The Anti-Tumor Effect of the Newly Developed LAT1 Inhibitor JPH203 in Colorectal Carcinoma, According to a Comprehensive Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. LAT Expression Based on UCSC Cancer Genomics Browser Analysis
2.2. Patients
2.3. Immunohistochemistry for LAT1 Expression in Human CRC Tissue
2.4. Classification of Human Colon Cancer Tissue According to Microsatellite Status
2.5. Evaluation of Human CRC Stromal Volume
2.6. RNA Extraction and Quantitative PCR to Evaluate LAT1 Expression in CRC Cell Lines
2.7. Cell Lines
2.8. JPH203 Effects on Cancer Cell Proliferation In Vitro
2.9. JPH203 Effects in CT26 and MSC Co-Culture System
2.10. Treatment Experiment Using JPH203 in an Orthotopic Implantation Tumor Model
2.11. Necropsy Procedures and Histological Evaluation
2.12. Double Immunofluorescence for E-Cadherin and Vimentin
2.13. RNA Sequencing and Gene Set Enrichment Analysis (GSEA)
2.14. Functional Enrichment Analysis
2.15. Reagents
2.16. Statistical Analysis
3. Results
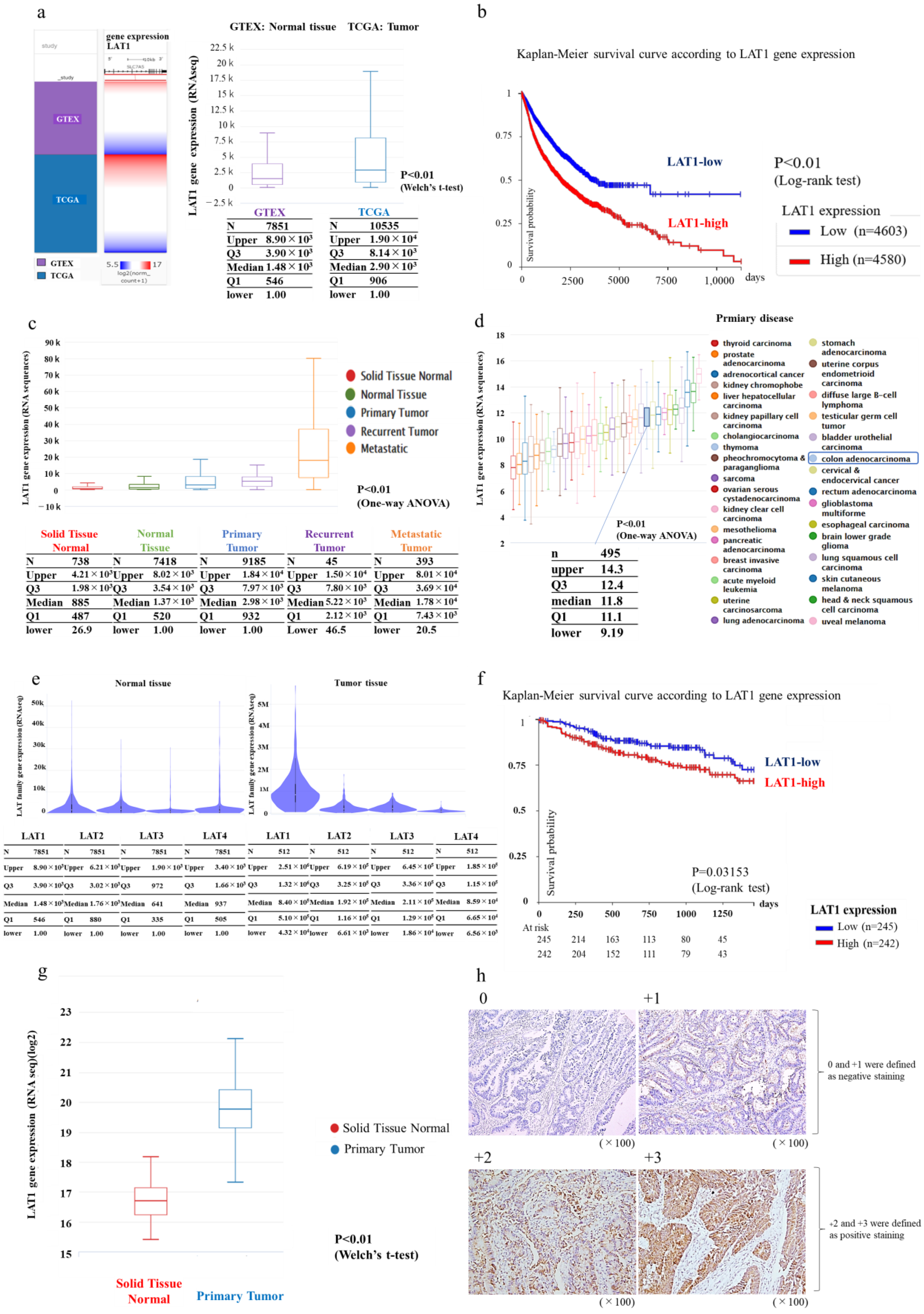
3.1. LAT1 Expression in Humans According to Database Analysis Using Xena
3.2. Immunostaining in Human CRC Specimens
3.3. LAT1 Expression in Colon Cancer Cell Lines
3.4. Proliferation Assay Using Colon Cancer Cell Lines with JPH203 In Vitro
3.5. JPH203 Administration Effect in a Congenic Mouse Tumor Model with Abundant Stroma
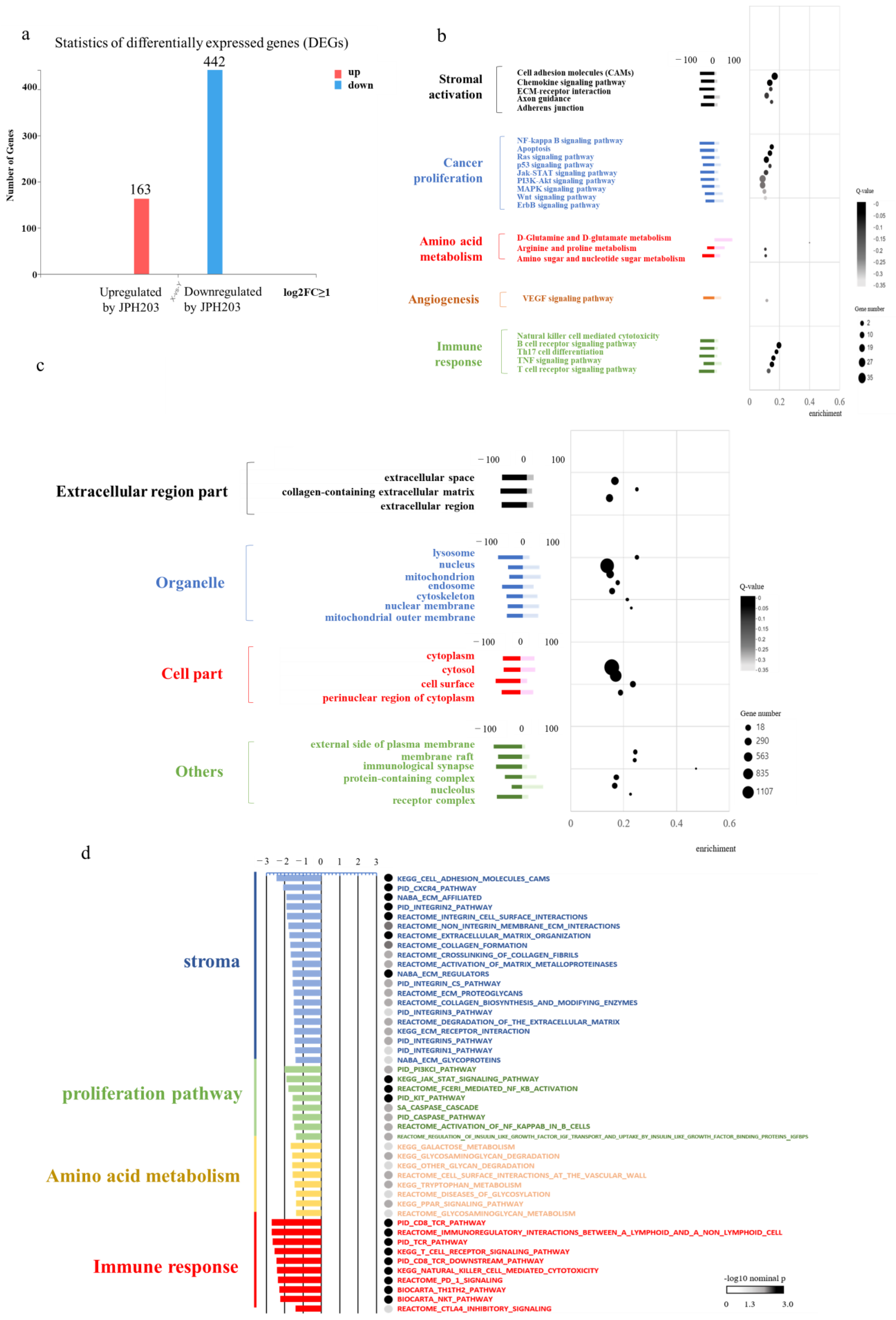
3.6. RNA Sequence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kanai, Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharmacol. Ther. 2022, 230, 107964. [Google Scholar] [CrossRef] [PubMed]
- Kaira, K.; Oriuchi, N.; Takahashi, T.; Nakagawa, K.; Ohde, Y.; Okumura, T.; Murakami, H.; Shukuya, T.; Kenmotsu, H.; Naito, T.; et al. LAT1 expression is closely associated with hypoxic markers and mTOR in resected non-small cell lung cancer. Am. J. Transl. Res. 2011, 3, 468–478. [Google Scholar] [PubMed]
- Kaira, K.; Sunose, Y.; Arakawa, K.; Ogawa, T.; Sunaga, N.; Shimizu, K.; Tominaga, H.; Oriuchi, N.; Itoh, H.; Nagamori, S.; et al. Prognostic significance of L-type amino-acid transporter 1 expression in surgically resected pancreatic cancer. Br. J. Cancer 2012, 107, 632–638. [Google Scholar] [CrossRef]
- Maeda, S.; Nakazato, Y.; Hayashi, K.; Nishihira, M.; Inoue, T.; Araki, O.; Karube, Y.; Kobayashi, S.; Chida, M. L-Type amino acid transporter 1 immunoreactivity as a possible diagnostic and prognostic marker of thymic carcinoma. Tohoku J. Exp. Med. 2018, 246, 167–174. [Google Scholar] [CrossRef]
- Kaira, K.; Sunose, Y.; Ohshima, Y.; Ishioka, N.S.; Arakawa, K.; Ogawa, T.; Sunaga, N.; Shimizu, K.; Tominaga, H.; Oriuchi, N.; et al. Clinical significance of L-type amino acid transporter 1 expression as a prognostic marker and potential of new targeting therapy in biliary tract cancer. BMC Cancer 2013, 13, 482. [Google Scholar] [CrossRef]
- Kim, D.K.; Ahn, S.G.; Park, J.C.; Kanai, Y.; Endou, H.; Yoon, J.H. Expression of L-type amino acid transporter 1 (LAT1) and 4F2 heavy chain (4F2hc) in oral squamous cell carcinoma and its precusor lesions. Anticancer Res. 2004, 24, 1671–1676. [Google Scholar]
- Kaira, K.; Takahashi, T.; Murakami, H.; Shukuya, T.; Kenmotsu, H.; Naito, T.; Oriuchi, N.; Kanai, Y.; Endo, M.; Kondo, H.; et al. Relationship between LAT1 expression and response to platinum-based chemotherapy in non-small cell lung cancer patients with postoperative recurrence. Anticancer Res. 2011, 31, 3775–3782. [Google Scholar]
- Okano, N.; Hana, K.; Naruge, D.; Kawai, K.; Kobayashi, T.; Nagashima, F.; Endou, H.; Furuse, J. Biomarker analyses in patients with advanced solid tumors treated with the LAT1 inhibitor JPH203. In Vivo 2020, 34, 2595–2606. [Google Scholar] [CrossRef]
- Kim, C.S.; Moon, I.S.; Park, J.H.; Shin, W.C.; Chun, H.S.; Lee, S.Y.; Kook, J.K.; Kim, H.J.; Park, J.C.; Endou, H.; et al. Inhibition of L-type amino acid transporter modulates the expression of cell cycle regulatory factors in KB oral cancer cells. Biol. Pharm. Bull. 2010, 33, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, H.; Kaira, K.; Motegi, Y.; Yokobori, T.; Takada, T.; Katoh, R.; Osone, K.; Takahashi, R.; Katayama, C.; Oyama, T.; et al. Role of amino acid transporter expression as a prognostic marker in patients with surgically resected colorectal cancer. Anticancer Res. 2019, 39, 2535–2543. [Google Scholar] [CrossRef]
- Li, N.; Li, X.; Huang, S.; Shen, S.; Wang, X. miR-126 inhibits colon cancer proliferation and invasion through targeting IRS1, SLC7A5 and TOM1 gene. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2013, 38, 809–817. [Google Scholar]
- Nawashiro, H.; Otani, N.; Shinomiya, N.; Fukui, S.; Ooigawa, H.; Shima, K.; Matsuo, H.; Kanai, Y.; Endou, H. L-type amino acid transporter 1 as a potential molecular target in human astrocytic tumors. Int. J. Cancer 2006, 119, 484–492. [Google Scholar] [CrossRef]
- Ohshima, Y.; Kaira, K.; Yamaguchi, A.; Oriuchi, N.; Tominaga, H.; Nagamori, S.; Kanai, Y.; Yokobori, T.; Miyazaki, T.; Asao, T.; et al. Efficacy of system l amino acid transporter 1 inhibition as a therapeutic target in esophageal squamous cell carcinoma. Cancer Sci. 2016, 107, 1499–1505. [Google Scholar] [CrossRef]
- Häfliger, P.; Graff, J.; Rubin, M.; Stooss, A.; Dettmer, M.S.; Altmann, K.H.; Gertsch, J.; Charles, R.P. The LAT1 inhibitor JPH203 reduces growth of thyroid carcinoma in a fully immunocompetent mouse model. J. Exp. Clin. Cancer Res. 2018, 37, 234. [Google Scholar] [CrossRef]
- Choi, D.W.; Kim, D.K.; Kanai, Y.; Wempe, M.F.; Endou, H.; Kim, J.-K. JPH203, a selective L-type amino acid transporter 1 inhibitor, induces mitochondria-dependent apoptosis in Saos2 human osteosarcoma cells. Korean J. Physiol. Pharmacol. 2017, 21, 599–607. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Vilar, E.; Gruber, S.B. Microsatellite instability in colorectal cancer-the stable evidence. Nat. Rev. Clin. Oncol. 2010, 7, 153–162. [Google Scholar] [CrossRef]
- Naito, T.; Yuge, R.; Kitadai, Y.; Takigawa, H.; Higashi, Y.; Kuwai, T.; Kuraoka, K.; Tanaka, S.; Chayama, K. Mesenchymal stem cells induce tumor stroma formation and epithelial-mesenchymal transition through SPARC expression in colorectal cancer. Oncol. Rep. 2021, 45, 104. [Google Scholar] [CrossRef]
- Maimaiti, M.; Sakamoto, S.; Yamada, Y.; Sugiura, M.; Rii, J.; Takeuchi, N.; Imamura, Y.; Furihata, T.; Ando, K.; Higuchi, K.; et al. Expression of L-type amino acid transporter 1 as a molecular target for prognostic and therapeutic indicators in bladder carcinoma. Sci. Rep. 2020, 10, 1292. [Google Scholar] [CrossRef]
- Takigawa, H.; Kitadai, Y.; Shinagawa, K.; Yuge, R.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Multikinase inhibitor regorafenib inhibits the growth and metastasis of colon cancer with abundant stroma. Cancer Sci. 2016, 107, 601–608. [Google Scholar] [CrossRef]
- Takigawa, H.; Kitadai, Y.; Shinagawa, K.; Yuge, R.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Mesenchymal stem cells induce epithelial to mesenchymal transition in colon cancer cells through direct cell-to-cell contact. Neoplasia 2017, 19, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Yorita, N.; Yuge, R.; Takigawa, H.; Ono, A.; Kuwai, T.; Kuraoka, K.; Kitadai, Y.; Tanaka, S.; Chayama, K. Stromal reaction inhibitor and immune-checkpoint inhibitor combination therapy attenuates excluded-type colorectal cancer in a mouse model. Cancer Lett. 2021, 498, 111–120. [Google Scholar] [CrossRef]
- Chen, C.H.; Kuo, C.Y.; Chen, S.H.; Mao, S.H.; Chang, C.Y.; Shalumon, K.T.; Chen, J.P. Thermosensitive injectable hydrogel for simultaneous intraperitoneal delivery of doxorubicin and prevention of peritoneal adhesion. Int. J. Mol. Sci. 2018, 19, 1373. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanda, Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transpl. 2013, 48, 452–458. [Google Scholar] [CrossRef]
- Häfliger, P.; Charles, R.-P. The L-type amino acid transporter LAT1—An emerging target in cancer. Int. J. Mol. Sci. 2019, 20, 2428. [Google Scholar] [CrossRef]
- del Amo, E.M.; Urtti, A.; Yliperttula, M. Pharmacokinetic role of L-type amino acid transporters LAT1 and LAT2. Eur. J. Pharm. Sci. 2008, 35, 161–174. [Google Scholar] [CrossRef]
- Rossier, G.; Meier, C.; Bauch, C.; Summa, V.; Sordat, B.; Verrey, F.; Kühn, L.C. LAT2, a new basolateral 4F2hc/CD98-associated amino acid transporter of kidney and intestine. J. Biol. Chem. 1999, 274, 34948–34954. [Google Scholar] [CrossRef]
- Kersemans, V.; Cornelissen, B.; Kersemans, K.; Bauwens, M.; Dierckx, R.A.; De Spiegeleer, B.; Mertens, J.; Slegers, G. 123/125I-labelled 2-iodo-L: -phenylalanine and 2-iodo-D: -phenylalanine: Comparative uptake in various tumour types and biodistribution in mice. Eur. J. Nucl. Med. Mol. Imaging 2006, 33, 919–927. [Google Scholar] [CrossRef]
- Sahin, I.H.; Akce, M.; Alese, O.; Shaib, W.; Lesinski, G.B.; El-Rayes, B.; Wu, C. Immune checkpoint inhibitors for the treatment of MSI-H/MMR-D colorectal cancer and a perspective on resistance mechanisms. Br. J. Cancer 2019, 121, 809–818. [Google Scholar] [CrossRef]
- Jo, W.-S.; Carethers, J.M. Chemotherapeutic implications in microsatellite unstable colorectal cancer. Cancer Biomark. 2006, 2, 51–60. [Google Scholar] [CrossRef]
- Oda, K.; Hosoda, N.; Endo, H.; Saito, K.; Tsujihara, K.; Yamamura, M.; Sakata, T.; Anzai, N.; Wempe, M.F.; Kanai, Y.; et al. l-Type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 2010, 101, 173–179. [Google Scholar] [CrossRef]
- Furuse, J.; Ikeda, M.; Ueno, M.; Furukawa, M.; Morizane, C.; Takehara, T.; Nishina, T.; Todaka, A.; Okano, N.; Hara, K.; et al. Nanvuranlat, an L-type amino acid transporter (LAT1) inhibitor for patients with pretreated advanced refractory biliary tract cancer (BTC): Primary endpoint results of a randomized, double-blind, placebo-controlled phase 2 study. J. Clin. Oncol. 2023, 41, 494. [Google Scholar] [CrossRef]
- Ngan, C.Y.; Yamamoto, H.; Seshimo, I.; Tsujino, T.; Man-i, M.; Ikeda, J.I.; Konishi, K.; Takemasa, I.; Ikeda, M.; Sekimoto, M.; et al. Quantitative evaluation of vimentin expression in tumour stroma of colorectal cancer. Br. J. Cancer 2007, 96, 986–992. [Google Scholar] [CrossRef]
- Enomoto, K.; Sato, F.; Tamagawa, S.; Gunduz, M.; Onoda, N.; Uchino, S.; Muragaki, Y.; Hotomi, M. A novel therapeutic approach for anaplastic thyroid cancer through inhibition of LAT1. Sci. Rep. 2019, 9, 14616. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, E.S.; Balaji, U.; Freinkman, E.; McCue, P.; Witkiewicz, A.K. Unique metabolic features of pancreatic cancer stroma: Relevance to the tumor compartment, prognosis, and invasive potential. Oncotarget 2016, 7, 78396–78411. [Google Scholar] [CrossRef]
- Vu, T.; Datta, P.K. Regulation of EMT in Colorectal Cancer: A Culprit in Metastasis. Cancers 2017, 9, 171. [Google Scholar] [CrossRef]
- Mizukoshi, K.; Okazawa, Y.; Haeno, H.; Koyama, Y.; Sulidan, K.; Komiyama, H.; Saeki, H.; Ohtsuji, N.; Ito, Y.; Kojima, Y.; et al. Metastatic seeding of human colon cancer cell clusters expressing the hybrid epithelial/mesenchymal state. Int. J. Cancer 2020, 146, 2547–2562. [Google Scholar] [CrossRef]
- Yuge, R.; Kitadai, Y.; Shinagawa, K.; Onoyama, M.; Tanaka, S.; Yasui, W.; Chayama, K. mTOR and PDGF pathway blockade inhibits liver metastasis of colorectal cancer by modulating the tumor microenvironment. Am. J. Pathol. 2015, 185, 399–408. [Google Scholar] [CrossRef]



| LAT1 Expression | ||||
|---|---|---|---|---|
| Positive | Negative | p * | ||
| Number of patients | 106 (68.8) | 48 (31.2) | ||
| Age (years) | 68.8 ± 12.0 | 66.3 ± 11.9 | 0.222 | |
| Sex | Male | 65 (61.3) | 26 (54.2) | 0.480 |
| Location | Right side colon | 34 (32.1) | 20 (41.7) | 0.277 |
| Left side colon | 72 (67.9) | 28 (58.3) | ||
| Histological Type | tub1/2 | 95 (89.6) | 44 (91.7) | 0.778 |
| Por/muc | 11 (10.4) | 4 (8.3) | ||
| Stage | I/II | 46 (43.4) | 32 (66.7) | 0.009 |
| III/IV | 60 (56.6) | 16 (33.3) | ||
| T | 1/2 | 26 (24.5) | 22 (45.8) | 0.014 |
| 3/4 | 80 (75.5) | 26 (54.2) | ||
| N | N0 | 48 (45.3) | 32 (66.7) | 0.015 |
| N1/2/3 | 58 (54.7) | 16 (33.3) | ||
| M | 0 | 85 (82.3) | 42 (79.4) | 0.361 |
| 1 | 21 (17.7) | 6 (20.6) | ||
| Budding grade | 1 | 21 (50.0) | 9 (37.5) | 0.732 |
| 2, 3 | 21 (50.0) | 15 (62.5) | ||
| Microsatellite instability | MSS | 98 (92.5) | 43 (89.6) | 0.545 |
| MSI-high | 8 (7.5) | 5 (10.4) | ||
| KRAS mutation | + | 42 (39.6) | 19 (39.6) | 1 |
| BRAF mutation | + | 2 (1.9) | 4 (8.3) | 0.076 |
| Group | Tumor Incident | Body Weight -Before (g) | Body Weight -After (g) | Tumor Weight (g) | Tumor Volume (mm3) | Lymph Node Metastasis |
|---|---|---|---|---|---|---|
| Control (n = 12) | 12/12 | 21.9 (21.0–23.5) | 20.7 (19.0–21.2) | 1.15 (0.27–1.58) | 312.2 (108–650) | 7/12 |
| JPH203 (n = 11) | 11/11 | 22.1 (21.5–22.8) | 20.4 (18.1–22.6) | 0.17 (0.15–0.27) | 44.5 (32–108) | 1/11 |
| p-value * | 1.00 | 0.37 | 0.49 | <0.001 | <0.001 | 0.027 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otani, R.; Takigawa, H.; Yuge, R.; Shimizu, D.; Ariyoshi, M.; Miyamoto, R.; Kadota, H.; Hiyama, Y.; Hayashi, R.; Urabe, Y.; et al. The Anti-Tumor Effect of the Newly Developed LAT1 Inhibitor JPH203 in Colorectal Carcinoma, According to a Comprehensive Analysis. Cancers 2023, 15, 1383. https://doi.org/10.3390/cancers15051383
Otani R, Takigawa H, Yuge R, Shimizu D, Ariyoshi M, Miyamoto R, Kadota H, Hiyama Y, Hayashi R, Urabe Y, et al. The Anti-Tumor Effect of the Newly Developed LAT1 Inhibitor JPH203 in Colorectal Carcinoma, According to a Comprehensive Analysis. Cancers. 2023; 15(5):1383. https://doi.org/10.3390/cancers15051383
Chicago/Turabian StyleOtani, Rina, Hidehiko Takigawa, Ryo Yuge, Daisuke Shimizu, Misa Ariyoshi, Ryo Miyamoto, Hiroki Kadota, Yuichi Hiyama, Ryohei Hayashi, Yuji Urabe, and et al. 2023. "The Anti-Tumor Effect of the Newly Developed LAT1 Inhibitor JPH203 in Colorectal Carcinoma, According to a Comprehensive Analysis" Cancers 15, no. 5: 1383. https://doi.org/10.3390/cancers15051383
APA StyleOtani, R., Takigawa, H., Yuge, R., Shimizu, D., Ariyoshi, M., Miyamoto, R., Kadota, H., Hiyama, Y., Hayashi, R., Urabe, Y., Ishikawa, A., Oue, N., Kitadai, Y., Oka, S., & Tanaka, S. (2023). The Anti-Tumor Effect of the Newly Developed LAT1 Inhibitor JPH203 in Colorectal Carcinoma, According to a Comprehensive Analysis. Cancers, 15(5), 1383. https://doi.org/10.3390/cancers15051383






