer
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, S.; Nagpal, M.; Singh, P.; Chauhan, P.; Zaidi, M.A. Tumor Markers: A Diagnostic Tool. Natl. J. Maxillofac. Surg. 2016, 7, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Virji, M.A.; Mercer, D.W.; Herberman, R.B. Tumor Markers in Cancer Diagnosis and Prognosis. CA Cancer J. Clin. 1988, 38, 104–126. [Google Scholar] [CrossRef] [PubMed]
- Kabel, A.M. Tumor Markers of Breast Cancer: New Prospectives. J. Oncol. Sci. 2017, 3, 5–11. [Google Scholar] [CrossRef]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of Immunohistochemistry. J. Pharm. BioAllied Sci. 2012, 4, S307–S309. [Google Scholar] [CrossRef] [PubMed]
- Ignatov, T.; Gorbunow, F.; Eggemann, H.; Ortmann, O.; Ignatov, A. Loss of HER2 after HER2-Targeted Treatment. Breast Cancer Res. Treat. 2019, 175, 401–408. [Google Scholar] [CrossRef]
- Sun, J.Y.; Zhang, D.; Wu, S.; Xu, M.; Zhou, X.; Lu, X.J.; Ji, J. Resistance to PD-1/PD-L1 Blockade Cancer Immunotherapy: Mechanisms, Predictive Factors, and Future Perspectives. Biomark. Res. 2020, 8, 35. [Google Scholar] [CrossRef]
- Hamid, O.; Bruno, R.; Fasso, M.; O’Hear, C.; Wu, B. Safety, Clinical Activity, and Biological Correlates of Response in Patients with Metastatic Melanoma: Results from a Phase I Trial of Atezolizumab-Response. Clin. Cancer Res. 2020, 26, 2436. [Google Scholar] [CrossRef]
- Konings, H.; Stappers, S.; Geens, M.; De Winter, B.Y.; Lamote, K.; Van Meerbeeck, J.P.; Specenier, P.; Vanderveken, O.M.; Ledeganck, K.J. A Literature Review of the Potential Diagnostic Biomarkers of Head and Neck Neoplasms. Front. Oncol. 2020, 10, 1020. [Google Scholar] [CrossRef]
- Brunner, M.; Ng, B.C.; Veness, M.J.; Clark, J.R. Assessment of the New Nodal Classification for Cutaneous Squamous Cell Carcinoma and Its Effect on Patient Stratification. Head Neck 2014, 37, 336–339. [Google Scholar] [CrossRef]
- Gore, S.M.; Shaw, D.; Martin, R.C.W.; Kelder, W.; Roth, K.; Uren, R.; Gao, K.; Davies, S.; Ashford, B.G.; Ngo, Q.; et al. Prospective Study of Sentinel Node Biopsy for High-Risk Cutaneous Squamous Cell Carcinoma of the Head and Neck. Head Neck 2015, 38, E884–E889. [Google Scholar] [CrossRef]
- Bois, F.; Noirot, C.; Dietemann, S.; Mainta, I.C.; Zilli, T.; Garibotto, V.; A Walter, M. [68Ga]Ga-PSMA-11 in Prostate Cancer: A Comprehensive Review. Am. J. Nucl. Med. Mol. Imaging 2020, 10, 349–374. [Google Scholar]
- Venables, Z.; Nijsten, T.; Wong, K.; Autier, P.; Broggio, J.; Deas, A.; Harwood, C.; Hollestein, L.; Langan, S.; Morgan, E.; et al. Epidemiology of Basal and Cutaneous Squamous Cell Carcinoma in the UK. 2013–15: A Cohort Study. Br. J. Dermatol. 2019, 181, 474–482. [Google Scholar] [CrossRef]
- Que, S.K.T.; Zwald, F.O.; Schmults, C.D. Cutaneous Squamous Cell Carcinoma: Incidence, Risk Factors, Diagnosis, and Staging. J. Am. Acad. Dermatol. 2018, 78, 237–247. [Google Scholar] [CrossRef]
- Green, A.; Olsen, C. Cutaneous Squamous Cell Carcinoma: An Epidemiological Review. Br. J. Dermatol. 2017, 177, 373–381. [Google Scholar] [CrossRef]
- Johnson, D.E.; Burtness, B.; Leemans, C.R.; Lui, V.W.Y.; Bauman, J.E.; Grandis, J.R. Head and Neck Squamous Cell Carcinoma. Nat. Rev. Dis. Prim. 2020, 6, 92. [Google Scholar] [CrossRef]
- Ilic, D.; Djulbegovic, M.; Jung, J.H.; Hwang, E.C.; Zhou, Q.; Cleves, A.; Agoritsas, T.; Dahm, P. Prostate Cancer Screening with Prostate-Specific Antigen (PSA) Test: A Systematic Review and Meta-Analysis. BMJ 2018, 362, k3519. [Google Scholar] [CrossRef]
- Paintal, A. Pathology of Head and Neck Neoplasms. 2022, Uptodate. Shah, S., Ed. Available online: www.uptodate.com/contents/pathology-of-head-and-neck-neoplasms (accessed on 9 September 2022).
- Sanderson, R.J.; Ironside, J.A.D. Squamous Cell Carcinomas of the Head and Neck. BMJ 2002, 325, 822. [Google Scholar] [CrossRef]
- Economopoulou, P.; De Bree, R.; Kotsantis, I.; Psyrri, A. Diagnostic Tumor Markers in Head and Neck Squamous Cell Carcinoma (HNSCC) in the Clinical Setting. Front. Oncol. 2019, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Rodust, P.; Stockfleth, E.; Ulrich, C.; Leverkus, M.; Eberle, J. UV-Induced Squamous Cell Carcinoma—A Role for Antiapoptotic Signalling Pathways. Br. J. Dermatol. 2009, 161, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hisamatsu, K.; Suzui, N.; Hara, A.; Tomita, H.; Miyazaki, T. A Review of HPV-Related Head and Neck Cancer. J. Clin. Med. 2018, 7, 241. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Asgari, M. Cutaneous Squamous Cell Carcinoma: Epidemiology and Risk Factors. 2021, Uptodate. Corona, R., Ed. Available online: www.uptodate.com/contents/cutaneous-squamous-cell-carcinoma-epidemiology-and-risk-factors (accessed on 9 September 2022).
- Rose, R.; Boon, A.; Forman, D.; Merchant, W.; Bishop, R.; Newton-Bishop, J. An Exploration of Reported Mortality from Cutaneous Squamous Cell Carcinoma Using Death Certification and Cancer Registry Data. Br. J. Dermatol. 2013, 169, 682–686. [Google Scholar] [CrossRef]
- Vigneswaran, N.; Williams, M.D. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac. Surg. Clin. N. Am. 2014, 26, 123–141. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Eigentler, T.K.; Leiter, U.; Häfner, H.-M.; Garbe, C.; Röcken, M.; Breuninger, H. Survival of Patients with Cutaneous Squamous Cell Carcinoma: Results of a Prospective Cohort Study. J. Investig. Dermatol. 2017, 137, 2309–2315. [Google Scholar] [CrossRef]
- Hedberg, M.L.; Berry, C.T.; Moshiri, A.S.; Xiang, Y.; Yeh, C.J.; Attilasoy, C.; Capell, B.C.; Seykora, J.T. Molecular Mechanisms of Cutaneous Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 3478. [Google Scholar] [CrossRef]
- Schmults, C.D.; Karia, P.S.; Carter, J.B.; Han, J.; Qureshi, A.A. Factors Predictive of Recurrence and Death from Cutaneous Squamous Cell Carcinoma: A 10-Year, Single-Institution Cohort Study. JAMA Dermatol. 2013, 149, 541–547. [Google Scholar] [CrossRef]
- Rees, J.R.; Zens, M.S.; Celaya, M.O.; Riddle, B.L.; Karagas, M.R.; Peacock, J.L. Survival after Squamous Cell and Basal Cell Carcinoma of the Skin: A Retrospective Cohort Analysis. Int. J. Cancer 2015, 137, 878–888. [Google Scholar] [CrossRef]
- Pandeya, N.; Olsen, C.M.; Whiteman, D.C. The Incidence and Multiplicity Rates of Keratinocyte Cancers in Australia. Med. J. Aust. 2017, 207, 339–343. [Google Scholar] [CrossRef]
- Condorelli, A.G.; Dellambra, E.; Logli, E.; Zambruno, G.; Castiglia, D. Epidermolysis Bullosa-Associated Squamous Cell Carcinoma: From Pathogenesis to Therapeutic Perspectives. Int. J. Mol. Sci. 2019, 20, 5707. [Google Scholar] [CrossRef]
- Has, C.; Bauer, J.W.; Bodemer, C.; Bolling, M.C.; Bruckner-Tuderman, L.; Diem, A.; Fine, J.-D.; Heagerty, A.; Hovnanian, A.; Marinkovich, M.P.; et al. Consensus Reclassification of Inherited Epidermolysis Bullosa and Other Disorders with Skin Fragility. Br. J. Dermatol. 2020, 183, 614–627. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare. Skin Cancer in Australia; Australian Government: Canberra, Australia, 2016.
- Fine, J.-D.; Johnson, L.B.; Weiner, M.; Li, K.-P.; Suchindran, C. Epidermolysis Bullosa and the Risk of Life-Threatening Cancers: The National EB Registry Experience, 1986–2006. J. Am. Acad. Dermatol. 2009, 60, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Tam, S.; Yao, C.M.K.L.; Amit, M.; Gajera, M.; Luo, X.; Treistman, R.; Khanna, A.; Aashiq, M.; Nagarajan, P.; Bell, D.; et al. Association of Immunosuppression with Outcomes of Patients with Cutaneous Squamous Cell Carcinoma of the Head and Neck. JAMA Otolaryngol. Neck Surg. 2020, 146, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Lekalakala, P.T.; Khammissa, R.A.G.; Kramer, B.; Ayo-Yusuf, O.A.; Lemmer, J.; Feller, L. Oculocutaneous Albinism and Squamous Cell Carcinoma of the Skin of the Head and Neck in Sub-Saharan Africa. J. Skin Cancer 2015, 2015, 167847. [Google Scholar] [CrossRef] [PubMed]
- Mellerio, J.; Robertson, S.; Bernardis, C.; Diem, A.; Fine, J.; George, R.; Goldberg, D.; Halmos, G.; Harries, M.; Jonkman, M.; et al. Management of Cutaneous Squamous Cell Carcinoma in Patients with Epidermolysis Bullosa: Best Clinical Practice Guidelines. Br. J. Dermatol. 2015, 174, 56–67. [Google Scholar] [CrossRef]
- Australian Institute of Health and Welfare. Head and Neck Cancers in Australia; Australian Government: Canberra, Australia, 2014.
- Annertz, K.; Rosenquist, K.; Andersson, G.; Jacobsson, H.; Hansson, B.G.; Wennerberg, J. High-Risk HPV and Survival in Patients with Oral and Oropharyngeal Squamous Cell—Follow up of a Population-Based Study. Acta Oto-Laryngol. 2014, 134, 843–851. [Google Scholar] [CrossRef]
- Beckham, T.; Leeman, J.E.; Xie, P.; Li, X.; Goldman, D.A.; Zhang, Z.; Sherman, E.; McBride, S.; Riaz, N.; Lee, N.; et al. Long-Term Survival in Patients with Metastatic Head and Neck Squamous Cell Carcinoma Treated with Metastasis-Directed Therapy. Br. J. Cancer 2019, 121, 897–903. [Google Scholar] [CrossRef]
- Patel, S.G.; Shah, J.P. TNM Staging of Cancers of the Head and Neck: Striving for Uniformity among Diversity. CA Cancer J. Clin. 2005, 55, 242–258. [Google Scholar] [CrossRef]
- Minaei, E.; Mueller, S.A.; Ashford, B.; Thind, A.S.; Mitchell, J.; Perry, J.R.; Genenger, B.; Clark, J.R.; Gupta, R.; Ranson, M. Cancer Progression Gene Expression Profiling Identifies the Urokinase Plasminogen Activator Receptor as a Biomarker of Metastasis in Cutaneous Squamous Cell Carcinoma. Front. Oncol. 2022, 12, 1188. [Google Scholar] [CrossRef]
- Wimmer, M.; Zauner, R.; Ablinger, M.; Piñón-Hofbauer, J.; Guttmann-Gruber, C.; Reisenberger, M.; Lettner, T.; Niklas, N.; Proell, J.; Sajinovic, M.; et al. A Cancer Stem Cell-Like Phenotype is Associated with miR-10b Expression in Aggressive Squamous Cell Carcinomas. Cell Commun. Signal. 2020, 18, 61. [Google Scholar] [CrossRef]
- Chi, L.-H.; Wu, A.T.H.; Hsiao, M.; Li, Y.-C. A Transcriptomic Analysis of Head and Neck Squamous Cell Carcinomas for Prognostic Indications. J. Pers. Med. 2021, 11, 782. [Google Scholar] [CrossRef]
- Pillai, J.; Chincholkar, T.; Dixit, R.; Pandey, M. A Systematic Review of Proteomic Biomarkers in Oral Squamous Cell Cancer. World J. Surg. Oncol. 2021, 19, 1–28. [Google Scholar] [CrossRef]
- Song, Q.; Yang, Y.; Jiang, D.; Qin, Z.; Xu, C.; Wang, H.; Huang, J.; Chen, L.; Luo, R.; Zhang, X.; et al. Proteomic Analysis Reveals Key Differences between Squamous Cell Carcinomas and Adenocarcinomas across Multiple Tissues. Nat. Commun. 2022, 13, 4167. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A Web Server for Cancer and Normal Gene Expression Profiling and Interactive Analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Fukushi, J.-I.; Inatani, M.; Yamaguchi, Y.; Stallcup, W.B. Expression of NG2 Proteoglycan during Endochondral and Intramembranous Ossification. Dev. Dyn. 2003, 228, 143–148. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Huret, J.-L.; Ahmad, M.; Arsaban, M.; Bernheim, A.; Cigna, J.; Desangles, F.; Guignard, J.-C.; Jacquemot-Perbal, M.-C.; Labarussias, M.; Leberre, V.; et al. Atlas of Genetics and Cytogenetics in Oncology and Haematology in 2013. Nucleic Acids Res. 2012, 41, D920–D924. [Google Scholar] [CrossRef]
- Carithers, L.J.; Ardlie, K.; Barcus, M.; Branton, P.A.; Britton, A.; Buia, S.A.; Compton, C.C.; DeLuca, D.S.; Peter-Demchok, J.; Gelfand, E.T.; et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv. Biobank. 2015, 13, 311–319. [Google Scholar] [CrossRef]
- Grako, K.A.; Stallcup, W.B. Participation of the NG2 Proteoglycan in Rat Aortic Smooth Muscle Cell Responses to Platelet-Derived Growth Factor. Exp. Cell Res. 1995, 221, 231–240. [Google Scholar] [CrossRef]
- Campoli, M.; Ferrone, S.; Wang, X. Functional and Clinical Relevance of Chondroitin Sulfate Proteoglycan 4. Adv. Cancer Res. 2010, 109, 73–121. [Google Scholar] [CrossRef]
- Ozerdem, U.; Grako, K.A.; Dahlin-Huppe, K.; Monosov, E.; Stallcup, W.B. NG2 Proteoglycan is Expressed Exclusively by Mural Cells during Vascular Morphogenesis. Dev. Dyn. 2001, 222, 218–227. [Google Scholar] [CrossRef]
- Ilieva, K.M.; Cheung, A.; Mele, S.; Chiaruttini, G.; Crescioli, S.; Griffin, M.; Nakamura, M.; Spicer, J.F.; Tsoka, S.; Lacy, K.E.; et al. Chondroitin Sulfate Proteoglycan 4 and Its Potential As an Antibody Immunotherapy Target across Different Tumor Types. Front. Immunol. 2018, 8, 1911. [Google Scholar] [CrossRef] [PubMed]
- Rolih, V.; Barutello, G.; Iussich, S.; Maria, R.D.; Quaglino, E.; Buracco, P.; Cavallo, F.; Riccardo, F. CSPG4: A Prototype Oncoantigen for Translational Immunotherapy Studies. J. Transl. Med. 2017, 15, 151. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.S.; Imai, K.; Natali, P.G.; Ferrone, S. Distribution and Molecular Characterization of a Cell-Surface and a Cytoplasmic Antigen Detectable in Human Melanoma Cells with Monoclonal Antibodies. Int. J. Cancer 1981, 28, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Pluschke, G.; Vanek, M.; Evans, A.; Dittmar, T.; Schmid, P.; Itin, P.; Filardo, E.J.; Reisfeld, R.A. Molecular Cloning of a Human Melanoma-Associated Chondroitin Sulfate Proteoglycan. Proc. Natl. Acad. Sci. USA 1996, 93, 9710–9715. [Google Scholar] [CrossRef] [PubMed]
- Stallcup, W.B. The NG2 Proteoglycan: Past Insights and Future Prospects. J. Neurocytol. 2002, 31, 423–435. [Google Scholar] [CrossRef]
- Price, M.A.; Wanshura, L.E.C.; Yang, J.; Carlson, J.; Xiang, B.; Li, G.; Ferrone, S.; Dudek, A.Z.; Turley, E.A.; McCarthy, J.B. CSPG4, a Potential Therapeutic Target, Facilitates Malignant Progression of Melanoma. Pigment Cell Melanoma Res. 2011, 24, 1148–1157. [Google Scholar] [CrossRef]
- Stallcup, W.B. The NG2 Antigen, a Putative Lineage Marker: Immunofluorescent Localization in Primary Cultures of Rat Brain. Dev. Biol. 1981, 83, 154–165. [Google Scholar] [CrossRef]
- Bai, Z.; Qu, Y.; Shi, L.; Li, X.; Yang, Z.; Ji, M.; Hou, P. Identification of a germline CSPG4 variation in a family with neurofibromatosis type 1-like phenotype. Cell Death Dis. 2021, 12, 765. [Google Scholar] [CrossRef]
- Levine, J. The Reactions and Role of NG2 Glia in Spinal Cord Injury. Brain Res. 2016, 1638, 199–208. [Google Scholar] [CrossRef]
- De Vrij, F.M.; Bouwkamp, C.G.; Gunhanlar, N.; Shpak, G.; Lendemeijer, B.; Baghdadi, M.; Gopalakrishna, S.; Ghazvini, M.; Li, T.M.; Quadri, M.; et al. Candidate CSPG4 Mutations and Induced Pluripotent Stem Cell Modeling Implicate Oligodendrocyte Progenitor Cell Dysfunction in Familial Schizophrenia. Mol. Psychiatry 2019, 24, 757–771. [Google Scholar] [CrossRef]
- Tamburini, E.; Dallatomasina, A.; Quartararo, J.; Cortelazzi, B.; Mangieri, D.; Lazzaretti, M.; Perris, R. Structural Deciphering of the NG2/CSPG4 Proteoglycan Multifunctionality. FASEB J. 2019, 33, 3112–3128. [Google Scholar] [CrossRef]
- Iida, J.; Wilhelmson, K.L.; Ng, J.; Lee, P.; Morrison, C.; Tam, E.; Overall, C.M.; McCarthy, J.B. Cell Surface Chondroitin Sulfate Glycosaminoglycan in Melanoma: Role in the Activation of Pro-MMP-2 (Pro-Gelatinase A). Biochem. J. 2007, 403, 553–563. [Google Scholar] [CrossRef]
- Tillet, E.; Gential, B.; Garrone, R.; Stallcup, W.B. NG2 Proteoglycan Mediates Beta1 Integrin-Independent Cell Adhesion and Spreading on Collagen VI. J. Cell. Biochem. 2002, 86, 726–736. [Google Scholar] [CrossRef]
- Timpl, R.; Tisi, D.; Talts, J.F.; Andac, Z.; Sasaki, T.; Hohenester, E. Structure and Function of Laminin LG Modules. Matrix Biol. 2000, 19, 309–317. [Google Scholar] [CrossRef]
- Aumailley, M. The Laminin Family. Cell Adhes. Migr. 2013, 7, 48–55. [Google Scholar] [CrossRef]
- Nishihara, T.; Remacle, A.G.; Angert, M.; Shubayev, I.; Shiryaev, S.A.; Liu, H.; Dolkas, J.; Chernov, A.V.; Strongin, A.Y.; Shubayev, V.I. Matrix Metalloproteinase-14 Both Sheds Cell Surface Neuronal Glial Antigen 2 (NG2) Proteoglycan on Macrophages and Governs the Response to Peripheral Nerve Injury. J. Biol. Chem. 2015, 290, 3693–3707. [Google Scholar] [CrossRef]
- Tillet, E.; Ruggiero, F.; Nishiyama, A.; Stallcup, W.B. The Membrane-Spanning Proteoglycan NG2 Binds to Collagens V and VI through the Central Nonglobular Domain of Its Core Protein. J. Biol. Chem. 1997, 272, 10769–10776. [Google Scholar] [CrossRef]
- Iida, J.; Meijne, A.M.; Oegema, T.R.; Yednock, T.A.; Kovach, N.L.; Furcht, L.T.; McCarthy, J.B. A role of chondroitin sulfate glycosaminoglycan binding site in alpha4beta1 integrin-mediated melanoma cell adhesion. J. Biol. Chem. 1998, 273, 5955–5962. [Google Scholar] [CrossRef]
- Iida, J.; Pei, D.; Kang, T.; Simpson, M.A.; Herlyn, M.; Furcht, L.T.; McCarthy, J.B. Melanoma Chondroitin Sulfate Proteoglycan Regulates Matrix Metalloproteinase-Dependent Human Melanoma Invasion into Type I Collagen. J. Biol. Chem. 2001, 276, 18786–18794. [Google Scholar] [CrossRef]
- Wen, Y.; Makagiansar, I.T.; Fukushi, J.-I.; Liu, F.-T.; Fukuda, M.N.; Stallcup, W.B. Molecular Basis of Interaction between NG2 Proteoglycan and Galectin-3. J. Cell. Biochem. 2005, 98, 115–127. [Google Scholar] [CrossRef]
- Makagiansar, I.T.; Williams, S.; Mustelin, T.; Stallcup, W.B. Differential Phosphorylation of NG2 Proteoglycan by ERK and PKCalpha Helps Balance Cell Proliferation and Migration. J. Cell Biol. 2007, 178, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Makagiansar, I.T.; Williams, S.; Dahlin-Huppe, K.; Fukushi, J.-I.; Mustelin, T.; Stallcup, W.B. Phosphorylation of NG2 Proteoglycan by Protein Kinase C-α Regulates Polarized Membrane Distribution and Cell Motility. J. Biol. Chem. 2004, 279, 55262–55270. [Google Scholar] [CrossRef] [PubMed]
- Nicolosi, P.A.; Dallatomasina, A.; Perris, R. Theranostic Impact of NG2/CSPG4 Proteoglycan in Cancer. Theranostics 2015, 5, 530–544. [Google Scholar] [CrossRef]
- Cattaruzza, S.; Nicolosi, P.A.; Braghetta, P.; Pazzaglia, L.; Benassi, M.S.; Picci, P.; Lacrima, K.; Zanocco, D.; Rizzo, E.; Stallcupet, W.B.; et al. NG2/CSPG4-Collagen Type VI Interplays Putatively Involved in the Microenvironmental Control of Tumour Engraftment and Local Expansion. J. Mol. Cell Biol. 2013, 5, 176–193. [Google Scholar] [CrossRef]
- Benassi, M.S.; Pazzaglia, L.; Chiechi, A.; Alberghini, M.; Conti, A.; Cattaruzza, S.; Wassermann, B.; Picci, P.; Perris, R. NG2 Expression Predicts the Metastasis Formation in Soft-Tissue Sarcoma Patients. J. Orthop. Res. 2008, 27, 135–140. [Google Scholar] [CrossRef]
- Yang, J.; Price, M.A.; Neudauer, C.L.; Wilson, C.; Ferrone, S.; Xia, H.; Iida, J.; Simpson, M.A.; McCarthy, J.B. Melanoma Chondroitin Sulfate Proteoglycan Enhances FAK and ERK Activation by Distinct Mechanisms. J. Cell Biol. 2004, 165, 881–891. [Google Scholar] [CrossRef] [PubMed]
- Chekenya, M.; Krakstad, C.; Svendsen, A.; Netland, I.A.; Staalesen, V.; Tysnes, B.B.; Selheim, F.; Wang, J.; Sakariassen, P.Ø.; Sandal, T.; et al. The Progenitor Cell Marker NG2/MPG Promotes Chemoresistance by Activation of Integrin-Dependent PI3K/Akt Signaling. Oncogene 2008, 27, 5182–5194. [Google Scholar] [CrossRef]
- Yang, J.; Price, M.A.; Li, G.Y.; Bar-Eli, M.; Salgia, R.; Jagedeeswaran, R.; Carlson, J.H.; Ferrone, S.; Turley, E.A.; McCarthy, J.B. Melanoma Proteoglycan Modifies Gene Expression to Stimulate Tumor Cell Motility, Growth, and Epithelial-to-Mesenchymal Transition. Cancer Res. 2009, 69, 7538–7547. [Google Scholar] [CrossRef]
- Eisenmann, K.M.; McCarthy, J.B.; Simpson, M.A.; Keely, P.J.; Guan, J.-L.; Tachibana, K.; Lim, L.; Manser, E.; Furcht, L.T.; Iida, J. Melanoma Chondroitin Sulphate Proteoglycan Regulates Cell Spreading through Cdc42, Ack-1 and p130cas. Nat. Cell Biol. 1999, 1, 507–513. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Hsu, S.C.; Nadesan, P.; Puviindran, V.; Stallcup, W.B.; Kirsch, D.G.; Alman, B.A. Effects of Chondroitin Sulfate Proteoglycan 4 (NG2/CSPG4) on Soft-Tissue Sarcoma Growth Depend on Tumor Developmental Stage. J. Biol. Chem. 2018, 293, 2466–2475. [Google Scholar] [CrossRef]
- Burg, M.A.; Grako, K.A.; Stallcup, W.B. Expression of the NG2 Proteoglycan Enhances the Growth and Metastatic Properties of Melanoma Cells. J. Cell. Physiol. 1998, 177, 299–312. [Google Scholar] [CrossRef]
- Nagase, H.; Visse, R.; Murphy, G. Structure and Function of Matrix Metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef]
- Löffek, S.; Schilling, O.; Franzke, C.W. Series Matrix Metalloproteinases in Lung Health and Disease: Biological Role of Matrix Metalloproteinases: A Critical Balance. Eur. Respir. J. 2011, 38, 191–208. [Google Scholar] [CrossRef]
- Safran, M.; Rosen, N.; Twik, M.; BarShir, R.; Stein, T.I.; Dahary, D.; Fishilevich, S.; Lancet, D. The GeneCards Suite. In Practical Guide to Life Science Databases; Abugessaisa, I., Kasukawa, T., Eds.; Springer Nature Singapore: Singapore, 2021; pp. 27–56. [Google Scholar]
- Agrawal, A.; Romero-Perez, D.; Jacobsen, J.; Villarreal, F.J.; Cohen, S.M. Zinc-Binding Groups Modulate Selective Inhibition of MMPs. ChemMedChem Chem. Enabling Drug Discov. 2008, 3, 812–820. [Google Scholar] [CrossRef]
- Couchman, J.R. Transmembrane Signaling Proteoglycans. Annu. Rev. Cell Dev. Biol. 2010, 26, 89–114. [Google Scholar] [CrossRef]
- Goretzki, L.; Burg, M.A.; Grako, K.A.; Stallcup, W.B. High-Affinity Binding of Basic Fibroblast Growth Factor and Platelet-derived Growth Factor-AA to the Core Protein of the NG2 Proteoglycan. J. Biol. Chem. 1999, 274, 16831–16837. [Google Scholar] [CrossRef]
- Nishiyama, A.; Lin, X.H.; Giese, N.; Heldin, C.H.; Stallcup, W.B. Interaction between NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells is required for optimal response to PDGF. J. Neurosci. Res. 1996, 43, 315–330. [Google Scholar] [CrossRef]
- Schlessinger, J.; Lax, I.; Lemmon, M. Regulation of Growth Factor Activation by Proteoglycans: What Is the Role of the Low Affinity Receptors? Cell 1995, 83, 357–360. [Google Scholar] [CrossRef]
- Yun, Y.-R.; Won, J.E.; Jeon, E.; Lee, S.; Kang, W.; Jo, H.; Jang, J.H.; Shin, U.S.; Kim, H.W. Fibroblast Growth Factors: Biology, Function, and Application for Tissue Regeneration. J. Tissue Eng. 2010, 1, 218142. [Google Scholar] [CrossRef]
- Andrae, J.; Gallini, R.; Betsholtz, C. Role of Platelet-Derived Growth Factors in Physiology and Medicine. Genes Dev. 2008, 22, 1276–1312. [Google Scholar] [CrossRef] [PubMed]
- Satyamoorthy, K.; Li, G.; Gerrero, M.R.; Brose, M.S.; Volpe, P.; Weber, B.L.; Van Belle, P.; Elder, D.E.; Herlyn, M. Constitutive Mitogen-Activated Protein Kinase Activation in Melanoma is Mediated by Both BRAF Mutations and Autocrine Growth Factor Stimulation. Cancer Res. 2003, 63, 756–759. [Google Scholar] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Raff, M.; Roberts, K.; Walter, P. Integrins. In Molecular Biology of the Cell, 4th ed.; Garland Science: New York, NY, USA, 2002. [Google Scholar]
- Joo, N.E.; Watanabe, T.; Chen, C.; Chekenya, M.; Stallcup, W.B.; Kapila, Y.L. NG2, a Novel Proapoptotic Receptor, Opposes Integrin Alpha 4 to Mediate Anoikis through PKCalpha-Dependent Suppression of FAK Phosphorylation. Cell Death Differ. 2008, 15, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Iida, J.; Skubitz, A.; Furcht, L.T.; A Wayner, E.; McCarthy, J.B. Coordinate Role for Cell Surface Chondroitin Sulfate Proteoglycan and Alpha 4 Beta 1 Integrin in Mediating Melanoma Cell Adhesion to Fibronectin. J. Cell Biol. 1992, 118, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Boyle, S.; Kopecki, Z. Mechanical Force and Actin Dynamics during Cutaneous Squamous Cell Carcinoma (cSCC) Progression: Opportunities for Novel Treatment Modalities. In Squamous Cell Carcinoma-Hallmark and Treatment Modalities; Daaboul, H., Ed.; IntechOpen Limited: London, UK, 2019. [Google Scholar]
- Kamal, J.M.; Qureshi, M.M.; Maruta, H. The CDC42-Specific Inhibitor Derived from ACK-1 Blocks v-Ha-Ras-Induced Transformation. Oncogene 1999, 18, 7787–7793. [Google Scholar] [CrossRef]
- Gehlsen, K.R.; Davis, G.E.; Sriramarao, P. Integrin Expression in Human Melanoma Cells with Differing Invasive and Metastatic Properties. Clin. Exp. Metastasis 1992, 10, 111–120. [Google Scholar] [CrossRef]
- Majumdar, M.; Vuori, K.; Stallcup, W.B. Engagement of the NG2 Proteoglycan Triggers Cell Spreading via Rac and p130cas. Cell. Signal. 2002, 15, 79–84. [Google Scholar] [CrossRef]
- Tang, F.; Lord, M.S.; Stallcup, W.B.; Whitelock, J.M. Cell Surface Chondroitin Sulphate Proteoglycan 4 (CSPG4) Binds to the Basement Membrane Heparan Sulphate Proteoglycan, Perlecan, and Is Involved in Cell Adhesion. J. Biochem. 2018, 163, 399–412. [Google Scholar] [CrossRef]
- Ribas, A.; Shin, D.S.; Zaretsky, J.; Frederiksen, J.; Cornish, A.; Avramis, E.; Seja, E.; Kivork, C.; Siebert, J.; Kaplan-Lefko, P.; et al. PD-1 Blockade Expands Intratumoral Memory T Cells. Cancer Immunol. Res. 2016, 4, 194–203. [Google Scholar] [CrossRef]
- Stallcup, W.B.; Huang, F.-J. A Role for the NG2 Proteoglycan in Glioma Progression. Cell Adhes. Migr. 2008, 2, 192–201. [Google Scholar] [CrossRef]
- Jones, F.S.; Jones, P.L. The Tenascin Family of ECM Glycoproteins: Structure, Function, and Regulation during Embryonic Development and Tissue Remodeling. Dev. Dyn. 2000, 218, 235–259. [Google Scholar] [CrossRef]
- Gates, M.A.; Fillmore, H.; Steindler, D.A. Chondroitin Sulfate Proteoglycan and Tenascin in the Wounded Adult Mouse Neostriatum In Vitro: Dopamine Neuron Attachment and Process Outgrowth. J. Neurosci. 1996, 16, 8005–8018. [Google Scholar] [CrossRef]
- Yoshida, T.; Akatsuka, T.; Imanaka-Yoshida, K.I. Tenascin-C and integrins in cancer. Cell Adhes. Migr. 2015, 9, 96–104. [Google Scholar] [CrossRef]
- Huang, W.; Chiquet-Ehrismann, R.; Moyano, J.V.; Garcia-Pardo, A.; Orend, G. Interference of Tenascin-C with Syndecan-4 Binding to Fibronectin Blocks Cell Adhesion and Stimulates Tumor Cell Proliferation. Cancer Res. 2001, 61, 8586–8594. [Google Scholar]
- Singh, P.; Carraher, C.; Schwarzbauer, J.E. Assembly of Fibronectin Extracellular Matrix. Annu. Rev. Cell Dev. Biol. 2010, 26, 397–419. [Google Scholar] [CrossRef]
- Sardone, F.; Santi, S.; Tagliavini, F.; Traina, F.; Merlini, L.; Squarzoni, S.; Cescon, M.; Wagener, R.; Maraldi, N.M.; Bonaldo, P.; et al. Collagen VI–NG2 Axis in Human Tendon Fibroblasts under Conditions Mimicking Injury Response. Matrix Biol. 2016, 55, 90–105. [Google Scholar] [CrossRef]
- Yang, J.; Price, M.A.; Wanshura, L.E.C.; He, J.; Yi, M.; Welch, D.R.; Li, G.; Conner, S.; Sachs, J.; Turley, E.A.; et al. CSPG4 Enhanced Melanoma Motility and Growth Requires a Cysteine in the Core Protein Transmembrane Domain. Melanoma Res. 2019, 29, 365–375. [Google Scholar] [CrossRef]
- Thiery, J.P. Epithelial–Mesenchymal Transitions in Tumour Progression. Nat. Cancer 2002, 2, 442–454. [Google Scholar] [CrossRef]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef]
- Ozerdem, U. Targeting Pericytes Diminishes Neovascularization in Orthotopic Uveal Melanoma in Nerve/Glial Antigen 2 Proteoglycan Knockout Mouse. Ophthalmic Res. 2006, 38, 251–254. [Google Scholar] [CrossRef]
- Huang, F.-J.; You, W.-K.; Bonaldo, P.; Seyfried, T.N.; Pasquale, E.B.; Stallcup, W.B. Pericyte Deficiencies Lead to Aberrant Tumor Vascularizaton in the Brain of the NG2 Null Mouse. Dev. Biol. 2010, 344, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Ozerdem, U.; Stallcup, W.B. Early Contribution of Pericytes to Angiogenic Sprouting and Tube Formation. Angiogenesis 2003, 6, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Ewald, A.J.; Stallcup, W.; Werb, Z.; Bergers, G. PDGFR beta + Perivascular Progenitor Cells in Tumours Regulate Pericyte Differentiation and Vascular Survival. Nat. Cell Biol. 2005, 7, 870–879. [Google Scholar] [CrossRef] [PubMed]
- Gibby, K.; You, W.K.; Kadoya, K.; Helgadottir, H.; Young, L.J.; Ellies, L.G.; Cardiff, R.D.; Stallcup, W.B. Early Vascular Deficits Are Correlated with Delayed Mammary Tumorigenesis in the MMTV-PyMT Transgenic Mouse Following Genetic Ablation of the NG2 Proteoglycan. Breast Cancer Res. 2012, 14, R67. [Google Scholar] [CrossRef] [PubMed]
- Sivan, U.; De Angelis, J.; Kusumbe, A.P. Role of Angiocrine Signals in Bone Development, Homeostasis and Disease. Open Biol. 2019, 9, 190144. [Google Scholar] [CrossRef]
- Schiffer, D.; Mellai, M.; Boldorini, R.; Bisogno, I.; Grifoni, S.; Corona, C.; Bertero, L.; Cassoni, P.; Casalone, C.; Annovazzi, L. The Significance of Chondroitin Sulfate Proteoglycan 4 (CSPG4) in Human Gliomas. Int. J. Mol. Sci. 2018, 19, 2724. [Google Scholar] [CrossRef]
- Stapor, P.C.; Sweat, R.S.; Dashti, D.C.; Betancourt, A.M.; Murfee, W.L. Pericyte Dynamics during Angiogenesis: New Insights from New Identities. J. Vasc. Res. 2014, 51, 163–174. [Google Scholar] [CrossRef]
- Girolamo, F.; Dallatomasina, A.; Rizzi, M.; Errede, M.; Wälchli, T.; Mucignat, M.T.; Frei, K.; Roncali, L.; Perris, R.; Virgintino, D. Diversified Expression of NG2/CSPG4 Isoforms in Glioblastoma and Human Foetal Brain Identifies Pericyte Subsets. PLoS ONE 2013, 8, e84883. [Google Scholar] [CrossRef]
- Ribatti, D.; Nico, B.; Crivellato, E. The Role of Pericytes in Angiogenesis. Int. J. Dev. Biol. 2011, 55, 261–268. [Google Scholar] [CrossRef]
- Xia, L.; Tan, S.; Zhou, Y.; Lin, J.; Wang, H.; Oyang, L.; Tian, Y.; Liu, L.; Su, M.; Wang, H.; et al. Role of the NFκB-Signaling Pathway in Cancer. OncoTargets Ther. 2018, 11, 2063–2073. [Google Scholar] [CrossRef]
- Cooney, C.A.; Jousheghany, F.; Yao-Borengasser, A.; Phanavanh, B.; Gomes, T.; Kieber-Emmons, A.M.; Siegel, E.R.; Suva, L.J.; Ferrone, S.; Kieber-Emmons, T.; et al. Chondroitin Sulfates Play a Major Role in Breast Cancer Metastasis: A Role for CSPG4 and Chst11 Gene Expression in Forming Surface P-Selectin Ligands in Aggressive Breast Cancer Cells. Breast Cancer Res. 2011, 13, R58. [Google Scholar] [CrossRef]
- Keleg, S.; Titov, A.; Heller, A.; Giese, T.; Tjaden, C.; Ahmad, S.S.; Gaida, M.M.; Bauer, A.S.; Werner, J.; Giese, N.A. Chondroitin Sulfate Proteoglycan CSPG4 as a Novel Hypoxia-Sensitive Marker in Pancreatic Tumors. PLoS ONE 2014, 9, e100178. [Google Scholar] [CrossRef]
- Petrovici, K.; Graf, M.; Hecht, K.; Reif, S.; Pfister, K.; Schmetzer, H. Use of NG2 (7.1) in AML as a Tumor Marker and Its Association with a Poor Prognosis. Cancer Genom. Proteom. 2010, 7, 173–180. [Google Scholar]
- Behm, F.; Smith, F.; Raimondi, S.; Pui, C.; Bernstein, I. Human Homologue of the Rat Chondroitin Sulfate Proteoglycan, NG2, Detected by Monoclonal Antibody 7.1, Identifies Childhood Acute Lymphoblastic Leukemias with t(4;11)(q21;q23) or t(11;19)(q23;p13) and MLL Gene Rearrangements. Blood 1996, 87, 1134–1139. [Google Scholar] [CrossRef]
- Vergilis, I.J.; Szarek, M.; Ferrone, S.; Reynolds, S.R. Presence and Prognostic Significance of Melanoma-Associated Antigens CYT-MAA and HMW-MAA in Serum of Patients with Melanoma. J. Investig. Dermatol. 2005, 125, 526–531. [Google Scholar] [CrossRef]
- Ulmer, A.; Fierlbeck, G. Circulating Tumor Cells and Detection of the Melanoma-Associated Antigen HMW-MAA in the Serum of Melanoma Patients. J. Investig. Dermatol. 2006, 126, 915. [Google Scholar] [CrossRef][Green Version]
- Warta, R.; Herold-Mende, C.; Chaisaingmongkol, J.; Popanda, O.; Möck, A.; Mogler, C.; Osswald, F.; Herpel, E.; Küstner, S.; Eckstein, V.; et al. Reduced Promoter Methylation and Increased Expression of CSPG4 Negatively Influences Survival of HNSCC Patients. Int. J. Cancer 2014, 135, 2727–2734. [Google Scholar] [CrossRef]
- Online Mendelian Inheritance in Man. Available online: https://omim.org/ (accessed on 11 November 2020).
- Nishiyama, A.; Dahlin, K.; Stallcup, W. The Expression of NG2 Proteoglycan in the Developing Rat Limb. Development 1991, 111, 933–944. [Google Scholar] [CrossRef]
- Grako, K.; Ochiya, T.; Barritt, D.; Nishiyama, A.; Stallcup, W. PDGF (Alpha)-Receptor is Unresponsive to PDGF-AA in Aortic Smooth Muscle Cells from the NG2 Knockout Mouse. J. Cell Sci. 1999, 112, 905–915. [Google Scholar] [CrossRef]
- Midwood, K.S.; Salter, D.M. Expression of NG2/Human Melanoma Proteoglycan in Human Adult Articular Chondrocytes. Osteoarthr. Cartil. 1998, 6, 297–305. [Google Scholar] [CrossRef]
- Ghali, L.; Wong, S.-T.; Tidman, N.; Quinn, A.; Philpott, M.P.; Leigh, I.M. Epidermal and Hair Follicle Progenitor Cells Express Melanoma-Associated Chondroitin Sulfate Proteoglycan Core Protein. J. Investig. Dermatol. 2004, 122, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.; Jensen, U.B.; Broad, S.; Leigh, I.; Watt, F.M. Role of Melanoma Chondroitin Sulphate Proteoglycan in Patterning Stem Cells in Human Interfollicular Epidermis. Development 2003, 130, 6049–6063. [Google Scholar] [CrossRef] [PubMed]
- Giangreco, A.; Goldie, S.J.; Failla, V.; Saintigny, G.; Watt, F.M. Human Skin Aging Is Associated with Reduced Expression of the Stem Cell Markers Beta1 Integrin and MCSP. J. Investig. Dermatol. 2010, 130, 604–608. [Google Scholar] [CrossRef] [PubMed]
- Trotter, J.; Karram, K.; Nishiyama, A. NG2 Cells: Properties, Progeny and Origin. Brain Res. Rev. 2010, 63, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Kozanoglu, I.; Boga, C.; Ozdogu, H.; Sozer, O.; Maytalman, E.; Yazici, A.C.; Sahin, F.I. Human Bone Marrow Mesenchymal Cells Express NG2: Possible Increase in Discriminative Ability of Flow Cytometry during Mesenchymal Stromal Cell Identification. Cytotherapy 2009, 11, 527–533. [Google Scholar] [CrossRef]
- Fukushi, J.; Makagiansar, I.T.; Stallcup, W.B. NG2 Proteoglycan Promotes Endothelial Cell Motility and Angiogenesis via Engagement of Galectin-3 and Alpha3beta1 Integrin. Mol. Biol. Cell 2004, 15, 3580–3590. [Google Scholar] [CrossRef]
- Cho, R.J.; Alexandrov, L.B.; Breems, N.Y.D.; Atanasova, V.S.; Farshchian, M.; Purdom, E.; Nguyen, T.N.; Coarfa, C.; Rajapakshe, K.; Prisco, M.; et al. APOBEC Mutation Drives Early-Onset Squamous Cell Carcinomas in Recessive Dystrophic Epidermolysis Bullosa. Sci. Transl. Med. 2018, 10, eaas9668. [Google Scholar] [CrossRef]
- Nishi, H.; Inoue, Y.; Kageshita, T.; Takata, M.; Ihn, H. The Expression of Human High Molecular Weight Melanoma-Associated Antigen in Acral Lentiginous Melanoma. Biosci. Trends 2010, 4, 86–89. [Google Scholar]
- Kageshita, T.; Kimura, T.; Yoshii, A.; Hirai, S.; Ono, T.; Ferrone, S. Antigenic Profile of Mucosal Melanoma Lesions. Int. J. Cancer 1994, 56, 370–374. [Google Scholar] [CrossRef]
- Beard, R.E.; Zheng, Z.; Lagisetty, K.H.; Burns, W.R.; Tran, E.; Hewitt, S.M.; Abate-Daga, D.; Rosati, S.F.; A Fine, H.; Ferrone, S.; et al. Multiple Chimeric Antigen Receptors Successfully Target Chondroitin Sulfate Proteoglycan 4 in Several Different Cancer Histologies and Cancer Stem Cells. J. Immunother. Cancer 2014, 2, 25. [Google Scholar] [CrossRef]
- Schwab, J.H.; Boland, P.J.; Agaram, N.P.; Socci, N.D.; Guo, T.; O’Toole, G.C.; Wang, X.; Ostroumov, E.; Hunter, C.J.; Block, J.A.; et al. Chordoma and Chondrosarcoma Gene Profile: Implications for Immunotherapy. Cancer Immunol. Immunother. 2008, 58, 339–349. [Google Scholar] [CrossRef]
- Wang, X.; Osada, T.; Wang, Y.; Yu, L.; Sakakura, K.; Katayama, A.; McCarthy, J.B.; Brufsky, A.; Chivukula, M.; Khoury, T.; et al. CSPG4 Protein as a New Target for the Antibody-Based Immunotherapy of Triple-Negative Breast Cancer. JNCI J. Natl. Cancer Inst. 2010, 102, 1496–1512. [Google Scholar] [CrossRef]
- Rivera, Z.; Ferrone, S.; Wang, X.; Jube, S.; Yang, H.; Pass, H.I.; Kanodia, S.; Gaudino, G.; Carbone, M. CSPG4 As a Target of Antibody-Based Immunotherapy for Malignant Mesothelioma. Clin. Cancer Res. 2012, 18, 5352–5363. [Google Scholar] [CrossRef]
- Riccardo, F.; Tarone, L.; Iussich, S.; Giacobino, D.; Arigoni, M.; Sammartano, F.; Morello, E.; Martano, M.; Gattino, F.; DE Maria, R.; et al. Identification of CSPG4 As a Promising Target for Translational Combinatorial Approaches in Osteosarcoma. Ther. Adv. Med. Oncol. 2019, 11, 1758835919855491. [Google Scholar] [CrossRef]
- Paijens, S.T.; Vledder, A.; de Bruyn, M.; Nijman, H.W. Tumor-Infiltrating Lymphocytes in the Immunotherapy Era. Cell. Mol. Immunol. 2020, 18, 842–859. [Google Scholar] [CrossRef]
- Stanton, S.E.; Disis, M.L. Clinical Significance of Tumor-Infiltrating Lymphocytes in Breast Cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef]
- Kamphorst, A.O.; Pillai, R.N.; Yang, S.; Nasti, T.H.; Akondy, R.S.; Wieland, A.; Sica, G.L.; Yu, K.; Koenig, L.; Patel, N.T.; et al. Proliferation of PD-1+ CD8 T cells in peripheral blood after PD-1-targeted therapy in lung cancer patients. Proc. Natl. Acad. Sci. USA 2017, 114, 4993–4998. [Google Scholar] [CrossRef]
- Hsu, N.C.; Nien, P.-Y.; Yokoyama, K.K.; Chu, P.-Y.; Hou, M.-F. High Chondroitin Sulfate Proteoglycan 4 Expression Correlates with Poor Outcome in Patients with Breast Cancer. Biochem. Biophys. Res. Commun. 2013, 441, 514–518. [Google Scholar] [CrossRef]
- Mellick, A.S.; Plummer, P.N.; Nolan, D.J.; Gao, D.; Bambino, K.; Hahn, M.; Catena, R.; Turner, V.; McDonnell, K.; Benezra, R.; et al. Using the Transcription Factor Inhibitor of DNA Binding 1 to Selectively Target Endothelial Progenitor Cells Offers Novel Strategies to Inhibit Tumor Angiogenesis and Growth. Cancer Res. 2010, 70, 7273–7282. [Google Scholar] [CrossRef]
- Plummer, P.N.; Freeman, R.; Taft, R.J.; Vider, J.; Sax, M.; Umer, B.A.; Gao, D.; Johns, C.; Mattick, J.S.; Wilton, S.D.; et al. MicroRNAs Regulate Tumor Angiogenesis Modulated by Endothelial Progenitor Cells. Cancer Res. 2013, 73, 341–352. [Google Scholar] [CrossRef]
- Gao, D.; Nolan, D.J.; Mellick, A.S.; Bambino, K.; McDonnell, K.; Mittal, V. Endothelial Progenitor Cells Control the Angiogenic Switch in Mouse Lung Metastasis. Science 2008, 319, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, G.; Oeffner, F.; von Messling, V.; Tschernig, T.; Grone, H.-J.; Klenk, H.D.; Herrler, G. Cloning and Characterization of Gp36, a Human Mucin-Type Glycoprotein Preferentially Expressed in Vascular Endothelium. Biochem. J. 1999, 341, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Lacy, S.E.; Bönnemann, C.G.; Buzney, E.A.; Kunkel, L.M. Identification of FLRT1, FLRT2, and FLRT3: A Novel Family of Transmembrane Leucine-Rich Repeat Proteins. Genomics 1999, 62, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, L.; Christiano, A.M.; Airenne, T.; Haakana, H.; Tryggvason, K.; Uitto, J. Mutations in the Gamma-2 Chain Gene (LAMC2) of Kalinin/Laminin 5 in the Junctional Forms of Epidermolysis Bullosa. Nat. Genet. 1994, 6, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Takada, Y.; Murphy, E.; Pil, P.; Chen, C.; Ginsberg, M.H.; Hemler, M.E. Molecular Cloning and Expression of the cDNA for Alpha-3 Subunit of Human Alpha-3/Beta-1 (VLA-3), an Integrin Receptor for Fibronectin, Laminin, and Collagen. J. Cell Biol. 1991, 115, 257–266. [Google Scholar] [CrossRef]
- Berns, D.S.; DeNardo, L.A.; Pederick, D.T.; Luo, L. Teneurin-3 Controls Topographic Circuit Assembly in the Hippocampus. Nature 2018, 554, 328–333. [Google Scholar] [CrossRef]
- Franzke, C.-W.; Tasanen, K.; Schäcke, H.; Zhou, Z.; Tryggvason, K.; Mauch, C.; Zigrino, P.; Sunnarborg, S.; Lee, D.C.; Fahrenholz, F.; et al. Transmembrane Collagen XVII, an Epithelial Adhesion Protein, Is Shed from the Cell Surface by ADAMs. EMBO J. 2002, 21, 5026–5035. [Google Scholar] [CrossRef]
- Nieto, A.M.; Sargent, M.G.; Wilkinson, D.G.; Cooke, J. Control of Cell Behavior during Vertebrate Development by Slug, a Zinc Finger Gene. Science 1994, 264, 835–839. [Google Scholar] [CrossRef]
- Adamson, M.; Dennis, C.; Delaney, S.; Christiansen, J.; Monkley, S.; Kozak, C.A.; Wainwright, B. Isolation and Genetic Mapping of Two Novel Members of the Murine Wnt Gene Family, Wnt11 and Wnt12, and the Mapping of Wnt5a and Wnt7a. Genomics 1994, 24, 9–13. [Google Scholar] [CrossRef]
- Suzuki, H.; Kaneko, M.K.; Kato, Y. Roles of Podoplanin in Malignant Progression of Tumor. Cells 2022, 11, 575. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Z.; Lu, N. A New Role for the PI3K/Akt Signaling Pathway in the Epithelial-Mesenchymal Transition. Cell Adhes. Migr. 2015, 9, 317–324. [Google Scholar] [CrossRef]
- Milenkovic, D.; Chaffaux, S.; Taourit, S.; Guérin, G. A Mutation in the LAMC2 Gene Causes the Herlitz Junctional Epidermolysis Bullosa (H-JEB) in Two French Draft Horse Breeds. Genet. Sel. Evol. 2003, 35, 1–56. [Google Scholar] [CrossRef]
- Pulkkinen, L.; Smith, F.J.D.; Shimizu, H.; Murata, S.; Yaoita, H.; Hachisuka, H.; Nishikawa, T.; McLean, W.H.I.; Uitto, J. Homozygous Deletion Mutations in the Plectin Gene (PLEC1) in Patients with Epidermolysis Bullosa Simplex Associated with Late-Onset Muscular Dystrophy. Hum. Mol. Genet. 1996, 5, 1539–1546. [Google Scholar] [CrossRef]
- Ando, T.; Tai-Nagara, I.; Sugiura, Y.; Kusumoto, D.; Okabayashi, K.; Kido, Y.; Sato, K.; Saya, H.; Navankasattusas, S.; Li, D.Y.; et al. Tumor-Specific Interendothelial Adhesion Mediated by FLRT2 Facilitates Cancer Aggressiveness. J. Clin. Investig. 2022, 132, e153626. [Google Scholar] [CrossRef]
- Guo, X.; Song, C.; Fang, L.; Li, M.; Yue, L.; Sun, Q. FLRT2 Functions As Tumor Suppressor Gene Inactivated by Promoter Methylation in Colorectal Cancer. J. Cancer 2020, 11, 7329–7338. [Google Scholar] [CrossRef]
- Takeuchi, H.; Yokota, A.; Ohoka, Y.; Iwata, M. Cyp26b1 Regulates Retinoic Acid-Dependent Signals in T Cells and Its Expression Is Inhibited by Transforming Growth Factor-β. PLoS ONE 2011, 6, e16089. [Google Scholar] [CrossRef]
- Liu, L.; Brown, D.; McKee, M.; LeBrasseur, N.K.; Yang, D.; Albrecht, K.H.; Ravid, K.; Pilch, P.F. Deletion of Cavin/PTRF Causes Global Loss of Caveolae, Dyslipidemia, and Glucose Intolerance. Cell Metab. 2008, 8, 310–317. [Google Scholar] [CrossRef]
- Meester-Smoor, M.A.; Vermeij, M.; van Helmond, M.J.L.; Molijn, A.C.; van Wely, K.H.M.; Hekman, A.C.P.; Vermey-Keers, C.; Riegman, P.H.J.; Zwarthoff, E.C. Targeted Disruption of the Mn1 Oncogene Results in Severe Defects in Development of Membranous Bones of the Cranial Skeleton. Mol. Cell. Biol. 2005, 25, 4229–4236. [Google Scholar] [CrossRef]
- Masters, C.L.; Simms, G.; A Weinman, N.; Multhaup, G.; McDonald, B.L.; Beyreuther, K. Amyloid Plaque Core Protein in Alzheimer Disease and Down Syndrome. Proc. Natl. Acad. Sci. USA 1985, 82, 4245–4249. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptome Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cance. Cell 2017, 171, 1611–1624. [Google Scholar] [CrossRef]
- Ji, A.L.; Rubin, A.J.; Thrane, K.; Jiang, S.; Reynolds, D.L.; Meyers, R.M.; Guo, M.G.; George, B.M.; Mollbrink, A.; Bergenstråhle, J.; et al. Multimodal Analysis of Composition and Spatial Architecture in Human Squamous Cell Carcinoma. Cell 2020, 182, 497–514. [Google Scholar] [CrossRef] [PubMed]
- Masunaga, T.; Ogawa, J.; Akiyama, M.; Nishikawa, T.; Shimizu, H.; Ishiko, A. Compound Heterozygosity for Novel Splice Site Mutations of ITGA6 in Lethal Junctional Epidermolysis Bullosa with Pyloric Atresia. J. Dermatol. 2016, 44, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Ruzzi, L.; Gagnoux-Palacios, L.; Pinola, M.; Belli, S.; Meneguzzi, G.; D’Alessio, M.; Zambruno, G. A Homozygous Mutation in the Integrin Alpha6 Gene in Junctional Epidermolysis Bullosa with Pyloric Atresia. J. Clin. Investig. 1997, 99, 2826–2831. [Google Scholar] [CrossRef] [PubMed]
- McGrath, J.A.; Gatalica, B.; Christiano, A.M.; Li, K.; Owaribe, K.; McMillan, J.R.; Eady, R.A.; Uitto, J. Mutations in the 180-kD Bullous Pemphigoid Antigen (BPAG2), a Hemidesmosomal Transmembrane Collagen (COL17A1), in Generalized Atrophic Benign Epidermolysis Bullosa. Nat. Genet. 1995, 11, 83–86. [Google Scholar] [CrossRef] [PubMed]
- Kedar, V.; Fletcher, L.; Powell, L. Molecular Cloning of a Novel mRNA Highly Expressed in Haemochromatotic Human Liver and Proliferating Cells. Biochem. Biophys. Res. Commun. 1996, 226, 461–466. [Google Scholar] [CrossRef]
- Prekeris, R.; Klumperman, J.; Scheller, R.H. A Rab11/Rip11 Protein Complex Regulates Apical Membrane Trafficking via Recycling Endosomes. Mol. Cell 2000, 6, 1437–1448. [Google Scholar] [CrossRef]
- Wright, M.D.; Geary, S.M.; Fitter, S.; Moseley, G.W.; Lau, L.-M.; Sheng, K.-C.; Apostolopoulos, V.; Stanley, E.G.; Jackson, D.E.; Ashman, L.K. Characterization of Mice Lacking the Tetraspanin Superfamily Member CD151. Mol. Cell. Biol. 2004, 24, 5978–5988. [Google Scholar] [CrossRef]
- Margadant, C.; Charafeddine, R.A.; Sonnenberg, A. Unique and Redundant Functions of Integrins in the Epidermis. FASEB J. 2010, 24, 4133–4152. [Google Scholar] [CrossRef]
- Watt, F.M. Role of Integrins in Regulating Epidermal Adhesion, Growth and Differentiation. EMBO J. 2002, 21, 3919–3926. [Google Scholar] [CrossRef]
- De Rosa, L.; Seconetti, A.S.; De Santis, G.; Pellacani, G.; Hirsch, T.; Rothoeft, T.; Teig, N.; Pellegrini, G.; Bauer, J.W.; De Luca, M. Laminin 332-Dependent YAP Dysregulation Depletes Epidermal Stem Cells in Junctional Epidermolysis Bullosa. Cell Rep. 2019, 27, 2036–2049. [Google Scholar] [CrossRef]
- Liu, N.; Matsumura, H.; Kato, T.; Ichinose, S.; Takada, A.; Namiki, T.; Asakawa, K.; Morinaga, H.; Mohri, Y.; De Arcangelis, A.; et al. Stem Cell Competition Orchestrates Skin Homeostasis and Ageing. Nature 2019, 568, 344–350. [Google Scholar] [CrossRef]
- Westekemper, H.; Karimi, S.; Süsskind, D.; Anastassiou, G.; Freistühler, M.; Meller, D.; Zeschnigk, M.; Steuhl, K.-P.; Bornfeld, N.; Schmid, K.-W.; et al. Expression of MCSP and PRAME in Conjunctival Melanoma. Br. J. Ophthalmol. 2010, 94, 1322–1327. [Google Scholar] [CrossRef][Green Version]




| Author/Source | Cell/Tissue/Organ Type | Protein Levels | Median Expression ** | Type (A/E) * | ||
|---|---|---|---|---|---|---|
| The Human Protein Atlas and Uhlén et al. [49] Huret et al. [50] | Brain | Cerebral cortex | High | A | ||
| Cerebellum | High | |||||
| Hippocampus | High | |||||
| Caudate | Medium | |||||
| Endocrine | Thyroid | Medium | A | |||
| Parathyroid | Medium | |||||
| Adrenal gland | Medium | |||||
| Respiratory | Nasopharynx | High | A | |||
| Bronchus | High | |||||
| Lung | Medium | |||||
| Proximal digestive tract | Oral mucosa | Low | A | |||
| Salivary gland | Medium | |||||
| Esophagus | Medium | |||||
| Gastrointestinal tract | Stomach | Medium | A | |||
| Duodenum | High | |||||
| Small intestine | High | |||||
| Colon | Medium | |||||
| Rectum | Medium | |||||
| Liver | Medium | A | ||||
| Gallbladder | High | A | ||||
| pancreas | High | A | ||||
| Kidney | Medium | A | ||||
| Bladder | Medium | A | ||||
| Male reproductive organs | Medium | A | ||||
| Female reproductive organs | Vagina | Medium | ||||
| Ovary | Medium | |||||
| Fallopian tube | Medium | |||||
| Endometrium | High | |||||
| Cervix | Medium | |||||
| Placenta | Medium | |||||
| Breast | Medium | |||||
| Muscle | Heart muscle | Medium | A | |||
| Smooth | Low | |||||
| Skeletal | Medium | |||||
| Adipose tissue and soft tissue | Low-Medium | A | ||||
| Skin | Medium | A | ||||
| Lymphoid tissue | Bone marrow | Medium | A | |||
| Appendix | High | |||||
| Spleen | Medium | |||||
| Lymph node | Medium | |||||
| Tonsil | Medium | |||||
| Haematopoietic | Low | A | ||||
| *** GTEx data, Carithers et al. [51] | Lung | 37.81 | A | |||
| Vascular | Aorta | 175.6 | ||||
| Coronary artery | 132.6 | |||||
| Tibial artery | 246.2 | |||||
| Heart | Atrial appendage | 12.85 | ||||
| Left ventricle | 8.040 | |||||
| Brain | Amygdala | 10.48 | ||||
| Anterior cingulate cortex | 8.864 | |||||
| Caudate | 6.361 | |||||
| Cerebellar hemisphere | 3.078 | |||||
| Cerebellum | 3.845 | |||||
| Cortex | 7.710 | |||||
| Frontal cortex | 6.361 | |||||
| Hippocampus | 8.213 | |||||
| Hypothalamus | 8.861 | |||||
| Nucleus accumbens | 6.160 | |||||
| Putamen | 5.692 | |||||
| Spinal cord | 5.574 | |||||
| Substantia nigra | 8.926 | |||||
| Nerve (tibial) | 66.59 | |||||
| Pituitary | 2.565 | |||||
| Thyroid | 11.30 | |||||
| Liver | 0.5535 | |||||
| Pancreas | 1.326 | |||||
| Spleen | 9.568 | |||||
| Stomach | 10.87 | |||||
| Small intestine | 10.92 | |||||
| Colon | Sigmoid | 105.7 | ||||
| Transverse | 35.10 | |||||
| Esophagus | Gastroesophageal junction | 98.16 | ||||
| Mucosa | 7.547 | |||||
| Muscularis | 98.31 | |||||
| Cervix | Ectocervix | 29.65 | ||||
| Endocervix | 28.90 | |||||
| Female reproductive organs | Fallopian tube | 37.31 | ||||
| Ovary | 12.14 | |||||
| Uterus | 60.30 | |||||
| Vagina | 19.53 | |||||
| Breast | 23.63 | |||||
| Male reproductive organs | Prostate | 23.09 | ||||
| Testis | 3.153 | |||||
| Kidney | Medulla | 6.372 | ||||
| Cortex | 4.208 | |||||
| Adrenal gland | 2.933 | |||||
| Bladder | 59.06 | |||||
| Adipose | Subcutaneous | 51.61 | ||||
| Visceral | 33.41 | |||||
| Skeletal muscle | 23.65 | |||||
| Fibroblasts (cultured) | 14.35 | |||||
| Skin | Sun exposed | 13.22 | ||||
| Non sun exposed | 10.93 | |||||
| Whole blood | 0.09142 | |||||
| Nishiyama et al. [136] | Chondroblast precursors | E | ||||
| Fukushi et al. [48] | Chondroblast precursors | E | ||||
| Osteoblast precursors | ||||||
| Ozerdem et al. [54], Grako et al. [137] | Cardiomyocytes | E | ||||
| Pericytes | E & A | |||||
| Vascular smooth muscle | E | |||||
| Midwood et al. [138] | Chondrocytes | A | ||||
| Ghali et al. [139] | Epidermal and interfollicular progenitor cells | A | ||||
| Legg et al. [140], Giangreco et al. [141] | Interfollicular epidermis progenitor cells | A | ||||
| Schiffer et al. [123] | Oligodendrocyte precursor cells | A | ||||
| Trotter et al. [142] | Oligodendrocyte precursor cells | A | ||||
| Protoplasmic astrocytes | ||||||
| Neurons | ||||||
| Kozanoglu et al. [143] | Bone marrow mesenchymal cells | A | ||||
| Author/Source | Tumour Type | % CSPG4+ Lesions Compared to Total | Rel. Expression Compared to Normal | |
|---|---|---|---|---|
| Wilson et al. [57] | Melanoma | 98.3–100% | ||
| NMSC | cSCC | 37.5–50% | ||
| Basal cell carcinoma | ||||
| Nishi et al. [146] | Melanoma | Acral lentigous melanoma (ALM) | 53.6% | Increased; staining intensity for ALM was weaker than SSM |
| Superficial spreading melanoma (SSM) | 100% | |||
| Kageshita et al. [147] | Melanoma | Primary | 50% | |
| Metastatic | 83.3% | |||
| Beard et al. [148] | Melanoma | Increased | ||
| Triple Negative breast Cancer (TNBC) | Increased | |||
| Glioblastoma | Increased | |||
| Tang et al. [47] | Pancreatic adenocarcinoma | Increased | ||
| Pheochromocytoma and paraganglioma | ||||
| Esophageal carcinoma | ||||
| HNSCC | ||||
| Brain lower grade glioma | ||||
| Glioblastoma multiforme | ||||
| Kidney renal clear cell carcinoma | ||||
| Melanoma | ||||
| Uhlen et al. [49] | Renal cancer | |||
| Urothelial cancer | ||||
| Glioma | ||||
| Warta et al. [134] | HNSCC | Increased | ||
| Schwab et al. [149] | Chordoma and chondrosarcoma | Present | ||
| Wang et al. [150] | Breast cancer | ER−/PR−/HER2−(TNBC) | 72.7% | |
| ER+ | 28.6% | |||
| HER2+ | 16.7% | |||
| Behm et al. [131] | Acute lymphoblastic leukemia | 8.6% | ||
| Petrovici et al. [130] | Acute myeloid leukemia | 50% | ||
| Rivera et al. [151] | Malignant mesothelioma | 60.98% | ||
| Keleg et al. [129] | Pancreatic carcinoma | Adenosquamous carcinoma | Increased | |
| Anaplastic ductal adenocarcinoma | ||||
| Intraductal papillary mucinous neoplasm with associated invasive carcinoma | ||||
| Riccardo et al. [152] | Osteosarcoma | Increased | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Yong, J.; Zauner, R.; Wally, V.; Whitelock, J.; Sajinovic, M.; Kopecki, Z.; Liang, K.; Scott, K.F.; Mellick, A.S. Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers 2022, 14, 5564. https://doi.org/10.3390/cancers14225564
Chen K, Yong J, Zauner R, Wally V, Whitelock J, Sajinovic M, Kopecki Z, Liang K, Scott KF, Mellick AS. Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers. 2022; 14(22):5564. https://doi.org/10.3390/cancers14225564
Chicago/Turabian StyleChen, Kathryn, Joel Yong, Roland Zauner, Verena Wally, John Whitelock, Mila Sajinovic, Zlatko Kopecki, Kang Liang, Kieran Francis Scott, and Albert Sleiman Mellick. 2022. "Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma" Cancers 14, no. 22: 5564. https://doi.org/10.3390/cancers14225564
APA StyleChen, K., Yong, J., Zauner, R., Wally, V., Whitelock, J., Sajinovic, M., Kopecki, Z., Liang, K., Scott, K. F., & Mellick, A. S. (2022). Chondroitin Sulfate Proteoglycan 4 as a Marker for Aggressive Squamous Cell Carcinoma. Cancers, 14(22), 5564. https://doi.org/10.3390/cancers14225564







