Metabolomic Signatures of Scarff–Bloom–Richardson (SBR) Grade in Non-Metastatic Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Population
2.2. Patient Data Collection and Statistical Analysis
2.3. Sample Collection
2.4. Sample Preparation
2.5. LC-MS Analysis
2.6. Data Preprocessing and Metabolite Identification
2.7. Metabolite Selection
2.8. Statistical and Pathway Analyses
3. Results
3.1. Clinical and Tumor Characteristics
3.2. SBR Grade Metabolomic Signature Discriminated between High-Grade (Grade III) and Low-Grade (Grade I–II) Groups
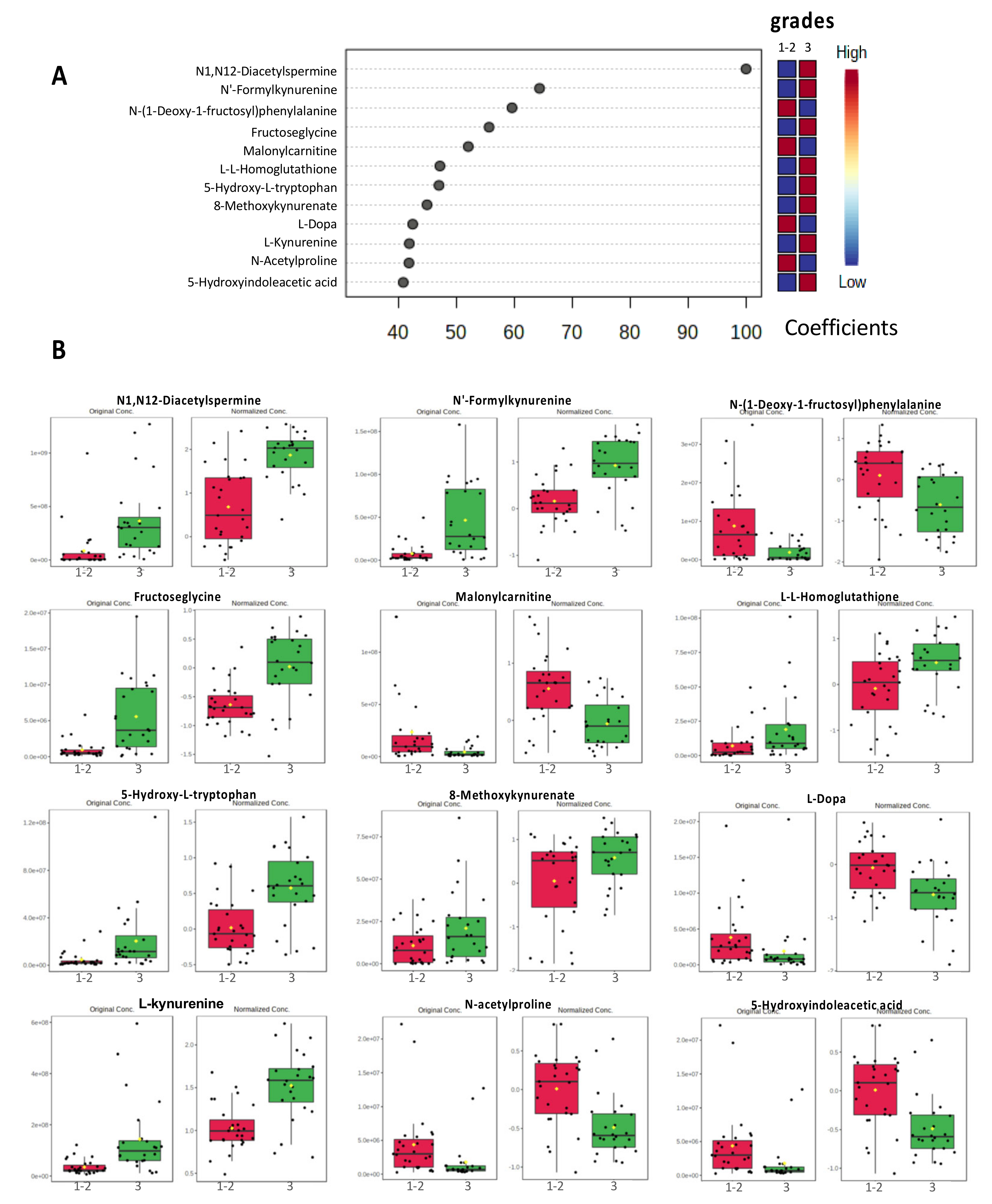
3.3. PLS-DA Models Identified a Discriminatory Signature with the Top 12 Metabolites
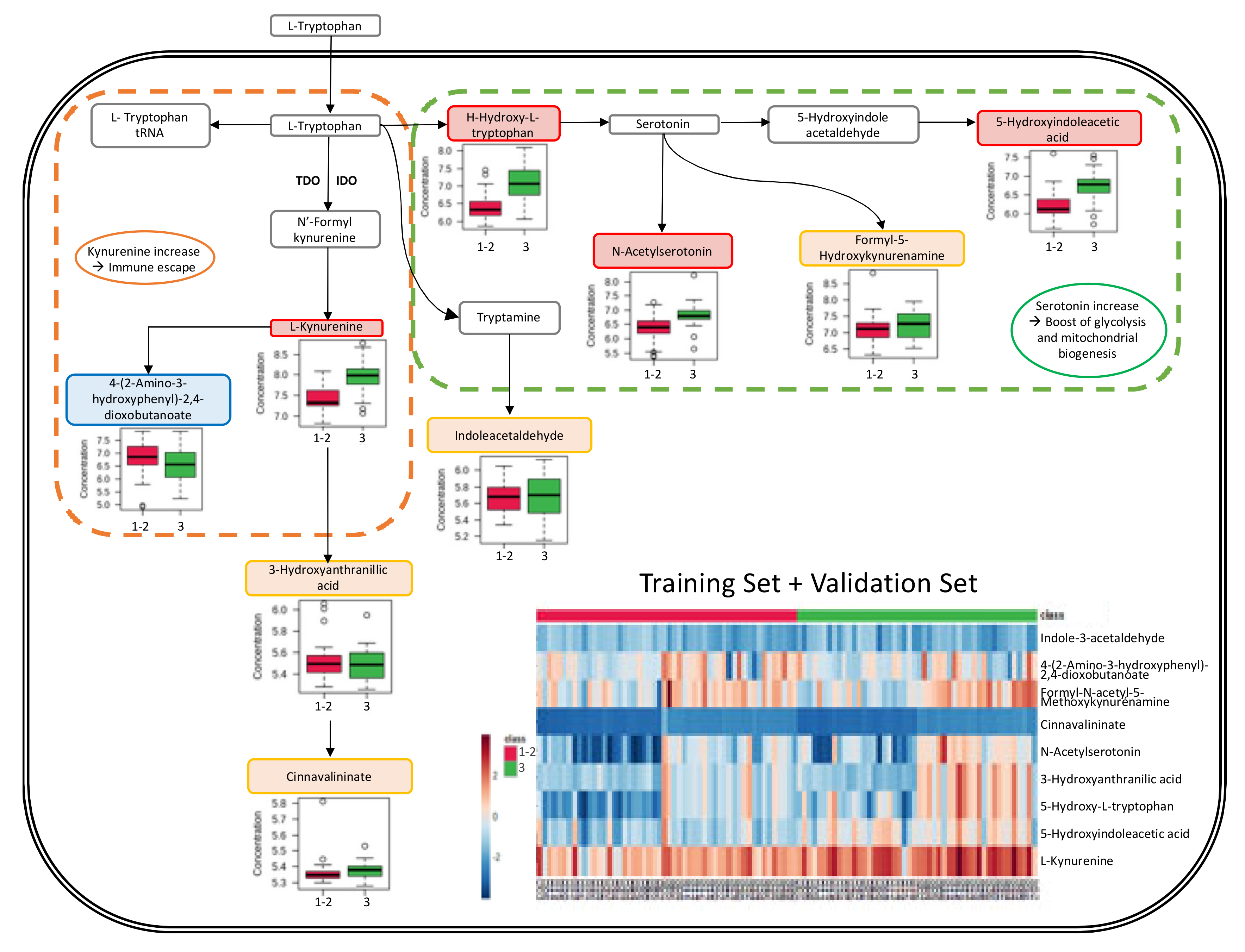
3.4. Metabolic Pathway Analysis
4. Discussion
4.1. Strengths and Weaknesses of the Study
4.2. N1,N12-Diacetylspermine Metabolite (DiAcSpm)
4.3. Kynurenine Synthesis via the Tryptophan Pathway
4.4. Serotonin Implications
4.5. Grade and Immune Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aboud, O.A.; Weiss, R.H. New Opportunities from the Cancer Metabolome. Clin. Chem. 2013, 59, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.; Santos, J.; Ribeiro, M.; Talarico, M.; Viana, L.; Derchain, S. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. [Google Scholar] [CrossRef] [PubMed]
- Mikó, E.; Kovács, T.; Sebő, É.; Tóth, J.; Csonka, T.; Ujlaki, G.; Sipos, A.; Szabó, J.; Méhes, G.; Bai, P. Microbiome—Microbial Metabolome—Cancer Cell Interactions in Breast Cancer—Familiar, but Unexplored. Cells 2019, 8, 293. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, R.; Zhang, L.; Li, Y.; Liu, B.; Kang, H.; Fan, Z.; Tian, Y.; Liu, S.; Li, T. Metabolomics research on potential role for 9-cis-retinoic acid in breast cancer progression. Cancer Sci. 2018, 109, 2315–2326. [Google Scholar] [CrossRef] [PubMed]
- Kapoore, R.V.; Coyle, R.; Staton, C.A.; Brown, N.J.; Vaidyanathan, S. Influence of washing and quenching in profiling the metabolome of adherent mammalian cells: A case study with the metastatic breast cancer cell line MDA-MB-231. Analyst 2017, 142, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Rethinking the war on cancer. Lancet 2014, 383, 558–563. [Google Scholar] [CrossRef]
- Subramani, R.; Poudel, S.; Smith, K.D.; Estrada, A.; Lakshmanaswamy, R. Metabolomics of Breast Cancer: A Review. Metabolites 2022, 12, 643. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.K.; Sharma, P.; Zia, A.; Siwan, D.; Nandave, D.; Nandave, M.; Gautam, R.K. Metabolomics and EMT Markers of Breast Cancer: A Crosstalk and Future Perspective. Pathophysiology 2022, 29, 17. [Google Scholar] [CrossRef]
- Chen, Z.; Li, Z.; Li, H.; Jiang, Y. Metabolomics: A promising diagnostic and therapeutic implement for breast cancer. OncoTargets Ther. 2019, 12, 6797–6811. [Google Scholar] [CrossRef]
- Gong, Y.; Ji, P.; Yang, Y.-S.; Xie, S.; Yu, T.-J.; Xiao, Y.; Jin, M.-L.; Ma, D.; Guo, L.-W.; Pei, Y.-C.; et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021, 33, 51–64.e9. [Google Scholar] [CrossRef] [PubMed]
- Dougan, M.M.; Li, Y.; Chu, L.W.; Haile, R.W.; Whittemore, A.S.; Han, S.S.; Moore, S.C.; Sampson, J.N.; Andrulis, I.L.; John, E.M.; et al. Metabolomic profiles in breast cancer: A pilot case-control study in the breast cancer family registry. BMC Cancer 2018, 18, 532. [Google Scholar] [CrossRef]
- Kanaan, Y.M.; Sampey, B.P.; Beyene, D.; Esnakula, A.K.; Naab, T.J.; Ricks-Santi, L.J.; Dasi, S.; Day, A.; Blackman, K.W.; Frederick, W.; et al. Metabolic profile of triple-negative breast cancer in African-American women reveals potential biomarkers of aggressive disease. Cancer Genom. Proteom. 2014, 11, 279–294. [Google Scholar]
- Kisanga, E.R.; Mellgren, G.; Lien, E.A. Excretion of hydroxylated metabolites of tamoxifen in human bile and urine. Anticancer Res. 2005, 25, 4487–4492. [Google Scholar]
- Visekruna, A.; Luu, M. The Role of Short-Chain Fatty Acids and Bile Acids in Intestinal and Liver Function, Inflammation, and Carcinogenesis. Front. Cell Dev. Biol. 2021, 9, 703218. [Google Scholar] [CrossRef] [PubMed]
- Arnone, A.A.; Cline, J.M.; Soto-Pantoja, D.R.; Cook, K.L. Investigating the role of endogenous estrogens, hormone replacement therapy, and blockade of estrogen receptor-α activity on breast metabolic signaling. Breast Cancer Res. Treat. 2021, 190, 53–67. [Google Scholar] [CrossRef]
- Scarff, R.; Torloni, H. Histological Typing of Breast Tumors; International Histological Classification of Tumours; World Health Organization: Geneva, Switerland, 1968; Volume 2, pp. 13–20. [Google Scholar]
- Bloom, H.J.G.; Richardson, W.W. Histological Grading and Prognosis in Breast Cancer: A Study of 1409 Cases of which 359 have been Followed for 15 Years. Br. J. Cancer 1957, 11, 359–377. [Google Scholar] [CrossRef]
- Prasad Maharjan, R.; Ferenci, T. Global metabolite analysis: The influence of extraction methodology on metabolome profiles of Escherichia coli. Anal. Biochem. 2003, 313, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Jing, L.; Guigonis, J.-M.; Borchiellini, D.; Durand, M.; Pourcher, T.; Ambrosetti, D. LC-MS based metabolomic profiling for renal cell carcinoma histologic subtypes. Sci. Rep. 2019, 9, 15635. [Google Scholar] [CrossRef]
- Hichri, M.; Vassaux, G.; Guigonis, J.-M.; Juhel, T.; Graslin, F.; Guglielmi, J.; Pourcher, T.; Cambien, B. Proteomic Analysis of Iodinated Contrast Agent-Induced Perturbation of Thyroid Iodide Uptake. J. Cell. Mol. 2020, 9, 329. [Google Scholar] [CrossRef] [PubMed]
- Suissa, L.; Guigonis, J.-M.; Graslin, F.; Doche, E.; Osman, O.; Chau, Y.; Sedat, J.; Lindenthal, S.; Pourcher, T. Metabolome of Cerebral Thrombi Reveals an Association between High Glycemia at Stroke Onset and Good Clinical Outcome. Metabolites 2020, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Suissa, L.; Flachon, V.; Guigonis, J.-M.; Olivieri, C.-V.; Burel-Vandenbos, F.; Guglielmi, J.; Ambrosetti, D.; Gérard, M.; Franken, P.; Darcourt, J.; et al. Urinary ketone body loss leads to degeneration of brain white matter in elderly SLC5A8-deficient mice. J. Cereb. Blood Flow Metab. 2020, 40, 1709–1723. [Google Scholar] [CrossRef]
- Suissa, L.; Guigonis, J.-M.; Graslin, F.; Robinet-Borgomano, E.; Chau, Y.; Sedat, J.; Lindenthal, S.; Pourcher, T. Combined Omic Analyzes of Cerebral Thrombi: A New Molecular Approach to Identify Cardioembolic Stroke Origin. Stroke 2021, 52, 2892–2901. [Google Scholar] [CrossRef]
- Castillo-Rivera, F.; Ondo-Méndez, A.; Guglielmi, J.; Guigonis, J.-M.; Jing, L.; Lindenthal, S.; Gonzalez, A.; López, D.; Cambien, B.; Pourcher, T. Tumor microenvironment affects exogenous sodium/iodide symporter expression. Transl. Oncol. 2021, 14, 100937. [Google Scholar] [CrossRef] [PubMed]
- Katajamaa, M.; Orešič, M. Processing methods for differential analysis of LC/MS profile data. BMC Bioinform. 2005, 6, 179. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef] [PubMed]
- Occelli, C.; Guigonis, J.-M.; Lindenthal, S.; Cagnard, A.; Graslin, F.; Brglez, V.; Seitz-Polski, B.; Dellamonica, J.; Levraut, J.; Pourcher, T. Untargeted plasma metabolomic fingerprinting highlights several biomarkers for the diagnosis and prognosis of coronavirus disease 19. Front. Med. 2022, 9, 995069. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.I.; Jamal, Q.; Iqbal, A.; Shaikh, F.; Somroo, S.; Musharraf, S.G. Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci. Rep. 2017, 7, 1715. [Google Scholar] [CrossRef]
- Kato, M.; Onishi, H.; Matsumoto, K.; Motoshita, J.; Tsuruta, N.; Higuchi, K.; Katano, M. Prognostic significance of urine N1, N12-diacetylspermine in patients with non-small cell lung cancer. Anticancer Res. 2014, 34, 3053–3059. [Google Scholar] [PubMed]
- Xu, J.; Chen, Y.; Olopade, O.I. MYC and Breast Cancer. Genes Cancer 2010, 1, 629–640. [Google Scholar] [CrossRef]
- Fallah, Y.; Brundage, J.; Allegakoen, P.; Shajahan-Haq, A.N. MYC-Driven Pathways in Breast Cancer Subtypes. Biomolecules 2017, 7, 53. [Google Scholar] [CrossRef]
- Gatza, M.L.; Lucas, J.E.; Barry, W.T.; Kim, J.W.; Wang, Q.; Crawford, M.D.; Datto, M.B.; Kelley, M.; Mathey-Prevot, B.; Potti, A.; et al. A pathway-based classification of human breast cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 6994–6999. [Google Scholar] [CrossRef] [PubMed]
- Zimmerli, D.; Brambillasca, C.S.; Talens, F.; Bhin, J.; Linstra, R.; Romanens, L.; Bhattacharya, A.; Joosten, S.E.P.; Da Silva, A.M.; Padrao, N.; et al. MYC promotes immune-suppression in triple-negative breast cancer via inhibition of interferon signaling. Nat. Commun. 2022, 13, 6579. [Google Scholar] [CrossRef] [PubMed]
- Bello-Fernandez, C.; Packham, G.; Cleveland, J.L. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc. Natl. Acad. Sci. USA 1993, 90, 7804–7808. [Google Scholar] [CrossRef]
- Bachmann, A.S.; Geerts, D. Polyamine synthesis as a target of MYC oncogenes. J. Biol. Chem. 2018, 293, 18757–18769. [Google Scholar] [CrossRef]
- Funakoshi-Tago, M.; Sumi, K.; Kasahara, T.; Tago, K. Critical Roles of Myc-ODC Axis in the Cellular Transformation Induced by Myeloproliferative Neoplasm-Associated JAK2 V617F Mutant. PLoS ONE 2013, 8, e52844. [Google Scholar] [CrossRef] [PubMed]
- Casero, R.A.; Marton, L.J. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 2007, 6, 373–390. [Google Scholar] [CrossRef]
- Sugimoto, M.; Hiramatsu, K.; Kamei, S.; Kinoshita, K.; Hoshino, M.; Iwasaki, K.; Kawakita, M. Significance of urinaryN 1,N 8-diacetylspermidine andN 1,N 12-diacetylspermine as indicators of neoplastic diseases. J. Cancer Res. Clin. Oncol. 1995, 121, 317–319. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Sugimoto, M.; Kamei, S.; Hoshino, M.; Kinoshita, K.; Iwasaki, K.; Kawakita, M. Diagnostic and prognostic usefulness of N 1, N 8 -diacetylspermidine and N 1, N 12 -diacetylspermine in urine as novel markers of malignancy. J. Cancer Res. Clin. Oncol. 1997, 123, 539–545. [Google Scholar] [CrossRef]
- Hiramatsu, K.; Takahashi, K.; Yamaguchi, T.; Matsumoto, H.; Miyamoto, H.; Tanaka, S.; Tanaka, C.; Tamamori, Y.; Imajo, M.; Kawaguchi, M.; et al. N 1, N 12-Diacetylspermine as a Sensitive and Specific Novel Marker for Early- and Late-Stage Colorectal and Breast Cancers. Clin. Cancer Res. 2005, 11, 2986–2990. [Google Scholar] [CrossRef]
- Cervelli, M.; Bellavia, G.; Fratini, E.; Amendola, R.; Polticelli, F.; Barba, M.; Federico, R.; Signore, F.; Gucciardo, G.; Grillo, R.; et al. Spermine oxidase (SMO) activity in breast tumor tissues and biochemical analysis of the anticancer spermine analogues BENSpm and CPENSpm. BMC Cancer 2010, 10, 555. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Liang, X.; Scott, G.K.; Chang, C.-H.; Baldwin, M.A.; Thomas, T.; Benz, C.C.; Weinstein, I.B. Polyamine inhibition of estrogen receptor (ER) DNA-binding and ligand-binding functions. Breast Cancer Res. Treat. 1998, 48, 243–257. [Google Scholar] [CrossRef]
- Fahrmann, J.F.; Vykoukal, J.; Fleury, A.; Tripathi, S.; Dennison, J.B.; Murage, E.; Wang, P.; Yu, C.-Y.; Capello, M.; Creighton, C.J.; et al. Association between Plasma Diacetylspermine and Tumor Spermine Synthase with Outcome in Triple-Negative Breast Cancer. J. Natl. Cancer Inst. 2020, 112, 607–616. [Google Scholar] [CrossRef]
- Platten, M.; Wick, W.; Van den Eynde, B.J. Tryptophan Catabolism in Cancer: Beyond IDO and Tryptophan Depletion. Cancer Res. 2012, 72, 5435–5440. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Gong, J.; Liu, Y. Indoleamine 2, 3-dioxygenase regulation of immune response (Review). Mol. Med. Rep. 2018, 17, 4867–4873. [Google Scholar] [CrossRef]
- Muller, A.J.; Sharma, M.D.; Chandler, P.R.; DuHadaway, J.B.; Everhart, M.E.; Johnson, B.A.; Kahler, D.J.; Pihkala, J.; Soler, A.P.; Munn, D.H.; et al. Chronic inflammation that facilitates tumor progression creates local immune suppression by inducing indoleamine 2,3 dioxygenase. Proc. Natl. Acad. Sci. USA 2008, 105, 17073–17078. [Google Scholar] [CrossRef]
- Brandacher, G.; Perathoner, A.; Ladurner, R.; Schneeberger, S.; Obrist, P.; Winkler, C.; Werner, E.R.; Werner-Felmayer, G.; Weiss, H.G.; G√∂bel, G.; et al. Prognostic value of indoleamine 2,3-dioxygenase expression in colorectal cancer: Effect on tumor-infiltrating T cells. Clin. Cancer Res. 2006, 12, 1144–1151. [Google Scholar] [CrossRef]
- Ino, K.; Yamamoto, E.; Shibata, K.; Kajiyama, H.; Yoshida, N.; Terauchi, M.; Nawa, A.; Nagasaka, T.; Takikawa, O.; Kikkawa, F. Inverse Correlation between Tumoral Indoleamine 2,3-Dioxygenase Expression and Tumor-Infiltrating Lymphocytes in Endometrial Cancer: Its Association with Disease Progression and Survival. Clin. Cancer Res. 2008, 14, 2310–2317. [Google Scholar] [CrossRef]
- Ino, K.; Yoshida, N.; Kajiyama, H.; Shibata, K.; Yamamoto, E.; Kidokoro, K.; Takahashi, N.; Terauchi, M.; Nawa, A.; Nomura, S.; et al. Indoleamine 2,3-dioxygenase is a novel prognostic indicator for endometrial cancer. Br. J. Cancer 2006, 95, 1555–1561. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Nikaido, T.; Ochiai, K.; Takakura, S.; Saito, M.; Aoki, Y.; Ishii, N.; Yanaihara, N.; Yamada, K.; Takikawa, O.; et al. Indoleamine 2,3-Dioxygenase Serves as a Marker of Poor Prognosis in Gene Expression Profiles of Serous Ovarian Cancer Cells. Clin. Cancer Res. 2005, 11, 6030–6039. [Google Scholar] [CrossRef]
- Nakamura, T.; Shima, T.; Saeki, A.; Hidaka, T.; Nakashima, A.; Takikawa, O.; Saito, S. Expression of indoleamine 2,3-dioxygenase and the recruitment of Foxp3-expressing regulatory T cells in the development and progression of uterine cervical cancer. Cancer Sci. 2007, 98, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Witkiewicz, A.; Williams, T.K.; Cozzitorto, J.; Durkan, B.; Showalter, S.L.; Yeo, C.J.; Brody, J.R. Expression of Indoleamine 2,3-Dioxygenase in Metastatic Pancreatic Ductal Adenocarcinoma Recruits Regulatory T Cells to Avoid Immune Detection. J. Am. Coll. Surg. 2008, 206, 849–854. [Google Scholar] [CrossRef]
- Brody, J.R.; Costantino, C.L.; Berger, A.C.; Sato, T.; Lisanti, M.P.; Yeo, C.J.; Emmons, R.V.; Witkiewicz, A.K. Expression of indoleamine 2,3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle 2009, 8, 1930–1934. [Google Scholar] [CrossRef] [PubMed]
- Seeber, A.; Klinglmair, G.; Fritz, J.; Steinkohl, F.; Zimmer, K.; Aigner, F.; Horninger, W.; Gastl, G.; Zelger, B.; Brunner, A.; et al. High IDO -1 expression in tumor endothelial cells is associated with response to immunotherapy in metastatic renal cell carcinoma. Cancer Sci. 2018, 109, 1583–1591. [Google Scholar] [CrossRef]
- Creelan, B.C.; Antonia, S.J.; Bepler, G.; Garrett, T.J.; Simon, G.R.; Soliman, H.H. Indoleamine 2,3-dioxygenase activity and clinical outcome following induction chemotherapy and concurrent chemoradiation in Stage III non-small cell lung cancer. OncoImmunology 2013, 2, e23428. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Huang, L.; Jin, J.-Y.; Jolly, S.; Zang, Y.; Wu, H.; Yan, L.; Pi, W.; Li, L.; Mellor, A.L.; et al. IDO Immune Status after Chemoradiation May Predict Survival in Lung Cancer Patients. Cancer Res. 2018, 78, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Botticelli, A.; Cerbelli, B.; Lionetto, L.; Zizzari, I.; Salati, M.; Pisano, A.; Federica, M.; Simmaco, M.; Nuti, M.; Marchetti, P. Can IDO activity predict primary resistance to anti-PD-1 treatment in NSCLC? J. Transl. Med. 2018, 16, 219. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Zhu, S.; Li, M.; Li, F.; Wei, F.; Liu, J.; Ren, X. High Indoleamine 2,3-Dioxygenase Is Correlated with Microvessel Density and Worse Prognosis in Breast Cancer. Front. Immunol. 2018, 9, 724. [Google Scholar] [CrossRef]
- D’Amato, N.C.; Rogers, T.J.; Gordon, M.A.; Greene, L.I.; Cochrane, D.R.; Spoelstra, N.S.; Nemkov, T.G.; D’Alessandro, A.; Hansen, K.C.; Richer, J.K. A TDO2-AhR Signaling Axis Facilitates Anoikis Resistance and Metastasis in Triple-Negative Breast Cancer. Cancer Res. 2015, 75, 4651–4664. [Google Scholar] [CrossRef] [PubMed]
- Terness, P.; Bauer, T.M.; Röse, L.; Dufter, C.; Watzlik, A.; Simon, H.; Opelz, G. Inhibition of Allogeneic T Cell Proliferation by Indoleamine 2,3-Dioxygenase–expressing Dendritic Cells. J. Exp. Med. 2002, 196, 447–457. [Google Scholar] [CrossRef]
- Frumento, G.; Rotondo, R.; Tonetti, M.; Damonte, G.; Benatti, U.; Ferrara, G.B. Tryptophan-derived Catabolites Are Responsible for Inhibition of T and Natural Killer Cell Proliferation Induced by Indoleamine 2,3-Dioxygenase. J. Exp. Med. 2002, 196, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Prendergast, G.C.; Malachowski, W.J.; Mondal, A.; Scherle, P.; Muller, A.J. Indoleamine 2,3-Dioxygenase and Its Therapeutic Inhibition in Cancer. Int. Rev. Cell Mol. Biol. 2018, 336, 175–203. [Google Scholar] [PubMed]
- Ye, Z.; Yue, L.; Shi, J.; Shao, M.; Wu, T. Role of IDO and TDO in Cancers and Related Diseases and the Therapeutic Implications. J. Cancer 2019, 10, 2771–2782. [Google Scholar] [CrossRef] [PubMed]
- Ramapriyan, R.; Caetano, M.S.; Barsoumian, H.B.; Mafra, A.C.P.; Zambalde, E.P.; Menon, H.; Tsouko, E.; Welsh, J.W.; Cortez, M.A. Altered cancer metabolism in mechanisms of immunotherapy resistance. Pharmacol. Ther. 2019, 195, 162–171. [Google Scholar] [CrossRef]
- Tang, X.; Lin, C.-C.; Spasojevic, I.; Iversen, E.S.; Chi, J.-T.; Marks, J.R. A joint analysis of metabolomics and genetics of breast cancer. Breast Cancer Res. 2014, 16, 415. [Google Scholar] [CrossRef]
- Olfati, Z.; Rigi, G.; Vaseghi, H.; Zamanzadeh, Z.; Sohrabi, M.; Hejazi, S.H. Evaluation of serotonin receptors (5HTR2A and 5HTR3A) mRNA expression changes in tumor of breast cancer patients. Med. J. Islam. Repub. Iran 2020, 34, 99. [Google Scholar] [CrossRef]
- Ballou, Y.; Rivas, A.; Belmont, A.; Patel, L.; Amaya, C.; Lipson, S.; Khayou, T.; Dickerson, E.; Nahleh, Z.; Bryan, B. 5-HT serotonin receptors modulate mitogenic signaling and impact tumor cell viability. Mol. Clin. Oncol. 2018, 9, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Gautam, J.; Banskota, S.; Regmi, S.C.; Ahn, S.; Jeon, Y.H.; Jeong, H.; Kim, S.J.; Nam, T.; Jeong, B.-S.; Kim, J.-A. Tryptophan hydroxylase 1 and 5-HT7 receptor preferentially expressed in triple-negative breast cancer promote cancer progression through autocrine serotonin signaling. Mol. Cancer 2016, 15, 75. [Google Scholar] [CrossRef]
- Balakrishna, P.; George, S.; Hatoum, H.; Mukherjee, S. Serotonin Pathway in Cancer. Int. J. Mol. Sci. 2021, 22, 1268. [Google Scholar] [CrossRef]
- Jose, J.; Tavares, C.D.J.; Ebelt, N.D.; Lodi, A.; Edupuganti, R.; Xie, X.; Devkota, A.K.; Kaoud, T.S.; Van Den Berg, C.L.; Anslyn, E.V.; et al. Serotonin Analogues as Inhibitors of Breast Cancer Cell Growth. ACS Med. Chem. Lett. 2017, 8, 1072–1076. [Google Scholar] [CrossRef]
- Badawy, A.A.-B. Tryptophan metabolism and disposition in cancer biology and immunotherapy. Biosci. Rep. 2022, 42, BSR20221682. [Google Scholar] [CrossRef] [PubMed]
- Sola-Penna, M.; Paixão, L.P.; Branco, J.R.; Ochioni, A.C.; Albanese, J.M.; Mundim, D.M.; Baptista-de-Souza, D.; Figueiredo, C.P.; Coelho, W.S.; Marcondes, M.C.; et al. Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. Br. J. Cancer 2020, 122, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed]
- Baganz, N.L.; Blakely, R.D. A Dialogue between the Immune System and Brain, Spoken in the Language of Serotonin. ACS Chem. Neurosci. 2013, 4, 48–63. [Google Scholar] [CrossRef]
- Herr, N.; Bode, C.; Duerschmied, D. The Effects of Serotonin in Immune Cells. Front. Cardiovasc. Med. 2017, 4, 48. [Google Scholar] [CrossRef] [PubMed]
- Koretzky, G.A. Multiple Roles of CD4 and CD8 in T Cell Activation. J. Immunol. 2010, 185, 2643–2644. [Google Scholar] [CrossRef] [PubMed]
- Baxevanis, C.N.; Fortis, S.P.; Perez, S.A. The balance between breast cancer and the immune system: Challenges for prognosis and clinical benefit from immunotherapies. Semin. Cancer Biol. 2021, 72, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Vazquez, M.I.; Catalan-Dibene, J.; Zlotnik, A. B cells responses and cytokine production are regulated by their immune microenvironment. Cytokine 2015, 74, 318–326. [Google Scholar] [CrossRef] [PubMed]
- Lal, A.; Chan, L.; DeVries, S.; Chin, K.; Scott, G.K.; Benz, C.C.; Chen, Y.-Y.; Waldman, F.M.; Hwang, E.S. FOXP3-positive regulatory T lymphocytes and epithelial FOXP3 expression in synchronous normal, ductal carcinoma in situ, and invasive cancer of the breast. Breast Cancer Res. Treat. 2013, 139, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Peyraud, F.; Guegan, J.-P.; Bodet, D.; Cousin, S.; Bessede, A.; Italiano, A. Targeting Tryptophan Catabolism in Cancer Immunotherapy Era: Challenges and Perspectives. Front. Immunol. 2022, 13, 807271. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.-A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef] [PubMed]



| Training Set | Validation Set | |||||
|---|---|---|---|---|---|---|
| (n = 51) | (n = 49) | |||||
| N/med | (%/SD) | N/med | (%/SD) | p | ||
| Age | p < 0.00001 (£) | |||||
| median | 65 | 51 | ||||
| min-max | 37–88 | 26–70 | ||||
| Histology | NS ($) | |||||
| DIC | 48 | (82.5%) | 45 | (91.8%) | ||
| LIC | 3 | (12.5%) | 3 | (6.1%) | ||
| other | 0 | (0.0%) | 1 | (2.0%) | ||
| Tumor size (mm) | 30 * | (21.9) | 40 ** | (22.4) | p < 0.00001 (£) | |
| T | p = 0.001 ($) | |||||
| T1 | 13 | (25.5%) | 3 | (6.1%) | ||
| T2 | 26 | (51.0%) | 37 | (75.5%) | ||
| T3 | 11 | (21.6%) | 3 | (6.1%) | ||
| T4 | 1 | (1.9%) | 5 | (10.2%) | ||
| unknown | 0 | (0.0%) | 1 | (2.0%) | ||
| N | p = 0.002 ($) | |||||
| N0 | 26 | (51.0%) | 14 | (28.6%) | ||
| N1 | 18 | (35.3%) | 34 | (69.4%) | ||
| N2 | 3 | (5.9%) | 1 | (2.0%) | ||
| N3 | 3 | (5.9%) | 0 | (0.0%) | ||
| unknown | 1 | (1.9%) | 0 | (0.0%) | ||
| SBR grading | NS ($) | |||||
| I | 5 | (9.8%) | 5 | (10.2%) | ||
| II | 22 | (43.1%) | 20 | (40.8%) | ||
| III | 24 | (47.1%) | 24 | (50.0%) | ||
| Ki67% | NS (£) | |||||
| median | 35 | (29.3) | 60 | (23.0) | ||
| ≤10% | 4 | (7.8%) | 1 | (2.0%) | ||
| Estrogen-receptor | NS (£/$) | |||||
| Mean | 50.2 | (47.9) | 65.4 | (43.6) | ||
| ≥10% of cells | 29 | (56.9%) | 28 | (57.1%) | ||
| Progesteron-receptor | NS (£/$) | |||||
| Mean | 40.3 | (42.5) | 43.4 | (38.3) | ||
| ≥10% of cells | 28 | (54.9%) | 31 | (63.3%) | ||
| HER2-positive receptor | NS ($) | |||||
| HER2 not amplified | 40 | (78.4%) | 41 | (83.7%) | ||
| HER2 amplified | 11 | (21.6%) | 8 | (16.3%) | ||
| NCT Number | Phase | Number of Patients | Trial Title | Intervention | Main Results |
|---|---|---|---|---|---|
| Pharmacological Inhibition of IDO-TDO/IDO Inhibitor | |||||
| NCT02178722 | I/II | 3 TNBC | Study to explore the safety, tolerability and efficacy of MK-3475 combined with INCB024360 in participants with selected cancers | Epacadostat 1 BID combined with pembrolizumab Q3W | Acceptable safety profile TNBC: ORR 10%; DCR 36% |
| NCT02471846 | I | 25 (17 TNBC) | A study of GDC-0919 and atezolizumab combination treatment in participants with locally advanced or metastatic solid tumors | Navoximod BID combined with atezolizumab Q3W | Advanced cancer: PR 9%; ORR 10%, SD 24%; Decreasing plasma Kyn with increasing doses |
| NCT02658890 | I/II | 627 advanced cancer | An investigational immuno-therapy study of BMS-986205 given combined with nivolumab and combined with both nivolumab and ipilimumab in cancers that are advanced or have spread | Linrodostat combined with immunotherapy (nivolumab or nivolumab+ipilimumab) | Acceptable safety profile No efficicacy results yet |
| NCT03343613 | I | 90 advanced cancer | A study of LY3381916 alone or combined with LY3300054 in participants with solid tumors | LY3381916 QD combined with LY3300054 (anti-PD-L1) Q2W | Best response: SD |
| NCT03328026 | I/II | 60 breast cancer | Study of SV-BR-1-GM combined with retifanlimab | Epacadostat + Retifanlimab (anti-PD1) + SV-BR-1-GM (vaccine) | Recruiting |
| Systemic depletion of Kyn/Kynureninase | |||||
| - | - | - | - | - | - |
| Blockade of AhR activation / synthetic AhR modulator | |||||
| NCT04200963 | I | 93 advanced cancer | A phase 1a/b study of IK-175 as a single agent and combined with nivolumab in patients with locally advanced or metastatic solid tumors and urothelial carcinoma | IK-175 combined with nivolumab | Recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bailleux, C.; Chardin, D.; Gal, J.; Guigonis, J.-M.; Lindenthal, S.; Graslin, F.; Arnould, L.; Cagnard, A.; Ferrero, J.-M.; Humbert, O.; et al. Metabolomic Signatures of Scarff–Bloom–Richardson (SBR) Grade in Non-Metastatic Breast Cancer. Cancers 2023, 15, 1941. https://doi.org/10.3390/cancers15071941
Bailleux C, Chardin D, Gal J, Guigonis J-M, Lindenthal S, Graslin F, Arnould L, Cagnard A, Ferrero J-M, Humbert O, et al. Metabolomic Signatures of Scarff–Bloom–Richardson (SBR) Grade in Non-Metastatic Breast Cancer. Cancers. 2023; 15(7):1941. https://doi.org/10.3390/cancers15071941
Chicago/Turabian StyleBailleux, Caroline, David Chardin, Jocelyn Gal, Jean-Marie Guigonis, Sabine Lindenthal, Fanny Graslin, Laurent Arnould, Alexandre Cagnard, Jean-Marc Ferrero, Olivier Humbert, and et al. 2023. "Metabolomic Signatures of Scarff–Bloom–Richardson (SBR) Grade in Non-Metastatic Breast Cancer" Cancers 15, no. 7: 1941. https://doi.org/10.3390/cancers15071941
APA StyleBailleux, C., Chardin, D., Gal, J., Guigonis, J.-M., Lindenthal, S., Graslin, F., Arnould, L., Cagnard, A., Ferrero, J.-M., Humbert, O., & Pourcher, T. (2023). Metabolomic Signatures of Scarff–Bloom–Richardson (SBR) Grade in Non-Metastatic Breast Cancer. Cancers, 15(7), 1941. https://doi.org/10.3390/cancers15071941








