Determination of the Cancer Genome Atlas (TCGA) Endometrial Cancer Molecular Subtypes Using the Variant Interpretation and Clinical Decision Support Software MH Guide
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
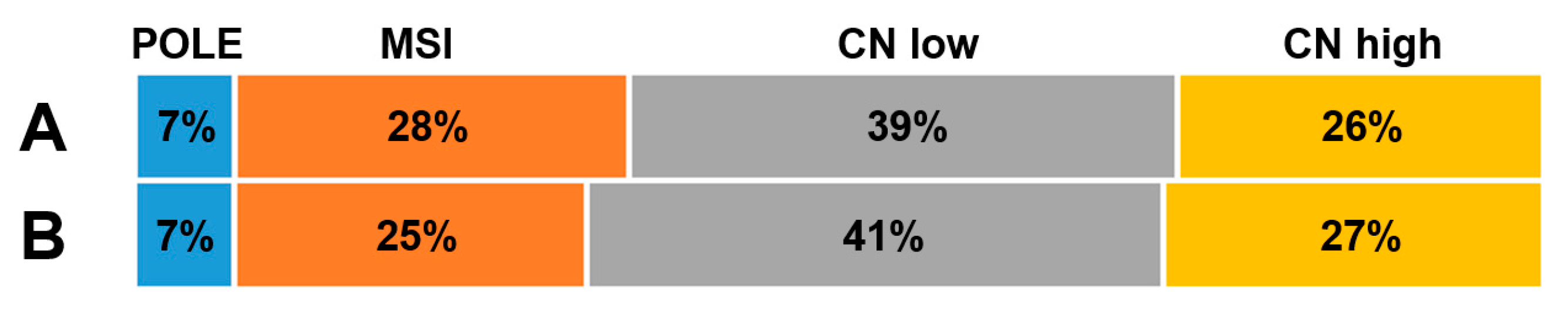
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Jansen, L.; Gondos, A.; Ressing, M.; Holleczek, B.; Katalinic, A.; Brenner, H. GEKID Cancer Survival Working Group. Survival of endometrial cancer patients in Germany in the early 21st century: A period analysis by age, histology, and stage. BMC Cancer 2012, 12, 128. [Google Scholar] [CrossRef] [PubMed]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Sherman, M.E. Theories of endometrial carcinogenesis: A multidisciplinary approach. Mod. Pathol. 2000, 13, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Emons, G.; Fleckenstein, G.; Hinney, B.; Huschmand, A.; Heyl, W. Hormonal interactions in endometrial cancer. Endocr. Relat. Cancer 2000, 7, 227–242. [Google Scholar] [CrossRef]
- Hecht, J.L.; Mutter, G.L. Molecular and pathologic aspects of endometrial carcinogenesis. J. Clin. Oncol. 2006, 24, 4783–4791. [Google Scholar] [CrossRef]
- Mustea, A.; Koensgen, D.; Belau, A.; Sehouli, J.; Lichtenegger, W.; Schneidewind, L.; Sommer, H.; Markmann, S.; Scharf, J.P.; Ehmke, P.; et al. Adjuvant sequential chemoradiation therapy in high-risk endometrial cancer: Results of a prospective, multicenter phase-II study of the NOGGO (North-Eastern German Society of Gynaecological Oncology). Cancer Chemother. Pharmacol. 2013, 72, 975–983. [Google Scholar] [CrossRef]
- Abd El-Wahed, M.M.; Abdou, A.G.; Al-Sharaky, D.R.; Kasem, H.A. Clinicopathological differences between type I and type II endometrial carcinoma. Menoufia. Med. J. 2017, 30, 946–951. [Google Scholar]
- Moore, K.N.; Fader, A.N. Uterine papillary serous carcinoma. Clin. Obstet. Gynecol. 2011, 54, 278–291. [Google Scholar] [CrossRef]
- Arend, R.C.; Jones, B.A.; Martinez, A.; Goodfellow, P. Endometrial cancer: Molecular markers and management of advanced stage disease. Gynecol. Oncol. 2018, 150, 569–580. [Google Scholar] [CrossRef]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jürgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved risk assessment by integrating molecular and clinicopathological factors in early-stage endometrial cancer-combined analysis of the PORTEC cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef]
- Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; Benz, C.C.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar]
- Huvila, J.; Orte, K.; Vainio, P.; Mettälä, T.; Joutsiniemi, T.; Hietanen, S. Molecular subtype diagnosis of endometrial carcinoma: Comparison of the next-generation sequencing panel and proactive molecular risk classifier for endometrial cancer classifier. Hum. Pathol. 2021, 111, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Leon-Castillo, A.; de Boer, S.M.; Powell, M.E.; Mileshkin, L.R.; Mackay, H.J.; Leary, A.; Nijman, H.W.; Singh, N.; Pollock, P.M.; Bessette, P.; et al. TransPORTEC consortium. Molecular classification of the PORTEC-3 trial for high-risk endometrial cancer: Impact on prognosis and benefit from adjuvant therapy. J. Clin. Oncol. 2020, 38, 3388–3397. [Google Scholar] [CrossRef] [PubMed]
- Buza, N. Immunohistochemistry in gynecologic carcinomas: Practical update with diagnostic and clinical considerations based on the 2020 WHO classification of tumors. Semin. Diagn. Pathol. 2022, 39, 58–77. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Hussein, Y.; Bandyopadhyay, S.; Cote, M.; Hassan, O.; Abdulfatah, E.; Alosh, B.; Guan, H.; Soslow, R.A.; Ali-Fehmi, R. Interobserver variability in the diagnosis of uterine high-grade endometrioid carcinoma. Arch. Pathol. Lab. Med. 2016, 140, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Stope, M.B.; Koensgen, D.; Weimer, J.; Paditz, M.; Burchardt, M.; Bauerschlag, D.; Mustea, A. The future therapy of endometrial cancer: MicroRNA’s functionality, capability, and putative clinical application. Arch. Gynecol. Obstet. 2016, 294, 889–895. [Google Scholar] [CrossRef]
- Latham, A.; Srinivasan, P.; Kemel, Y.; Shia, J.; Bandlamudi, C.; Mandelker, D.; Middha, S.; Hechtman, J.; Zehir, A.; Dubard-Gault, M.; et al. Microsatellite instability is associated with the presence of Lynch syndrome pan-cancer. J. Clin. Oncol. 2019, 37, 286–295. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Qian, L.; Deng, S.; Liu, L.; Xiao, W.; Zhou, Y. A review of the clinical characteristics and novel molecular subtypes of endometrioid ovarian cancer. Front. Oncol. 2021, 11, 668151. [Google Scholar] [CrossRef]
- de Boer, S.M.; Powell, M.E.; Mileshkin, L.; Katsaros, D.; Bessette, P.; Haie-Meder, C.; Ottevanger, P.B.; Ledermann, J.A.; Khaw, P.; Colombo, A.; et al. PORTEC study group. Adjuvant chemoradiotherapy versus radiotherapy alone for women with high-risk endometrial cancer (PORTEC-3): Final results of an international, open-label, multicentre, randomised, phase 3 trial. Lancet Oncol. 2018, 19, 295–309. [Google Scholar] [CrossRef]
- Wortman, B.G.; Bosse, T.; Nout, R.A.; Lutgens, L.C.H.W.; van der Steen-Banasik, E.M.; Westerveld, H.; van den Berg, H.; Slot, A.; De Winter, K.A.J.; Verhoeven-Adema, K.W.; et al. Molecular-integrated risk profile to determine adjuvant radiotherapy in endometrial cancer: Evaluation of the pilot phase of the PORTEC-4a trial. Gynecol. Oncol. 2018, 151, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Wortman, B.G.; Astreinidou, E.; Laman, M.S.; van der Steen-Banasik, E.M.; Lutgens, L.C.H.W.; Westerveld, H.; Koppe, F.; Slot, A.; van den Berg, H.A.; Nowee, M.E.; et al. Brachytherapy quality assurance in the PORTEC-4a trial for molecular-integrated risk profile guided adjuvant treatment of endometrial cancer. Radiother. Oncol. 2020, 155, 160–166. [Google Scholar] [CrossRef] [PubMed]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef] [PubMed]

| TCGA Classification | MH Guide Classification | ||||
|---|---|---|---|---|---|
| POLE | MSI | CN-Low | CN-High | ||
| POLE | 17/232 | 17 (100%) | 0 | 0 | 0 |
| MSI | 65/232 | 0 | 58 (89%) | 6 (9%) | 1 (2%) |
| CN-low | 90/232 | 0 | 0 | 80 (89%) | 10 (11%) |
| CN-high | 60/232 | 0 | 0 | 9 (15%) | 51 (85%) |
| Original TCGA Classification 36-Month PFS (n) | MH Guide Classification 36-Month PFS (n) | |
|---|---|---|
| POLE | 1.00 (17) | 1.00 (17) |
| MSI | 0.81 (61) | 0.79 (56) |
| CN-low | 0.87 (88) | 0.88 (91) |
| CN-high | 0.60 (54) | 0.58 (56) |
| Original TCGA Classification 36-Month OS (n) | MH Guide Classification 36-Month OS (n) | |
|---|---|---|
| POLE | 1.00 (17) | 1.00 (17) |
| MSI | 0.84 (65) | 0.87 (58) |
| CN-low | 0.95 (90) | 0.91 (95) |
| CN-high | 0.81 (60) | 0.82 (62) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mustea, A.; Ralser, D.J.; Egger, E.; Ziehm, U.; Vivas, S.; Brock, S.; Jackson, D.; Condic, M.; Meisel, C.; Otten, L.; et al. Determination of the Cancer Genome Atlas (TCGA) Endometrial Cancer Molecular Subtypes Using the Variant Interpretation and Clinical Decision Support Software MH Guide. Cancers 2023, 15, 2053. https://doi.org/10.3390/cancers15072053
Mustea A, Ralser DJ, Egger E, Ziehm U, Vivas S, Brock S, Jackson D, Condic M, Meisel C, Otten L, et al. Determination of the Cancer Genome Atlas (TCGA) Endometrial Cancer Molecular Subtypes Using the Variant Interpretation and Clinical Decision Support Software MH Guide. Cancers. 2023; 15(7):2053. https://doi.org/10.3390/cancers15072053
Chicago/Turabian StyleMustea, Alexander, Damian J. Ralser, Eva Egger, Ulrike Ziehm, Sonia Vivas, Stephan Brock, David Jackson, Mateja Condic, Christian Meisel, Lucia Otten, and et al. 2023. "Determination of the Cancer Genome Atlas (TCGA) Endometrial Cancer Molecular Subtypes Using the Variant Interpretation and Clinical Decision Support Software MH Guide" Cancers 15, no. 7: 2053. https://doi.org/10.3390/cancers15072053
APA StyleMustea, A., Ralser, D. J., Egger, E., Ziehm, U., Vivas, S., Brock, S., Jackson, D., Condic, M., Meisel, C., Otten, L., Laib, A., Cordova, M. C., Hartmann, R., Stein, M. A., Koensgen, D., & Stope, M. B. (2023). Determination of the Cancer Genome Atlas (TCGA) Endometrial Cancer Molecular Subtypes Using the Variant Interpretation and Clinical Decision Support Software MH Guide. Cancers, 15(7), 2053. https://doi.org/10.3390/cancers15072053







