The Interaction between Gut Microbiota and Host Amino Acids Metabolism in Multiple Myeloma
Abstract
Simple Summary
Abstract
1. Introduction
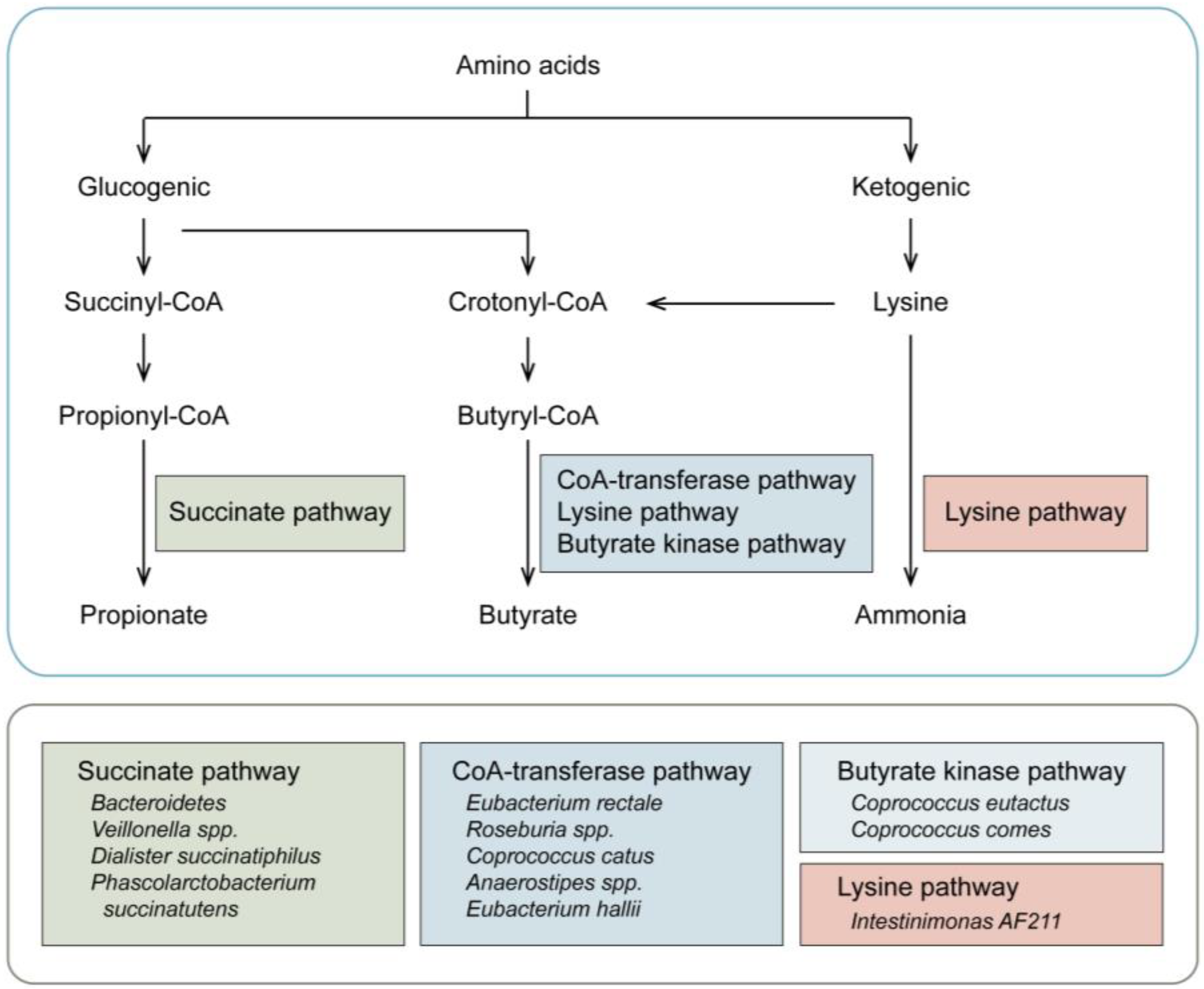
2. Gut Microbiota and Amino Acid Catabolism
3. Abnormal Amino Acid Metabolism and Multiple Myeloma
3.1. The Effects of Abnormal Amino Acid Metabolism on MM
3.2. The Effects of Abnormal Amino Acid Metabolism on MM Progression
3.3. The Effects of Abnormal Amino Acid Metabolism on MM Therapy
4. Gut Microbiota and Multiple Myeloma
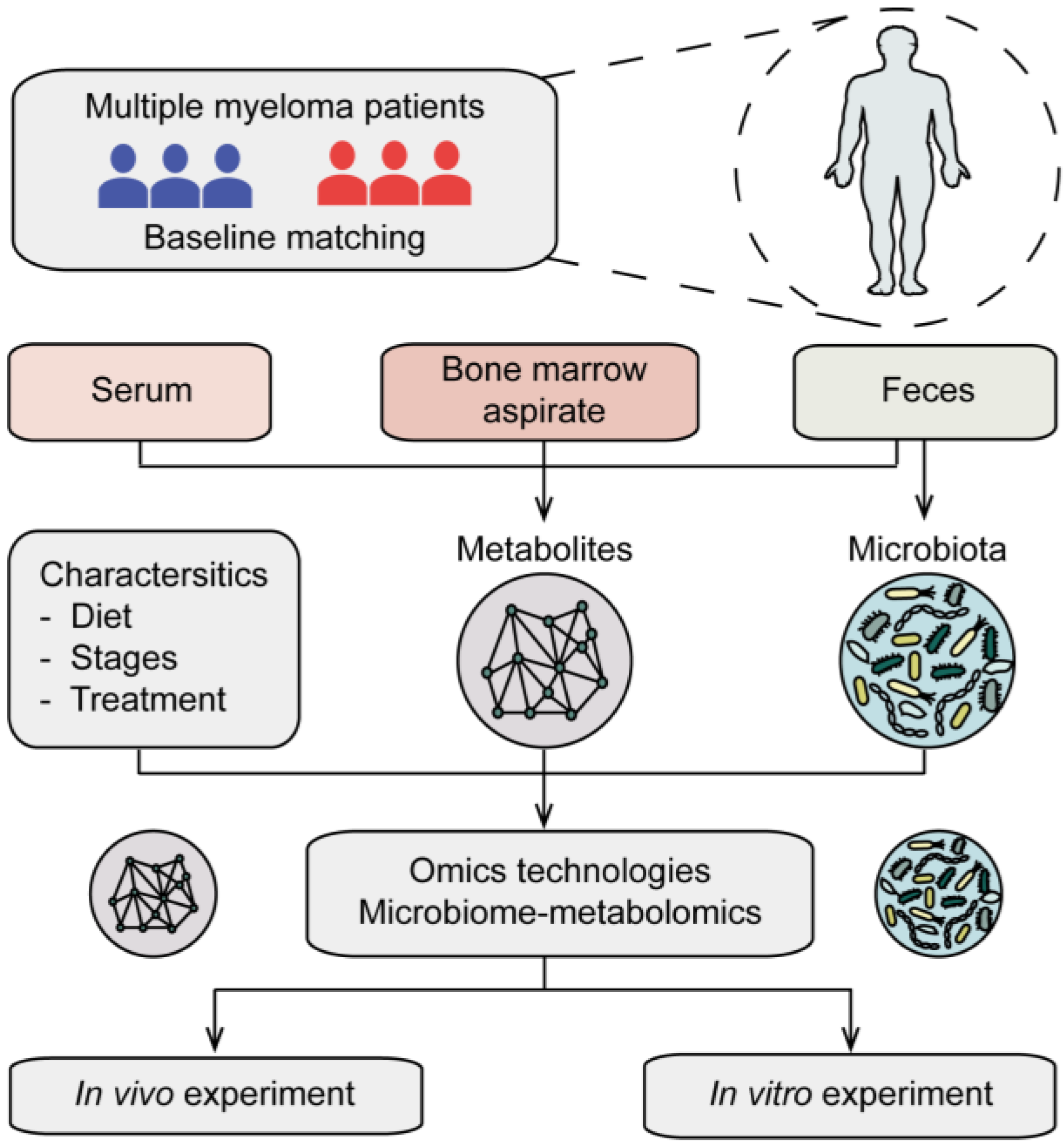
4.1. Gut Microbiota Composition and MM
4.2. Gut Microbiota and MM Progression
4.3. Gut Microbiota and MM Therapy
4.3.1. Gut Microbiota and Chemotherapy
4.3.2. Gut Microbiota and Hematopoietic Stem Cell Transplantation
4.3.3. Gut Microbiota and CAR-T Therapy
4.4. Gut Microbiota and Therapeutic Response
| Study (Year) | Patient Population | Microbiome Technique | Alteration of Microbiome | The Differential Microbiota | Findings |
|---|---|---|---|---|---|
| Gut microbiota and MM progression | |||||
| Jian X. 2020 [26] | Healthy: n = 18 MM patients: n = 19 | Metagenomic shotgun sequencing /qPCR | The lower diversity in MM patients compared to the healthy controls |
|
|
| Zhang B. 2019 [57] | Healthy: n = 17 MM patients: n = 61 | 16S rRNA /qPCR | The lower diversity in MM patients compared to the healthy |
|
|
| Calcinotto A. 2018 [61] | MM mice | 16S rRNA | NA |
|
|
| Gut microbiota and therapeutic response | |||||
| Huang EY. 2015 [64] | Mice | 16S rRNA | NA |
|
|
| Viaud S. 2013 [65] | Mice | 16S rRNA /qPCR | The disruption of the intestinal barrier and alteration of composition by CTX treatment |
|
|
| Pianko MJ. 2019 [79] | MM patients: n = 34 | 16S rRNA | No significant difference in the α diversity between MRD- and MRD+ patients |
|
|
| Gut microbiota and HSCT in MM | |||||
| Khan N. 2021 [71] | MM with auto-HSCT n = 272 | 16S rRNA | The lower diversity of the early pretransplant period and decreasing further during transplantation |
|
|
| D’Angelo C. 2023 [72] | MM with auto-HSCT n = 30 | 16S rRNA | A significant loss of diversity following transplant |
|
|
| El-Jurdi N. 2019 [73] | MM with auto-HSCT n = 15 | 16S rRNA | The decrease in diversity, and the recovery within 1 month after transplantation |
|
|
| Taur Y. 2012 [75] | MM with allo-HSCT n = 8 | 16S rRNA | The reduction of diversity during allo-HSCT |
|
|
| Hu Y. 2022 [78] | RRMM n = 43 | 16S rRNA | A significant decrease in diversity after the CAR-T therapy |
|
|
5. Gut Microbiota-Host Amino Acid Metabolism Interaction on Multiple Myeloma
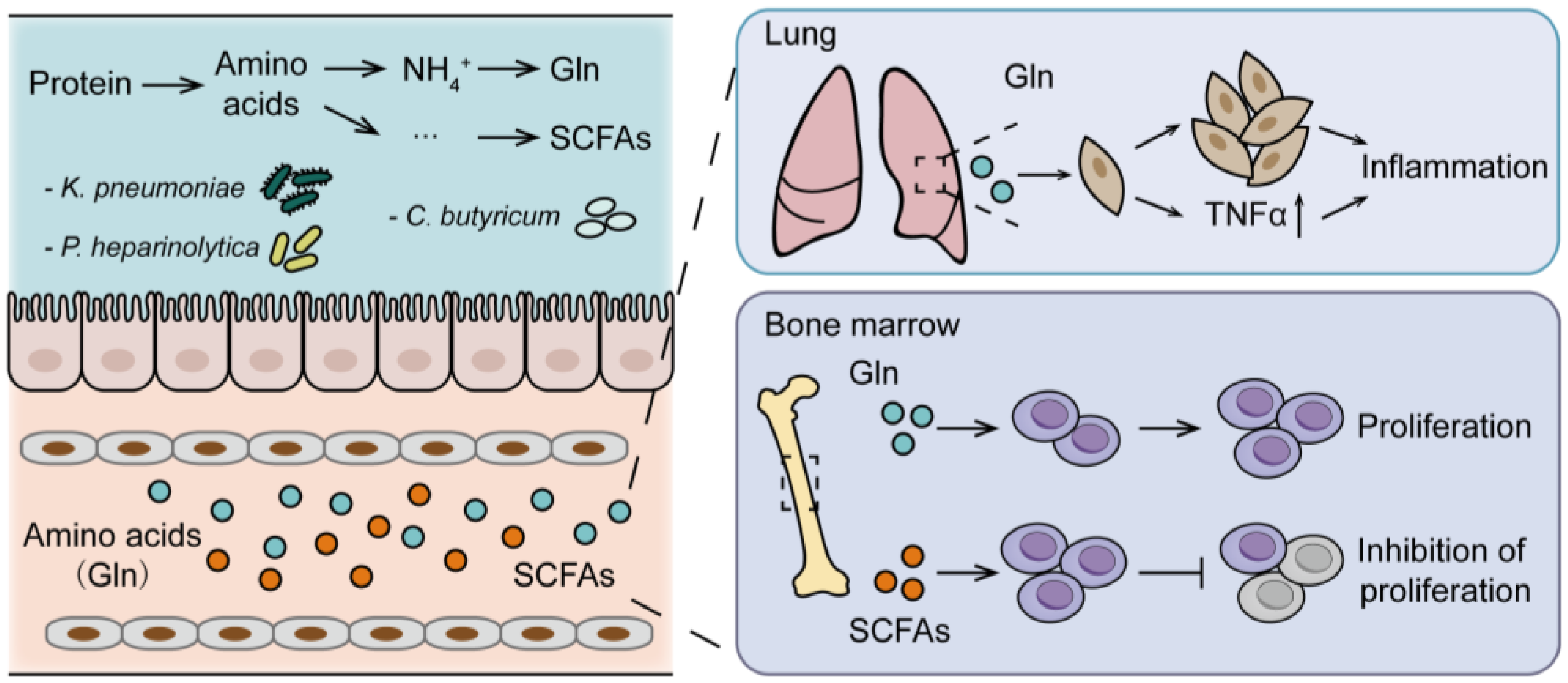
5.1. Gut Microbiota-Host Glutamine Metabolism Interaction on MM Progression
5.2. Gut Microbiota-Host Short-Chain Fatty Acids Metabolism Interaction on MM Progression
5.3. Clinical Translational Insights of the Microbiota-Host Amino Acid Metabolism Interactions
6. The Challenges of Gut Microbiota in Multiple Myeloma: Future Directions
6.1. Limitations of Microbiome Analysis and Studies
6.2. Challenges of the Microbiome in Future Clinical Translations
6.3. Future Directions
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- van de Donk, N.; Pawlyn, C.; Yong, K.L. Multiple myeloma. Lancet 2021, 397, 410–427. [Google Scholar] [CrossRef] [PubMed]
- Cowan, A.J.; Green, D.J.; Kwok, M.; Lee, S.; Coffey, D.G.; Holmberg, L.A.; Tuazon, S.; Gopal, A.K.; Libby, E.N. Diagnosis and Management of Multiple Myeloma: A Review. JAMA 2022, 327, 464–477. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.; Kumar, S.K.; San Miguel, J.; Davies, F.; Zamagni, E.; Bahlis, N.; Ludwig, H.; Mikhael, J.; Terpos, E.; Schjesvold, F.; et al. Treatment of relapsed and refractory multiple myeloma: Recommendations from the International Myeloma Working Group. Lancet Oncol. 2021, 22, e105–e118. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271–285. [Google Scholar] [CrossRef]
- Khanna, S.; Tosh, P.K. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin. Proc. 2014, 89, 107–114. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Tulstrup, M.V.; Christensen, E.G.; Carvalho, V.; Linninge, C.; Ahrné, S.; Højberg, O.; Licht, T.R.; Bahl, M.I. Antibiotic Treatment Affects Intestinal Permeability and Gut Microbial Composition in Wistar Rats Dependent on Antibiotic Class. PLoS ONE 2015, 10, e0144854. [Google Scholar] [CrossRef]
- Faith, J.J.; Guruge, J.L.; Charbonneau, M.; Subramanian, S.; Seedorf, H.; Goodman, A.L.; Clemente, J.C.; Knight, R.; Heath, A.C.; Leibel, R.L.; et al. The long-term stability of the human gut microbiota. Science 2013, 341, 1237439. [Google Scholar] [CrossRef]
- Dzutsev, A.; Goldszmid, R.S.; Viaud, S.; Zitvogel, L.; Trinchieri, G. The role of the microbiota in inflammation, carcinogenesis, and cancer therapy. Eur. J. Immunol. 2015, 45, 17–31. [Google Scholar] [CrossRef]
- Couturier-Maillard, A.; Secher, T.; Rehman, A.; Normand, S.; De Arcangelis, A.; Haesler, R.; Huot, L.; Grandjean, T.; Bressenot, A.; Delanoye-Crespin, A.; et al. NOD2-mediated dysbiosis predisposes mice to transmissible colitis and colorectal cancer. J. Clin. Investig. 2013, 123, 700–711. [Google Scholar] [CrossRef]
- Hu, B.; Elinav, E.; Huber, S.; Strowig, T.; Hao, L.; Hafemann, A.; Jin, C.; Wunderlich, C.; Wunderlich, T.; Eisenbarth, S.C.; et al. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 9862–9867. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Loo, T.M.; Kamachi, F.; Watanabe, Y.; Yoshimoto, S.; Kanda, H.; Arai, Y.; Nakajima-Takagi, Y.; Iwama, A.; Koga, T.; Sugimoto, Y.; et al. Gut Microbiota Promotes Obesity-Associated Liver Cancer through PGE(2)-Mediated Suppression of Antitumor Immunity. Cancer Discov. 2017, 7, 522–538. [Google Scholar] [CrossRef]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Cao, Y. Adipocyte and lipid metabolism in cancer drug resistance. J. Clin. Investig. 2019, 129, 3006–3017. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef] [PubMed]
- Singh, V.; Yeoh, B.S.; Chassaing, B.; Xiao, X.; Saha, P.; Aguilera Olvera, R.; Lapek, J.D., Jr.; Zhang, L.; Wang, W.B.; Hao, S.; et al. Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 2018, 175, 679–694.e22. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Xia, J.; Zhang, J.; Wu, X.; Du, W.; Zhu, Y.; Liu, X.; Liu, Z.; Meng, B.; Guo, J.; Yang, Q.; et al. Blocking glycine utilization inhibits multiple myeloma progression by disrupting glutathione balance. Nat. Commun. 2022, 13, 4007. [Google Scholar] [CrossRef]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef]
- Marcobal, A.; Kashyap, P.C.; Nelson, T.A.; Aronov, P.A.; Donia, M.S.; Spormann, A.; Fischbach, M.A.; Sonnenburg, J.L. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013, 7, 1933–1943. [Google Scholar] [CrossRef]
- Fan, P.; Li, L.; Rezaei, A.; Eslamfam, S.; Che, D.; Ma, X. Metabolites of Dietary Protein and Peptides by Intestinal Microbes and their Impacts on Gut. Curr. Protein Pept. Sci. 2015, 16, 646–654. [Google Scholar] [CrossRef]
- Andriamihaja, M.; Davila, A.M.; Eklou-Lawson, M.; Petit, N.; Delpal, S.; Allek, F.; Blais, A.; Delteil, C.; Tomé, D.; Blachier, F. Colon luminal content and epithelial cell morphology are markedly modified in rats fed with a high-protein diet. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G1030–G1037. [Google Scholar] [CrossRef]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef]
- Eklou-Lawson, M.; Bernard, F.; Neveux, N.; Chaumontet, C.; Bos, C.; Davila-Gay, A.M.; Tomé, D.; Cynober, L.; Blachier, F. Colonic luminal ammonia and portal blood L-glutamine and L-arginine concentrations: A possible link between colon mucosa and liver ureagenesis. Amino Acids 2009, 37, 751–760. [Google Scholar] [CrossRef]
- Mouillé, B.; Robert, V.; Blachier, F. Adaptative increase of ornithine production and decrease of ammonia metabolism in rat colonocytes after hyperproteic diet ingestion. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G344–G351. [Google Scholar] [CrossRef]
- Bui, T.P.; Ritari, J.; Boeren, S.; de Waard, P.; Plugge, C.M.; de Vos, W.M. Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 2015, 6, 10062. [Google Scholar] [CrossRef]
- O’Keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Russell, W.R.; Duncan, S.H.; Scobbie, L.; Duncan, G.; Cantlay, L.; Calder, A.G.; Anderson, S.E.; Flint, H.J. Major phenylpropanoid-derived metabolites in the human gut can arise from microbial fermentation of protein. Mol. Nutr. Food Res. 2013, 57, 523–535. [Google Scholar] [CrossRef]
- Ou, J.; Carbonero, F.; Zoetendal, E.G.; DeLany, J.P.; Wang, M.; Newton, K.; Gaskins, H.R.; O’Keefe, S.J. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am. J. Clin. Nutr. 2013, 98, 111–120. [Google Scholar] [CrossRef]
- Russell, W.R.; Gratz, S.W.; Duncan, S.H.; Holtrop, G.; Ince, J.; Scobbie, L.; Duncan, G.; Johnstone, A.M.; Lobley, G.E.; Wallace, R.J.; et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 2011, 93, 1062–1072. [Google Scholar] [CrossRef]
- Loh, Y.H.; Jakszyn, P.; Luben, R.N.; Mulligan, A.A.; Mitrou, P.N.; Khaw, K.T. N-Nitroso compounds and cancer incidence: The European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk Study. Am. J. Clin. Nutr. 2011, 93, 1053–1061. [Google Scholar] [CrossRef]
- Gill, C.I.; Rowland, I.R. Diet and cancer: Assessing the risk. Br. J. Nutr. 2002, 88 (Suppl. S1), S73–S87. [Google Scholar] [CrossRef]
- Hanfrey, C.C.; Pearson, B.M.; Hazeldine, S.; Lee, J.; Gaskin, D.J.; Woster, P.M.; Phillips, M.A.; Michael, A.J. Alternative spermidine biosynthetic route is critical for growth of Campylobacter jejuni and is the dominant polyamine pathway in human gut microbiota. J. Biol. Chem. 2011, 286, 43301–43312. [Google Scholar] [CrossRef] [PubMed]
- Pegg, A.E. Toxicity of polyamines and their metabolic products. Chem. Res. Toxicol. 2013, 26, 1782–1800. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, M.L.; Campilongo, R.; Casalino, M.; Micheli, G.; Colonna, B.; Prosseda, G. Polyamines: Emerging players in bacteria-host interactions. Int. J. Med. Microbiol. 2013, 303, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Zeng, P.; Li, Y.; Chai, Y.; Wu, C.; Gao, B. Nontargeted and targeted metabolomics approaches reveal the key amino acid alterations involved in multiple myeloma. PeerJ 2022, 10, e12918. [Google Scholar] [CrossRef]
- Chanukuppa, V.; More, T.H.; Taunk, K.; Taware, R.; Chatterjee, T.; Sharma, S.; Rapole, S. Serum metabolomic alterations in multiple myeloma revealed by targeted and untargeted metabolomics approaches: A pilot study. RSC Adv. 2019, 9, 29522–29532. [Google Scholar] [CrossRef]
- Grisolia, F.T.; Cohen, P.P. Amino acid analysis of serum proteins in multiple myeloma. Cancer Res. 1953, 13, 851–854. [Google Scholar]
- Nair, R.; Gupta, P.; Shanmugam, M. Mitochondrial metabolic determinants of multiple myeloma growth, survival, and therapy efficacy. Front. Oncol. 2022, 12, 1000106. [Google Scholar] [CrossRef]
- Steiner, N.; Müller, U.; Hajek, R.; Sevcikova, S.; Borjan, B.; Jöhrer, K.; Göbel, G.; Pircher, A.; Gunsilius, E. The metabolomic plasma profile of myeloma patients is considerably different from healthy subjects and reveals potential new therapeutic targets. PLoS ONE 2018, 13, e0202045. [Google Scholar] [CrossRef]
- Fei, F.; Ma, T.; Zhou, X.; Zheng, M.; Cao, B.; Li, J. Metabolic markers for diagnosis and risk-prediction of multiple myeloma. Life Sci. 2021, 265, 118852. [Google Scholar] [CrossRef]
- Wu, X.; Xia, J.; Zhang, J.; Zhu, Y.; Wu, Y.; Guo, J.; Chen, S.; Lei, Q.; Meng, B.; Kuang, C.; et al. Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br. J. Haematol. 2020, 190, 52–66. [Google Scholar] [CrossRef]
- Lv, J.; Sun, H.; Gong, L.; Wei, X.; He, Y.; Yu, Z.; Liu, L.; Yi, S.; Sui, W.; Xu, Y.; et al. Aberrant metabolic processes promote the immunosuppressive microenvironment in multiple myeloma. Front. Immunol. 2022, 13, 1077768. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, Q.; Zhu, Y.; Jian, X.; Guo, J.; Zhang, J.; Kuang, C.; Feng, X.; An, G.; Qiu, L.; et al. Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma. Cancers 2022, 14, 4188. [Google Scholar] [CrossRef]
- Zhang, J.; Shi, F.; Liu, X.; Wu, X.; Hu, C.; Guo, J.; Yang, Q.; Xia, J.; He, Y.; An, G.; et al. Proline promotes proliferation and drug resistance of multiple myeloma by downregulation of proline dehydrogenase. Br. J. Haematol. 2023. [Google Scholar] [CrossRef]
- Prelowska, M.K.; Mehlich, D.; Ugurlu, M.T.; Kedzierska, H.; Cwiek, A.; Kosnik, A.; Kaminska, K.; Marusiak, A.A.; Nowis, D. Inhibition of the l-glutamine transporter ASCT2 sensitizes plasma cell myeloma cells to proteasome inhibitors. Cancer Lett. 2021, 507, 13–25. [Google Scholar] [CrossRef]
- Soncini, D.; Minetto, P.; Martinuzzi, C.; Becherini, P.; Fenu, V.; Guolo, F.; Todoerti, K.; Calice, G.; Contini, P.; Miglino, M.; et al. Amino acid depletion triggered by l-asparaginase sensitizes MM cells to carfilzomib by inducing mitochondria ROS-mediated cell death. Blood Adv. 2020, 4, 4312–4326. [Google Scholar] [CrossRef]
- Bajpai, R.; Matulis, S.M.; Wei, C.; Nooka, A.K.; Von Hollen, H.E.; Lonial, S.; Boise, L.H.; Shanmugam, M. Targeting glutamine metabolism in multiple myeloma enhances BIM binding to BCL-2 eliciting synthetic lethality to venetoclax. Oncogene 2016, 35, 3955–3964. [Google Scholar] [CrossRef]
- Zhang, B.; Gu, J.; Liu, J.; Huang, B.; Li, J. Fecal Microbiota Taxonomic Shifts in Chinese Multiple Myeloma Patients Analyzed by Quantitative Polimerase Chain Reaction (QPCR) and 16S rRNA High-Throughput Sequencing. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 8269–8280. [Google Scholar] [CrossRef]
- Pepeljugoski, C.A. Analysis of Intestinal Microbiome in Multiple Myeloma Reveals Progressive Dysbiosis Compared to MGUS and Healthy Individuals. Available online: https://ash.confex.com/ash/2019/webprogram/Paper130643.html (accessed on 13 November 2019).
- Ahmed, N.; Ghannoum, M.; Gallogly, M.; de Lima, M.; Malek, E. Influence of gut microbiome on multiple myeloma: Friend or foe? J. Immunother. Cancer 2020, 8, e000576. [Google Scholar] [CrossRef]
- Kwon, O.; Lee, S.; Kim, J.H.; Kim, H.; Lee, S.W. Altered Gut Microbiota Composition in Rag1-deficient Mice Contributes to Modulating Homeostasis of Hematopoietic Stem and Progenitor Cells. Immune Netw. 2015, 15, 252–259. [Google Scholar] [CrossRef]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef] [PubMed]
- McAllister, F.; Bailey, J.M.; Alsina, J.; Nirschl, C.J.; Sharma, R.; Fan, H.; Rattigan, Y.; Roeser, J.C.; Lankapalli, R.H.; Zhang, H.; et al. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell 2014, 25, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Muranski, P.; Restifo, N.P. Essentials of Th17 cell commitment and plasticity. Blood 2013, 121, 2402–2414. [Google Scholar] [CrossRef] [PubMed]
- Huang, E.Y.; Inoue, T.; Leone, V.A.; Dalal, S.; Touw, K.; Wang, Y.; Musch, M.W.; Theriault, B.; Higuchi, K.; Donovan, S.; et al. Using corticosteroids to reshape the gut microbiome: Implications for inflammatory bowel diseases. Inflamm. Bowel Dis. 2015, 21, 963–972. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Alkharabsheh, O.; Sidiqi, M.H.; Aljama, M.A.; Gertz, M.A.; Frankel, A.E. The Human Microbiota in Multiple Myeloma and Proteasome Inhibitors. Acta Haematol. 2020, 143, 118–123. [Google Scholar] [CrossRef]
- Jain, T.; Sonbol, M.B.; Firwana, B.; Kolla, K.R.; Almader-Douglas, D.; Palmer, J.; Fonseca, R. High-Dose Chemotherapy with Early Autologous Stem Cell Transplantation Compared to Standard Dose Chemotherapy or Delayed Transplantation in Patients with Newly Diagnosed Multiple Myeloma: A Systematic Review and Meta-Analysis. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2019, 25, 239–247. [Google Scholar] [CrossRef]
- Routy, B.; Letendre, C.; Enot, D.; Chenard-Poirier, M.; Mehraj, V.; Seguin, N.C.; Guenda, K.; Gagnon, K.; Woerther, P.L.; Ghez, D.; et al. The influence of gut-decontamination prophylactic antibiotics on acute graft-versus-host disease and survival following allogeneic hematopoietic stem cell transplantation. Oncoimmunology 2017, 6, e1258506. [Google Scholar] [CrossRef]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Velardi, E.; Tsai, J.J.; Slingerland, A.E.; Smith, O.M.; Young, L.F.; Gupta, J.; et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med. 2016, 8, 339ra71. [Google Scholar] [CrossRef]
- D’Angelo, C. Impact of Antibiotics on Gut Microbiota Diversity and the Results of a Prospective Dietary Assessment in Patients with Multiple Myeloma Undergoing Autologous Hematopoietic Stem Cell Transplantation. Available online: https://ash.confex.com/ash/2019/webprogram/Paper123530.html (accessed on 13 November 2019).
- Khan, N.; Lindner, S.; Gomes, A.L.C.; Devlin, S.M.; Shah, G.L.; Sung, A.D.; Sauter, C.S.; Landau, H.J.; Dahi, P.B.; Perales, M.A.; et al. Fecal microbiota diversity disruption and clinical outcomes after auto-HCT: A multicenter observational study. Blood 2021, 137, 1527–1537. [Google Scholar] [CrossRef]
- D’Angelo, C.; Sudakaran, S.; Asimakopoulos, F.; Hematti, P.; El-Gamal, D.; Safdar, N.; Callander, N. Perturbation of the gut microbiome and association with outcomes following autologous stem cell transplantation in patients with multiple myeloma. Leuk. Lymphoma 2023, 64, 87–97. [Google Scholar] [CrossRef]
- El Jurdi, N.; Filali-Mouhim, A.; Salem, I.; Retuerto, M.; Dambrosio, N.M.; Baer, L.; Lazarus, H.M.; Caimi, P.; Cooper, B.; Tomlinson, B.; et al. Gastrointestinal Microbiome and Mycobiome Changes during Autologous Transplantation for Multiple Myeloma: Results of a Prospective Pilot Study. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2019, 25, 1511–1519. [Google Scholar] [CrossRef]
- Zhang, L.; Xiang, Y.; Li, Y.; Zhang, J. Gut microbiome in multiple myeloma: Mechanisms of progression and clinical applications. Front. Immunol. 2022, 13, 1058272. [Google Scholar] [CrossRef]
- Taur, Y.; Xavier, J.B.; Lipuma, L.; Ubeda, C.; Goldberg, J.; Gobourne, A.; Lee, Y.J.; Dubin, K.A.; Socci, N.D.; Viale, A.; et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2012, 55, 905–914. [Google Scholar] [CrossRef]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef]
- CD19 CAR T-cell Clinical Outcome Is Associated with the Gut Microbiome. Cancer Discov. 2022, 12, 1182. [CrossRef]
- Hu, Y.; Li, J.; Ni, F.; Yang, Z.; Gui, X.; Bao, Z.; Zhao, H.; Wei, G.; Wang, Y.; Zhang, M.; et al. CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat. Commun. 2022, 13, 5313. [Google Scholar] [CrossRef]
- Pianko, M.J.; Devlin, S.M.; Littmann, E.R.; Chansakul, A.; Mastey, D.; Salcedo, M.; Fontana, E.; Ling, L.; Tavitian, E.; Slingerland, J.B.; et al. Minimal residual disease negativity in multiple myeloma is associated with intestinal microbiota composition. Blood Adv. 2019, 3, 2040–2044. [Google Scholar] [CrossRef]
- Shah, U.A.; Maclachlan, K.H.; Derkach, A.; Salcedo, M.; Barnett, K.; Caple, J.; Blaslov, J.; Tran, L.; Ciardiello, A.; Burge, M.; et al. Sustained Minimal Residual Disease Negativity in Multiple Myeloma is Associated with Stool Butyrate and Healthier Plant-Based Diets. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 5149–5155. [Google Scholar] [CrossRef]
- Bolzoni, M.; Chiu, M.; Accardi, F.; Vescovini, R.; Airoldi, I.; Storti, P.; Todoerti, K.; Agnelli, L.; Missale, G.; Andreoli, R.; et al. Dependence on glutamine uptake and glutamine addiction characterize myeloma cells: A new attractive target. Blood 2016, 128, 667–679. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Kodama, M.; Oshikawa, K.; Shimizu, H.; Yoshioka, S.; Takahashi, M.; Izumi, Y.; Bamba, T.; Tateishi, C.; Tomonaga, T.; Matsumoto, M.; et al. A shift in glutamine nitrogen metabolism contributes to the malignant progression of cancer. Nat. Commun. 2020, 11, 1320. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bai, C.; Ruan, Y.; Liu, M.; Chu, Q.; Qiu, L.; Yang, C.; Li, B. Coordinative metabolism of glutamine carbon and nitrogen in proliferating cancer cells under hypoxia. Nat. Commun. 2019, 10, 201. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.; Toscani, D.; Marchica, V.; Taurino, G.; Costa, F.; Bianchi, M.G.; Andreoli, R.; Franceschi, V.; Storti, P.; Burroughs-Garcia, J.; et al. Myeloma Cells Deplete Bone Marrow Glutamine and Inhibit Osteoblast Differentiation Limiting Asparagine Availability. Cancers 2020, 12, 3267. [Google Scholar] [CrossRef] [PubMed]
- Mai, E.K.; Haas, E.M.; Lucke, S.; Lopprich, M.; Kunz, C.; Pritsch, M.; Knaup-Gregori, P.; Raab, M.S.; Schlenzka, J.; Bertsch, U.; et al. A systematic classification of death causes in multiple myeloma. Blood Cancer J. 2018, 8, 30. [Google Scholar] [CrossRef]
- Yao, C.K.; Muir, J.G.; Gibson, P.R. Review article: Insights into colonic protein fermentation, its modulation and potential health implications. Aliment. Pharmacol. Ther. 2016, 43, 181–196. [Google Scholar] [CrossRef]
- Allen-Vercoe, E.; Daigneault, M.; White, A.; Panaccione, R.; Duncan, S.H.; Flint, H.J.; O’Neal, L.; Lawson, P.A. Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. 1976. Anaerobe 2012, 18, 523–529. [Google Scholar] [CrossRef]
- Stoeva, M.K.; Garcia-So, J.; Justice, N.; Myers, J.; Tyagi, S.; Nemchek, M.; McMurdie, P.J.; Kolterman, O.; Eid, J. Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 2021, 13, 1907272. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci. USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef]
- Paulos, C.M.; Wrzesinski, C.; Kaiser, A.; Hinrichs, C.S.; Chieppa, M.; Cassard, L.; Palmer, D.C.; Boni, A.; Muranski, P.; Yu, Z.; et al. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Investig. 2007, 117, 2197–2204. [Google Scholar] [CrossRef]
- Drayson, M.T.; Bowcock, S.; Planche, T.; Iqbal, G.; Pratt, G.; Yong, K.; Wood, J.; Raynes, K.; Higgins, H.; Dawkins, B.; et al. Levofloxacin prophylaxis in patients with newly diagnosed myeloma (TEAMM): A multicentre, double-blind, placebo-controlled, randomised, phase 3 trial. Lancet Oncol. 2019, 20, 1760–1772. [Google Scholar] [CrossRef]
- Albrich, W.C.; Boggian, K. Levofloxacin prophylaxis in patients with myeloma. Lancet Oncol. 2020, 21, e68. [Google Scholar] [CrossRef]
- Teh, B.W.; Harrison, S.J.; Worth, L.J.; Thursky, K.A.; Slavin, M.A. Levofloxacin prophylaxis in patients with myeloma. Lancet Oncol. 2020, 21, e67. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- DeFilipp, Z.; Peled, J.U.; Li, S.; Mahabamunuge, J.; Dagher, Z.; Slingerland, A.E.; Del Rio, C.; Valles, B.; Kempner, M.E.; Smith, M.; et al. Third-party fecal microbiota transplantation following allo-HCT reconstitutes microbiome diversity. Blood Adv. 2018, 2, 745–753. [Google Scholar] [CrossRef]
- Henig, I.; Yehudai-Ofir, D.; Zuckerman, T. The clinical role of the gut microbiome and fecal microbiota transplantation in allogeneic stem cell transplantation. Haematologica 2021, 106, 933–946. [Google Scholar] [CrossRef]
- Shono, Y.; van den Brink, M.R.M. Gut microbiota injury in allogeneic haematopoietic stem cell transplantation. Nat. Rev. Cancer 2018, 18, 283–295. [Google Scholar] [CrossRef]
- Taur, Y.; Coyte, K.; Schluter, J.; Robilotti, E.; Figueroa, C.; Gjonbalaj, M.; Littmann, E.R.; Ling, L.; Miller, L.; Gyaltshen, Y.; et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 2018, 10, eaap9489. [Google Scholar] [CrossRef]
- Gough, E.; Shaikh, H.; Manges, A.R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2011, 53, 994–1002. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Li, X.; Shen, J.; Feng, Q. Fecal Microbiota Transplantation for the Treatment of Inflammatory Bowel Disease: An Update. Front. Pharmacol. 2020, 11, 574533. [Google Scholar] [CrossRef] [PubMed]
- Spindelboeck, W.; Schulz, E.; Uhl, B.; Kashofer, K.; Aigelsreiter, A.; Zinke-Cerwenka, W.; Mulabecirovic, A.; Kump, P.K.; Halwachs, B.; Gorkiewicz, G.; et al. Repeated fecal microbiota transplantations attenuate diarrhea and lead to sustained changes in the fecal microbiota in acute, refractory gastrointestinal graft-versus-host-disease. Haematologica 2017, 102, e210–e213. [Google Scholar] [CrossRef] [PubMed]
- van Lier, Y.F.; Davids, M.; Haverkate, N.J.E.; de Groot, P.F.; Donker, M.L.; Meijer, E.; Heubel-Moenen, F.; Nur, E.; Zeerleder, S.S.; Nieuwdorp, M.; et al. Donor fecal microbiota transplantation ameliorates intestinal graft-versus-host disease in allogeneic hematopoietic cell transplant recipients. Sci. Transl. Med. 2020, 12, eaaz8926. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Lee, D.H.; Fung, T.T.; Tabung, F.K.; Marinac, C.R.; Devore, E.E.; Rosner, B.A.; Ghobrial, I.M.; Colditz, G.A.; Giovannucci, E.L.; Birmann, B.M. Prediagnosis dietary pattern and survival in patients with multiple myeloma. Int. J. Cancer 2020, 147, 1823–1830. [Google Scholar] [CrossRef]
- Wheeler, M.L.; Limon, J.J.; Bar, A.S.; Leal, C.A.; Gargus, M.; Tang, J.; Brown, J.; Funari, V.A.; Wang, H.L.; Crother, T.R.; et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe 2016, 19, 865–873. [Google Scholar] [CrossRef]
- Kernbauer, E.; Ding, Y.; Cadwell, K. An enteric virus can replace the beneficial function of commensal bacteria. Nature 2014, 516, 94–98. [Google Scholar] [CrossRef]
- Hildebrand, F.; Nguyen, T.L.; Brinkman, B.; Yunta, R.G.; Cauwe, B.; Vandenabeele, P.; Liston, A.; Raes, J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013, 14, R4. [Google Scholar] [CrossRef]
- Baxter, N.T.; Zackular, J.P.; Chen, G.Y.; Schloss, P.D. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2014, 2, 20. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1, 6ra14. [Google Scholar] [CrossRef]
- Smith, K.; McCoy, K.D.; Macpherson, A.J. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin. Immunol. 2007, 19, 59–69. [Google Scholar] [CrossRef]
- Plantinga, T.S.; van Maren, W.W.; van Bergenhenegouwen, J.; Hameetman, M.; Nierkens, S.; Jacobs, C.; de Jong, D.J.; Joosten, L.A.; van’t Land, B.; Garssen, J.; et al. Differential Toll-like receptor recognition and induction of cytokine profile by Bifidobacterium breve and Lactobacillus strains of probiotics. Clin. Vaccine Immunol. 2011, 18, 621–628. [Google Scholar] [CrossRef]
- Kadowaki, N.; Ho, S.; Antonenko, S.; Malefyt, R.W.; Kastelein, R.A.; Bazan, F.; Liu, Y.J. Subsets of human dendritic cell precursors express different toll-like receptors and respond to different microbial antigens. J. Exp. Med. 2001, 194, 863–869. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Q.; Wei, Y.; Zhu, Y.; Guo, J.; Zhang, J.; He, Y.; Li, X.; Liu, J.; Zhou, W. The Interaction between Gut Microbiota and Host Amino Acids Metabolism in Multiple Myeloma. Cancers 2023, 15, 1942. https://doi.org/10.3390/cancers15071942
Yang Q, Wei Y, Zhu Y, Guo J, Zhang J, He Y, Li X, Liu J, Zhou W. The Interaction between Gut Microbiota and Host Amino Acids Metabolism in Multiple Myeloma. Cancers. 2023; 15(7):1942. https://doi.org/10.3390/cancers15071942
Chicago/Turabian StyleYang, Qin, Yumou Wei, Yinghong Zhu, Jiaojiao Guo, Jingyu Zhang, Yanjuan He, Xin Li, Jing Liu, and Wen Zhou. 2023. "The Interaction between Gut Microbiota and Host Amino Acids Metabolism in Multiple Myeloma" Cancers 15, no. 7: 1942. https://doi.org/10.3390/cancers15071942
APA StyleYang, Q., Wei, Y., Zhu, Y., Guo, J., Zhang, J., He, Y., Li, X., Liu, J., & Zhou, W. (2023). The Interaction between Gut Microbiota and Host Amino Acids Metabolism in Multiple Myeloma. Cancers, 15(7), 1942. https://doi.org/10.3390/cancers15071942






