Intestinal Microbes and Hematological Malignancies
Abstract
Simple Summary
Abstract
1. Introduction
2. Leukemia
2.1. Gut Microbes and Leukemia: Clinical Relevance
| Cancer | Detection Method | Sampling Materials | Main Findings | Study |
|---|---|---|---|---|
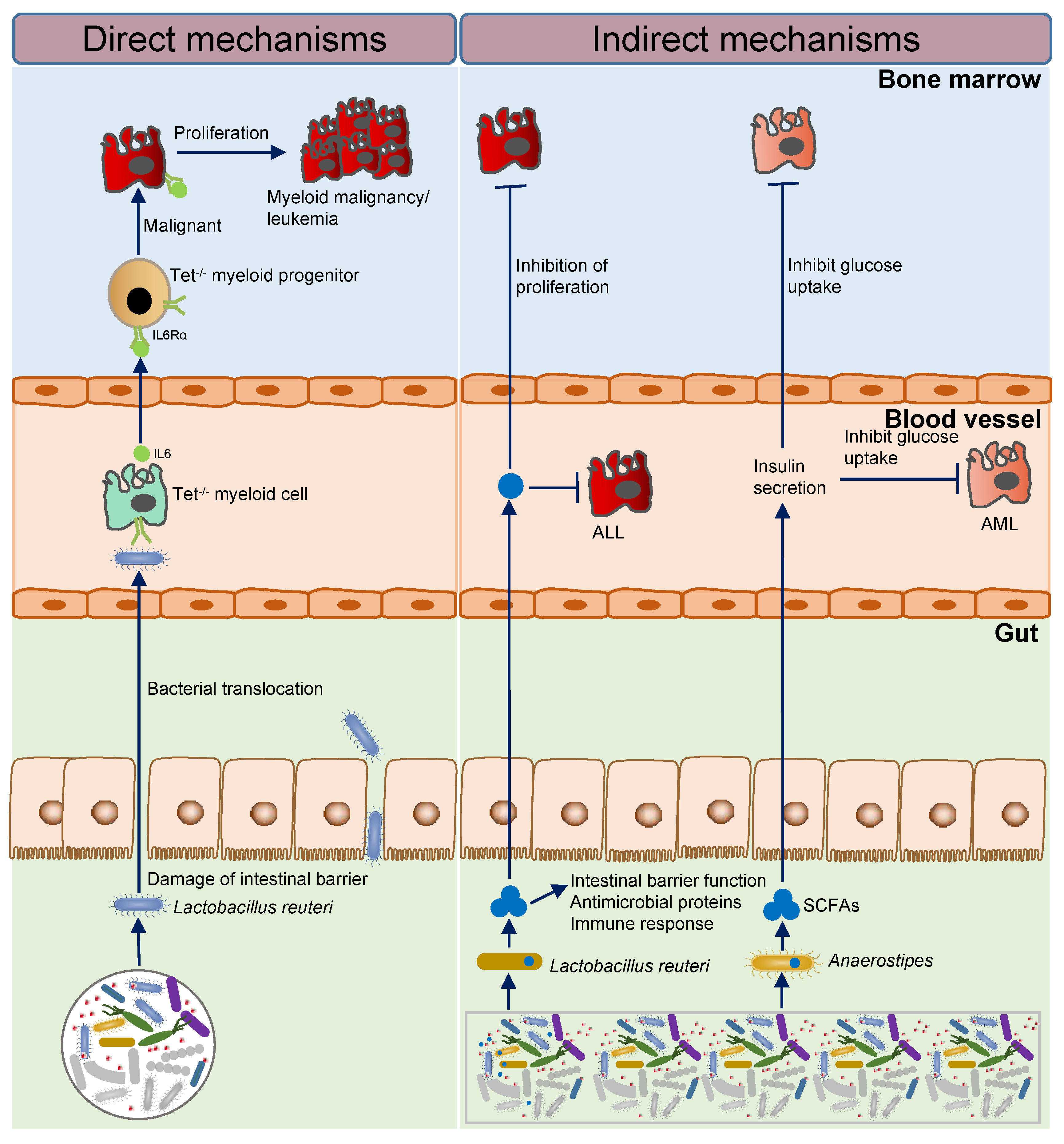
| PMP | 16S, qPCR | Mouse model | In TET 2 knockout mouse model, the lack of TET 2 can cause intestinal barrier damage, lead to intestinal microbial translocation, such as translocation of Lactobacillus reuteri, then cause immune activation and lead to an increase in hematopoietic stem cell self-renewal ability, so that its bias to the myeloid development eventually causes pre-leukaemic myeloproliferation (PMP), promoting the initiation of leukemia. | Meisel, M. et al., 2018 [15] |
| ALL | 16S | Human | Bacteroides genus is enriched in patients. | Chua, L.L. et al., 2020 [10] |
| ALL | 16S | Human | Enterococcaceae abundance is related to increased risk of granulocyte deficiency with fever and diarrhea; Streptococcidae abundance is associated with diarrhea. | Hakim, H. et al., 2018 [11] |
| ALL | 16S | Mouse model | The relative abundance of Lactobacillus reuteri is reduced in leukemic mice, and inhibits proliferation of supplement with Lactobacillus reuteri; Parabacteroides goldsteinii/ASF 519 abundance is enriched in leukemic mice, damages intestinal balance, and promotes progression. | Bindels, L.B. et al., 2016 [16] |
| ALL | 16S | Human | Faecalibacterium abundance shows a positive correlation with chronic diseases. | Thomas, R. et al., 2020 [13] |
| AL | 16S | Human | Bayesian analysis of longitudinal data demonstrated larger departures of microbial communities from the pre-chemotherapy baseline during repeat therapy compared to induction. This increased ecosystem instability during repeat therapy possibly impairs colonization resistance and increases vulnerability to Enterococcus outgrowth. | Rashidi, A. et al., 2019 [17] |
| AML | 16S | Human, mouse model | Anaerostipes, Lachnospiraceae, and Bacteroidales S24–7 are significantly reduced in leukemic mice, which inhibits progression and maintains intestinal balance by producing SCFAs, which promote insulin secretion and inhibit glucose uptake by AML cells. | Ye, H. et al., 2018 [18] |
| AML | 16S | Human | Negative correlation between efficacy and α-diversity. | Lee, S. et al., 2019 [12] |
| CLL | 16S | Human | Firmicutes and Bacteroidetes show a positive correlation between ratio and insulin resistance; Proteobacteria is enriched in patients. | Kawari, M. et al., 2019 [9] |
2.2. Intestinal Microbial Imbalance and the Initiation and Progression of Leukemia
2.2.1. Imbalance of Intestinal Flora Activates Inflammatory Factors to Promote the Initiation of Leukemia
2.2.2. Short-Chain Fatty Acid Producing Bacteria Inhibit the Progression of Leukemia
2.3. The Treatment Prospect for Leukemia
3. Lymphoma
3.1. Gut Microbes and Lymphoma: Clinical Relevance
| Cancer | Detection Method | Sampling Materials | Main Findings | Study |
|---|---|---|---|---|
| NHL | 16S | Human | Barnesiellaceae, Coriobacteriaceae, Faecalibacterium, Christensenella, Dehalobacterium, Desulfovibrio, and Sutterella negative correlation of BSI. | Montassier, E. et al., 2016 [30] |
| HL | 16S | Human | Streptococcus, Sellimonas, Candidatus, Stoquefichus, Veillonella, Faeclitalea, and unclassified S24-7 enriched in HD; Faecalibacterium and Eubacterium oxidoreducens group enriched in cured patients. | Huang, N. et al., 2017 [31] |
| NHL | Flow cytometry, 16S | Human | α-diversity decreased during therapy. | Schmiester, M. et al., 2022 [37] |
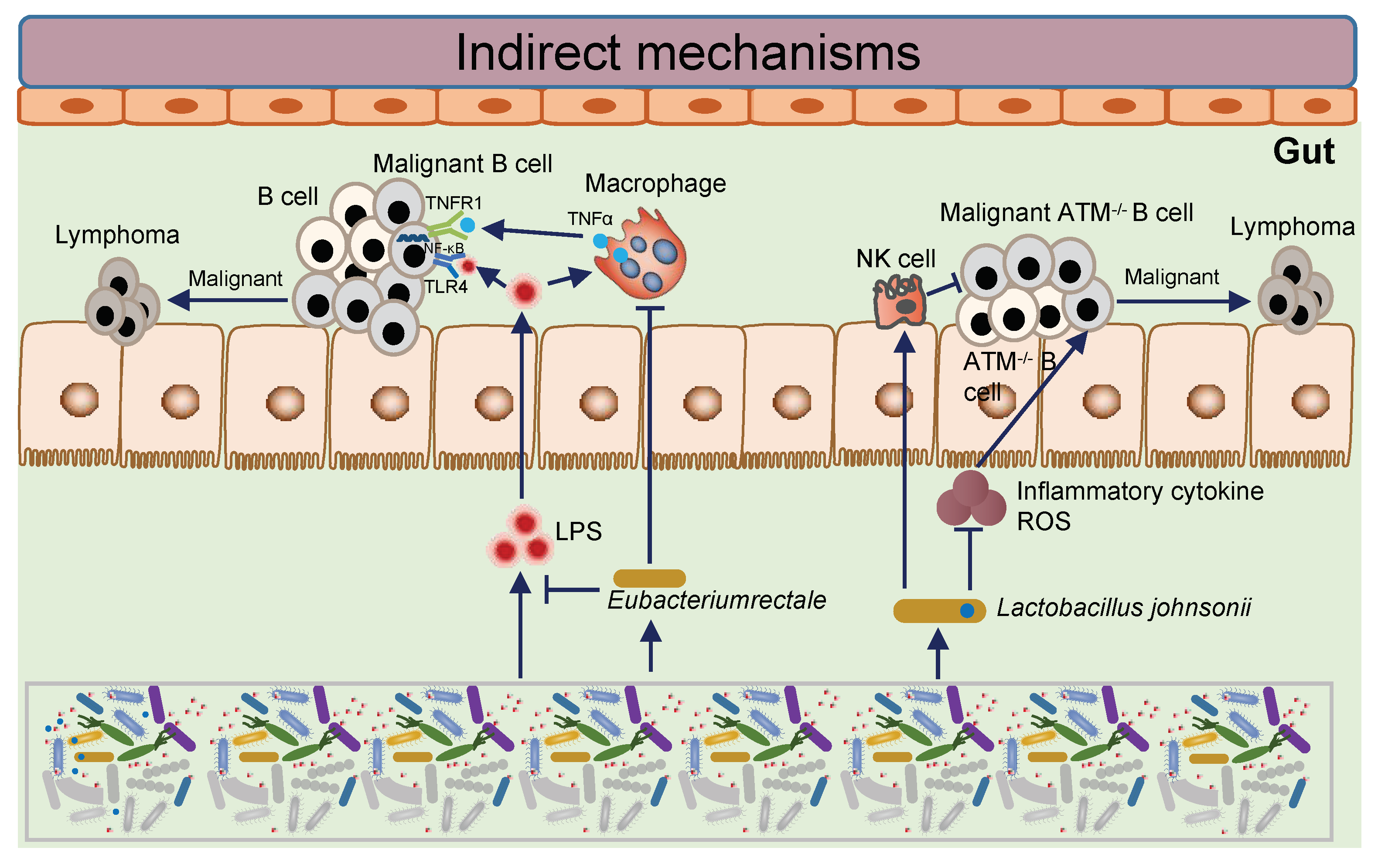
| NHL | Metagenomic | Human, mouse model | Eubacterium rectum decreased in patients, which inhibits intestinal inflammation and inactivates TLR4/MyD88 signaling to inactivate NF-kB pathway in B cells and inhibits B cell malignant. | Lu, H. et al., 2022 [38] |
| Lymphoma | Metagenomic | Human | A higher relative abundance of Bacteroides eggerthii, Ruminococcus lactaris, Eubacterium spp. CAG 180, Akkermansia muciniphila, and Erysipelatoclostridium ramosum increased the probability for CR prediction, whereas higher relative abundances of Bacteroides stercoris and others increased the probability for non-response prediction. | Christoph K Stein-Thoeringer et al., 2023 [32] |
| Lymphoma | 16S | Mouse model | Lactobacillus johnsonii, which was deficient in the more cancer-prone mouse colony, was causally tested for its capacity to confer reduced genotoxicity when restored by short-term oral transfer. | Mitsuko L. Yamamoto et al., 2013 [39] |
3.2. Intestinal Microbial Imbalance and the Lymphomagenesis
3.3. The Treatment Prospect for Lymphoma
4. Multiple Myeloma
4.1. Gut Microbes and Multiple Myeloma: Clinical Relevance
4.2. Intestinal Microbe Imbalance and the Progression of Multiple Myeloma
4.2.1. Alterations of Gut Microbiome Accelerate Multiple Myeloma Progression by Increasing the Relative Abundances of Nitrogen-Recycling Bacteria
4.2.2. The Imbalance of Intestinal Flora Activates Th17 Cells to Promote the Process of Multiple Myeloma
4.3. The Treatment Prospect for Multiple Myeloma
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Brody, H. Multiple myeloma. Nature 2011, 480, S33. [Google Scholar] [CrossRef]
- Harris, N.L.; Jaffe, E.S.; Diebold, J.; Flandrin, G.; Muller-Hermelink, H.K.; Vardiman, J.; Lister, T.A.; Bloomfield, C.D. The World Health Organization classification of hematological malignancies report of the Clinical Advisory Committee Meeting, Airlie House, Virginia, November 1997. Mod. Pathol. 2000, 13, 193–207. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 2016, 164, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Van der Hee, B.; Wells, J.M. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol. 2021, 29, 700–712. [Google Scholar] [CrossRef]
- Amoroso, C.; Perillo, F.; Strati, F.; Fantini, M.C.; Caprioli, F.; Facciotti, F. The Role of Gut Microbiota Biomodulators on Mucosal Immunity and Intestinal Inflammation. Cells 2020, 9, 1234. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Kawari, M.; Akhtar, M.; Sager, M.; Basbous, Z.; Baydoun, I.; Kabanja, J.; Darweesh, M.; Mokhtar, N.; Kanfar, S.; Mutahar, E.; et al. Alterations of Gut Microbiome in Untreated Chronic Lymphocytic Leukemia (CLL); Future Therapeutic Potentials. Blood 2019, 134, 5455. [Google Scholar] [CrossRef]
- Chua, L.L.; Rajasuriar, R.; Lim, Y.A.L.; Woo, Y.L.; Loke, P.; Ariffin, H. Temporal changes in gut microbiota profile in children with acute lymphoblastic leukemia prior to commencement-, during-, and post-cessation of chemotherapy. BMC Cancer 2020, 20, 151. [Google Scholar] [CrossRef]
- Hakim, H.; Dallas, R.; Wolf, J.; Tang, L.; Schultz-Cherry, S.; Darling, V.; Johnson, C.; Karlsson, E.A.; Chang, T.C.; Jeha, S.; et al. Gut Microbiome Composition Predicts Infection Risk During Chemotherapy in Children With Acute Lymphoblastic Leukemia. Clin. Infect. Dis. 2018, 67, 541–548. [Google Scholar] [CrossRef]
- Lee, S.; Ritchie, E.K.; Miah, S.; Andy, C.; Curcio, T.; Goudy, F.; Hovan, M.; Douglass, C.; Desai, P.; Samuel, M.B.; et al. Changes in Gut Microbial Diversity and Correlations with Clinical Outcomes in Patients with Newly Diagnosed Acute Myeloid Leukemia (AML) Receiving Intensive. Blood 2019, 134, 1336. [Google Scholar] [CrossRef]
- Thomas, R.; Wong, W.S.W.; Saadon, R.; Vilboux, T.; Deeken, J.; Niederhuber, J.; Hourigan, S.K.; Yang, E. Gut microbial composition difference between pediatric ALL survivors and siblings. Pediatr. Hematol. Oncol. 2020, 37, 475–488. [Google Scholar] [CrossRef]
- Lahteenmaki, K.; Wacklin, P.; Taskinen, M.; Tuovinen, E.; Lohi, O.; Partanen, J.; Matto, J.; Vettenranta, K. Haematopoietic stem cell transplantation induces severe dysbiosis in intestinal microbiota of paediatric ALL patients. Bone Marrow Transplant. 2017, 52, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Meisel, M.; Hinterleitner, R.; Pacis, A.; Chen, L.; Earley, Z.M.; Mayassi, T.; Pierre, J.F.; Ernest, J.D.; Galipeau, H.J.; Thuille, N.; et al. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature 2018, 557, 580–584. [Google Scholar] [CrossRef]
- Bindels, L.B.; Neyrinck, A.M.; Claus, S.P.; Le Roy, C.I.; Grangette, C.; Pot, B.; Martinez, I.; Walter, J.; Cani, P.D.; Delzenne, N.M. Synbiotic approach restores intestinal homeostasis and prolongs survival in leukaemic mice with cachexia. ISME J. 2016, 10, 1456–1470. [Google Scholar] [CrossRef]
- Rashidi, A.; Kaiser, T.; Shields-Cutler, R.; Graiziger, C.; Holtan, S.G.; Rehman, T.U.; Wasko, J.; Weisdorf, D.J.; Dunny, G.; Khoruts, A.; et al. Dysbiosis patterns during re-induction/salvage versus induction chemotherapy for acute leukemia. Sci. Rep. 2019, 9, 6083. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Adane, B.; Khan, N.; Alexeev, E.; Nusbacher, N.; Minhajuddin, M.; Stevens, B.M.; Winters, A.C.; Lin, X.; Ashton, J.M.; et al. Subversion of Systemic Glucose Metabolism as a Mechanism to Support the Growth of Leukemia Cells. Cancer Cell 2018, 34, 659–673.e6. [Google Scholar] [CrossRef]
- Kosmider, O.; Gelsi-Boyer, V.; Ciudad, M.; Racoeur, C.; Jooste, V.; Vey, N.; Quesnel, B.; Fenaux, P.; Bastie, J.N.; Beyne-Rauzy, O.; et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica 2009, 94, 1676–1681. [Google Scholar] [CrossRef]
- Tefferi, A.; Lim, K.H.; Levine, R. Mutation in TET2 in myeloid cancers. N. Engl. J. Med. 2009, 361, 1117. [Google Scholar] [CrossRef]
- Busque, L.; Patel, J.P.; Figueroa, M.E.; Vasanthakumar, A.; Provost, S.; Hamilou, Z.; Mollica, L.; Li, J.; Viale, A.; Heguy, A.; et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat. Genet. 2012, 44, 1179–1181. [Google Scholar] [CrossRef]
- Moran-Crusio, K.; Reavie, L.; Shih, A.; Abdel-Wahab, O.; Ndiaye-Lobry, D.; Lobry, C.; Figueroa, M.E.; Vasanthakumar, A.; Patel, J.; Zhao, X.; et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011, 20, 11–24. [Google Scholar] [CrossRef]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Quivoron, C.; Couronne, L.; Della Valle, V.; Lopez, C.K.; Plo, I.; Wagner-Ballon, O.; Do Cruzeiro, M.; Delhommeau, F.; Arnulf, B.; Stern, M.H.; et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011, 20, 25–38. [Google Scholar] [CrossRef]
- Argiles, J.M.; Busquets, S.; Stemmler, B.; Lopez-Soriano, F.J. Cancer cachexia: Understanding the molecular basis. Nat. Rev. Cancer 2014, 14, 754–762. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.J. Adaptive homeostasis. Mol. Aspects Med. 2016, 49, 1–7. [Google Scholar] [CrossRef]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Montassier, E.; Al-Ghalith, G.A.; Ward, T.; Corvec, S.; Gastinne, T.; Potel, G.; Moreau, P.; de la Cochetiere, M.F.; Batard, E.; Knights, D. Pretreatment gut microbiome predicts chemotherapy-related bloodstream infection. Genome Med. 2016, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Hwang, A.; Lagishetty, V.; Epeldegui, M.; Yu, Y.; Buchanan, L.; Yu, G.; Nathwani, B.N.; Millstein, J.; Mack, T.M.; et al. Differences in Fecal Microbiota in Long-Term Adolscent/Young Adult Hodgkin Lymphoma Survivors and Their Unaffected Twins. Blood 2017, 130, 4084. [Google Scholar]
- Stein-Thoeringer, C.K.; Saini, N.Y.; Zamir, E.; Blumenberg, V.; Schubert, M.L.; Mor, U.; Fante, M.A.; Schmidt, S.; Hayase, E.; Hayase, T.; et al. A non-antibiotic-disrupted gut microbiome is associated with clinical responses to CD19-CAR-T cell cancer immunotherapy. Nat. Med. 2023. [Google Scholar] [CrossRef]
- Shono, Y.; Docampo, M.D.; Peled, J.U.; Perobelli, S.M.; Velardi, E.; Tsai, J.J.; Slingerland, A.E.; Smith, O.M.; Young, L.F.; Gupta, J.; et al. Increased GVHD-related mortality with broad-spectrum antibiotic use after allogeneic hematopoietic stem cell transplantation in human patients and mice. Sci. Transl. Med. 2016, 8, 339ra371. [Google Scholar] [CrossRef] [PubMed]
- Keefe, D.M.; Schubert, M.M.; Elting, L.S.; Sonis, S.T.; Epstein, J.B.; Raber-Durlacher, J.E.; Migliorati, C.A.; McGuire, D.B.; Hutchins, R.D.; Peterson, D.E.; et al. Updated clinical practice guidelines for the prevention and treatment of mucositis. Cancer 2007, 109, 820–831. [Google Scholar] [CrossRef]
- Tuncer, H.H.; Rana, N.; Milani, C.; Darko, A.; Al-Homsi, S.A. Gastrointestinal and hepatic complications of hematopoietic stem cell transplantation. World J. Gastroenterol. 2012, 18, 1851–1860. [Google Scholar] [CrossRef]
- Cozen, W.; Yu, G.; Gail, M.; Nathwani, B.N.; Hwang, A.E.; Hamilton, A.S.; Mack, T.M.; Goedert, J.J. Fecal Microbiota Diversity in Survivors of Adolescent/Young Adult Hodgkin Lymphom. Blood 2012, 120, 1533. [Google Scholar] [CrossRef]
- Schmiester, M.; Maier, R.; Riedel, R.; Durek, P.; Frentsch, M.; Kolling, S.; Mashreghi, M.F.; Jenq, R.; Zhang, L.; Peterson, C.B.; et al. Flow cytometry can reliably capture gut microbial composition in healthy adults as well as dysbiosis dynamics in patients with aggressive B-cell non-Hodgkin lymphoma. Gut Microbes 2022, 14, 2081475. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Xu, X.; Fu, D.; Gu, Y.; Fan, R.; Yi, H.; He, X.; Wang, C.; Ouyang, B.; Zhao, P.; et al. Butyrate-producing Eubacterium rectale suppresses lymphomagenesis by alleviating the TNF-induced TLR4/MyD88/NF-kappaB axis. Cell Host Microbe 2022, 30, 1139–1150.e7. [Google Scholar] [CrossRef]
- Yamamoto, M.L.; Maier, I.; Dang, A.T.; Berry, D.; Liu, J.; Ruegger, P.M.; Yang, J.I.; Soto, P.A.; Presley, L.L.; Reliene, R.; et al. Intestinal bacteria modify lymphoma incidence and latency by affecting systemic inflammatory state, oxidative stress, and leukocyte genotoxicity. Cancer Res. 2013, 73, 4222–4232. [Google Scholar] [CrossRef] [PubMed]
- Meyn, M.S. Ataxia-telangiectasia, cancer and the pathobiology of the ATM gene. Clin. Genet. 1999, 55, 289–304. [Google Scholar] [CrossRef]
- Peterson, R.D.; Funkhouser, J.D.; Tuck-Muller, C.M.; Gatti, R.A. Cancer susceptibility in ataxia-telangiectasia. Leukemia 1992, 6 (Suppl. 1), 8–13. [Google Scholar]
- Ben Arush, M.W. Treatment of lymphoid malignancies in patients with ataxia-telangiectasia. Med. Pediatr. Oncol. 1999, 32, 479–480. [Google Scholar] [CrossRef]
- Zhan, F.; Huang, Y.; Colla, S.; Stewart, J.P.; Hanamura, I.; Gupta, S.; Epstein, J.; Yaccoby, S.; Sawyer, J.; Burington, B.; et al. The molecular classification of multiple myeloma. Blood 2006, 108, 2020–2028. [Google Scholar] [CrossRef] [PubMed]
- Jian, X.; Zhu, Y.; Ouyang, J.; Wang, Y.; Lei, Q.; Xia, J.; Guan, Y.; Zhang, J.; Guo, J.; He, Y.; et al. Alterations of gut microbiome accelerate multiple myeloma progression by increasing the relative abundances of nitrogen-recycling bacteria. Microbiome 2020, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yang, Q.; Zhu, Y.; Jian, X.; Guo, J.; Zhang, J.; Kuang, C.; Feng, X.; An, G.; Qiu, L.; et al. Intestinal Klebsiella pneumoniae Contributes to Pneumonia by Synthesizing Glutamine in Multiple Myeloma. Cancers 2022, 14, 4188. [Google Scholar] [CrossRef] [PubMed]
- Pianko, M.J.; Devlin, S.M.; Littmann, E.R.; Chansakul, A.; Mastey, D.; Salcedo, M.; Fontana, E.; Ling, L.; Tavitian, E.; Slingerland, J.B.; et al. Minimal residual disease negativity in multiple myeloma is associated with intestinal microbiota composition. Blood Adv. 2019, 3, 2040–2044. [Google Scholar] [CrossRef] [PubMed]
- Calcinotto, A.; Brevi, A.; Chesi, M.; Ferrarese, R.; Garcia Perez, L.; Grioni, M.; Kumar, S.; Garbitt, V.M.; Sharik, M.E.; Henderson, K.J.; et al. Microbiota-driven interleukin-17-producing cells and eosinophils synergize to accelerate multiple myeloma progression. Nat. Commun. 2018, 9, 4832. [Google Scholar] [CrossRef]
- Yang, Q.; Ouyang, J.; Sun, F.; Yang, J. Short-Chain Fatty Acids: A Soldier Fighting against Inflammation and Protecting from Tumorigenesis in People with Diabetes. Front. Immunol. 2020, 11, 590685. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, R.; Afaghi, A.; Babakhani, S.; Sohrabi, M.R.; Hosseini-Fard, S.R.; Babolhavaeji, K.; Khani Ali Akbari, S.; Yousefimashouf, R.; Karampoor, S. Role of microbiota-derived short-chain fatty acids in cancer development and prevention. Biomed. Pharmacother. 2021, 139, 111619. [Google Scholar] [CrossRef]
- Shah, U.A.; Maclachlan, K.H.; Derkach, A.; Salcedo, M.; Barnett, K.; Caple, J.; Blaslov, J.; Tran, L.; Ciardiello, A.; Burge, M.; et al. Sustained Minimal Residual Disease Negativity in Multiple Myeloma is Associated with Stool Butyrate and Healthier Plant-Based Diets. Clin. Cancer Res. 2022, 28, 5149–5155. [Google Scholar] [CrossRef]
- Cheriyath, V.; Kuhns, M.A.; Kalaycio, M.E.; Borden, E.C. Potentiation of apoptosis by histone deacetylase inhibitors and doxorubicin combination: Cytoplasmic cathepsin B as a mediator of apoptosis in multiple myeloma. Br. J. Cancer 2011, 104, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Mercado, A.I.; Del Valle Cano, A.; Fernandez, M.F.; Fontana, L. Gut Microbiota and Breast Cancer: The Dual Role of Microbes. Cancers 2023, 15, 443. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhang, J.; Wu, X.; Du, W.; Zhu, Y.; Liu, X.; Liu, Z.; Meng, B.; Guo, J.; Yang, Q.; et al. Blocking glycine utilization inhibits multiple myeloma progression by disrupting glutathione balance. Nat. Commun. 2022, 13, 4007. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Xia, J.; Zhang, J.; Zhu, Y.; Wu, Y.; Guo, J.; Chen, S.; Lei, Q.; Meng, B.; Kuang, C.; et al. Phosphoglycerate dehydrogenase promotes proliferation and bortezomib resistance through increasing reduced glutathione synthesis in multiple myeloma. Br. J. Haematol. 2020, 190, 52–66. [Google Scholar] [CrossRef] [PubMed]

| Cancer | Detection Method | Sampling Materials | Main Findings | Study |
|---|---|---|---|---|
| MM | 16S | Mouse model | Prevotella heparinolytica promotes Th17 differentiation, and Th17 migrates to the BM niche, and further contributes to the eosinophil-Th17-MM cells network. | Calcinotto, A. et al., 2018 [47] |
| 16S | Human | Eubacterium hallii is enriched in MRD negative patients. | Pianko, M.J. et al., 2019 [46] | |
| Metagenomic, qPCR | Human, mouse model | Clostridium butyricum, and Anaerostipes hadrus is enriched in HD, and Clostridium butyricum inhibits MM progression; Klebsiella, Streptococcus, etc. shows positive correlation of ISS stage; Glutamine synthesized by Klebsiella pneumoniae, and which belongs to nitrogen-recycling bacteria, and glutamine promotes proliferation of MM cells. | Jian, X. et al., 2020 [44] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Yang, Q.; Yang, Q.; He, Y.; Zhou, W. Intestinal Microbes and Hematological Malignancies. Cancers 2023, 15, 2284. https://doi.org/10.3390/cancers15082284
Zhu Y, Yang Q, Yang Q, He Y, Zhou W. Intestinal Microbes and Hematological Malignancies. Cancers. 2023; 15(8):2284. https://doi.org/10.3390/cancers15082284
Chicago/Turabian StyleZhu, Yinghong, Qiaohui Yang, Qin Yang, Yanjuan He, and Wen Zhou. 2023. "Intestinal Microbes and Hematological Malignancies" Cancers 15, no. 8: 2284. https://doi.org/10.3390/cancers15082284
APA StyleZhu, Y., Yang, Q., Yang, Q., He, Y., & Zhou, W. (2023). Intestinal Microbes and Hematological Malignancies. Cancers, 15(8), 2284. https://doi.org/10.3390/cancers15082284






