Canine-Inspired Chemometric Analysis of Volatile Organic Compounds in Urine Headspace to Distinguish Prostate Cancer in Mice and Men
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Instrumentation
2.2. Patient Recruitment
2.3. Murine Models of Prostate Cancer
2.4. Human and Mouse Urine Sample Processing
2.5. SPME GC-MS QTOF Protocols
2.6. Data Screening and Chemometric Analysis
3. Results
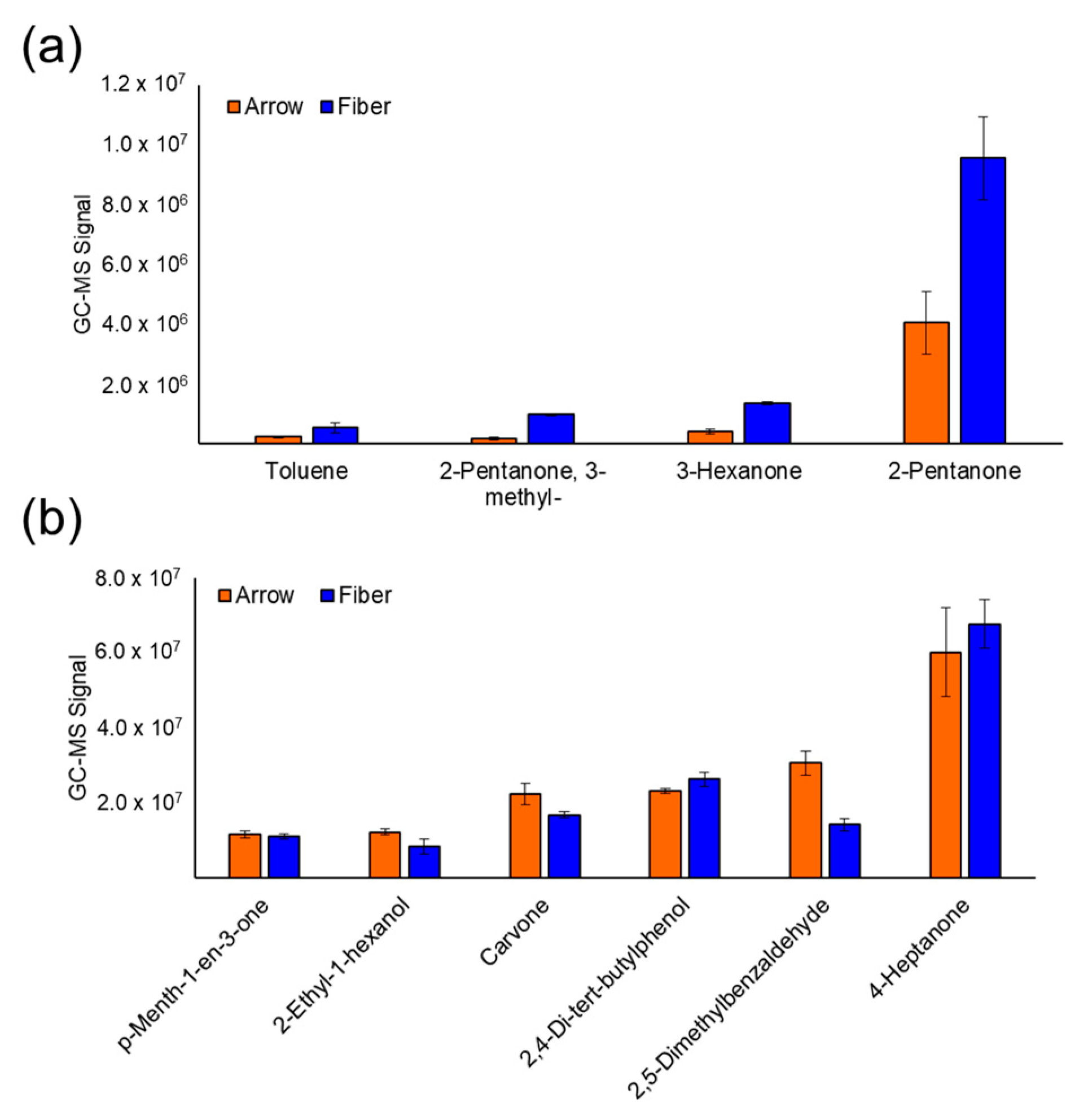
3.1. SPME Optimization
3.2. Patient Recruitment and Urine Collection
3.3. Mouse Urine VOC Analysis
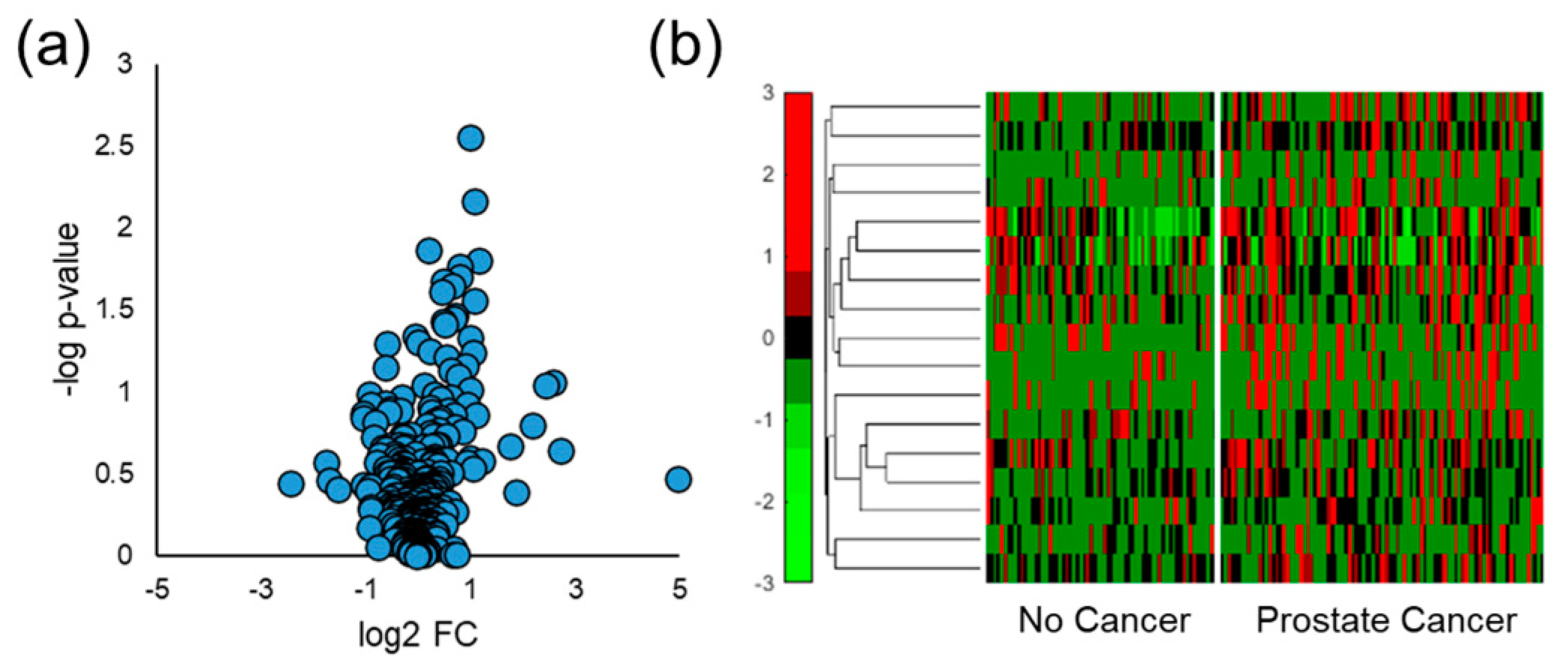
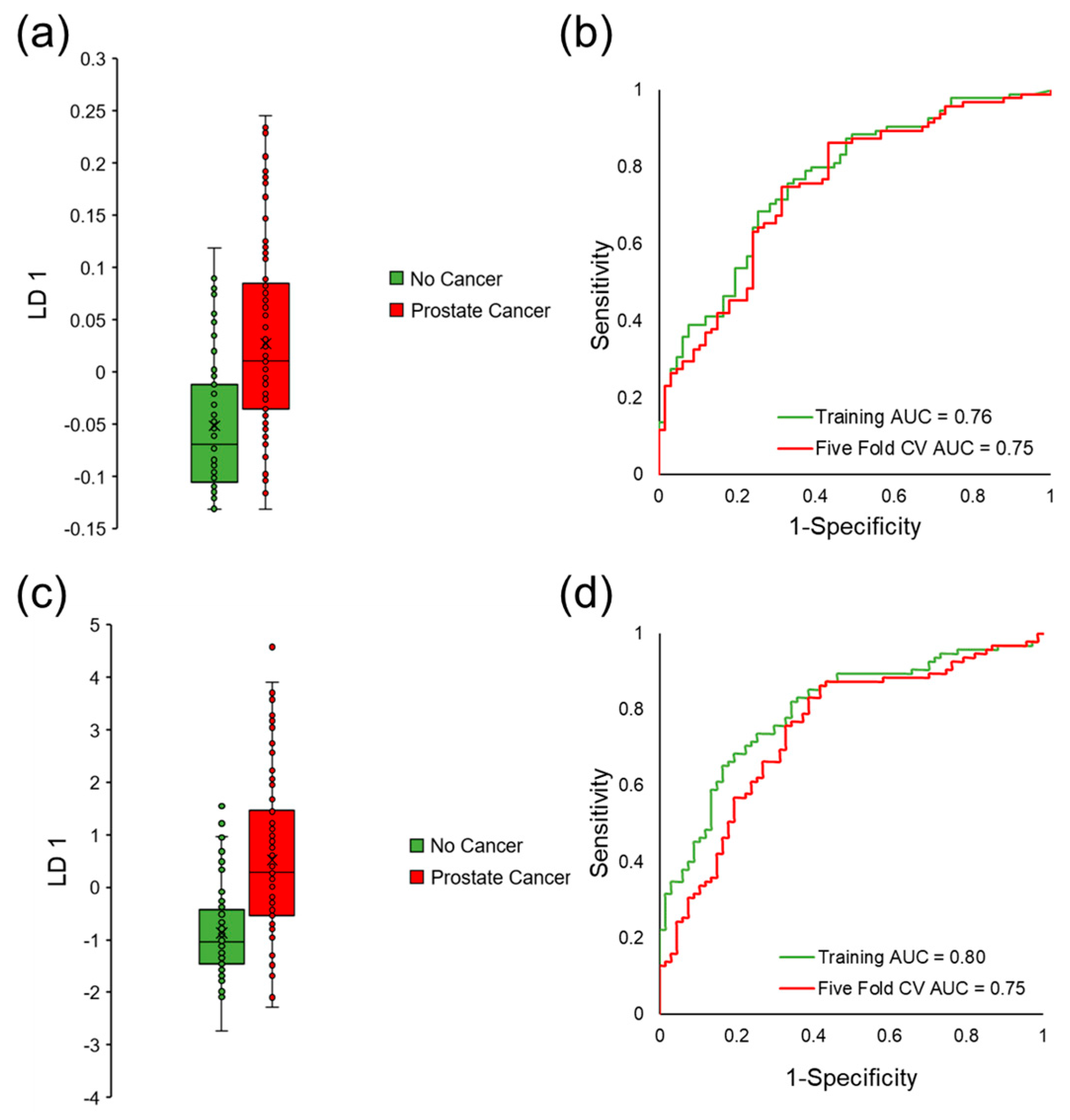
3.4. Distinguishing Prostate Cancer in Humans
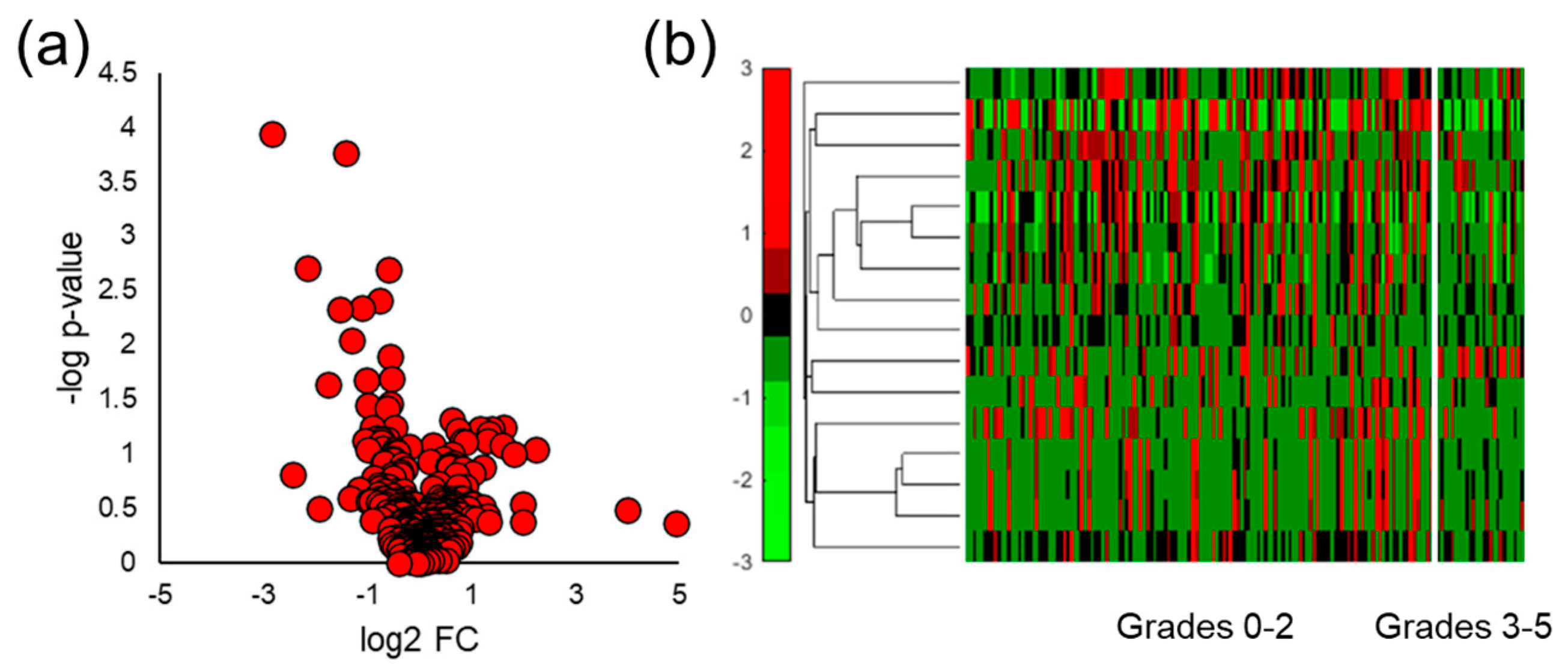
3.5. Stratifying Aggressive Prostate Cancer
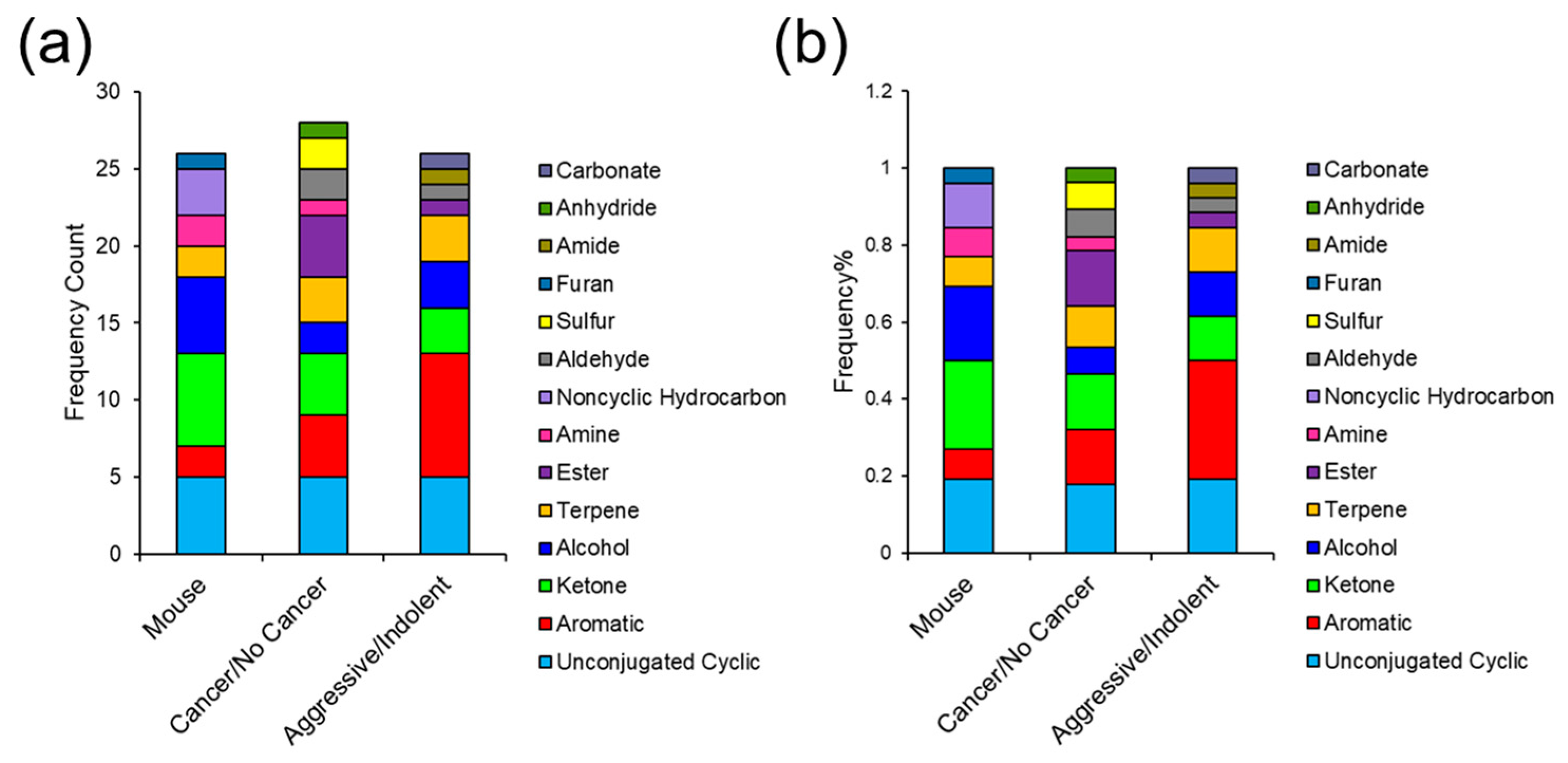
3.6. Comparing VOC Biomarkers in Mouse and Human Urine
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Catalona, W.J.; Smith, D.S.; Ratliff, T.L.; Dodds, K.M.; Coplen, D.E.; Yuan, J.J.; Petros, J.A.; Andriole, G.L. Andriole, Measurement of prostate-specific antigen in serum as a screening test for prostate cancer. N. Engl. J. Med. 1991, 324, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Hong, S.K. Clinical utility of current biomarkers for prostate cancer detection. Investig. Clin. Urol. 2021, 62, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kandirali, E.; Boran, C.; Serin, E.; Semercioz, A.; Metin, A. Metin, Association of extent and aggressiveness of inflammation with serum PSA levels and PSA density in asymptomatic patients. Urology 2007, 70, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Heijnsdijk, E.; Der Kinderen, A.; Wever, E.M.; Draisma, G.; Roobol, M.J.; De Koning, H.J. Overdetection, overtreatment and costs in prostate-specific antigen screening for prostate cancer. Br. J. Cancer 2009, 101, 1833–1838. [Google Scholar] [CrossRef]
- Rubin, M.; Demichelis, F. The Genomics of Prostate Cancer: Emerging understanding with technologic advances. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc 2018, 31, S1–S11. [Google Scholar] [CrossRef]
- Bolis, M.; Bossi, D.; Vallerga, A.; Ceserani, V.; Cavalli, M.; Impellizzieri, D.; Di Rito, L.; Zoni, E.; Mosole, S.; Elia, A.R.; et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat. Commun. 2021, 12, 7022. [Google Scholar] [CrossRef]
- Martignano, F.; Rossi, L.; Maugeri, A.; Gallà, V.; Conteduca, V.; De Giorgi, U.; Casadio, V.; Schepisi, G. Urinary RNA-based biomarkers for prostate cancer detection. Clin. Chim. Acta 2017, 473, 96–105. [Google Scholar] [CrossRef]
- Dhondt, B.; Geeurickx, E.; Tulkens, J.; Van Deun, J.; Vergauwen, G.; Lippens, L.; Miinalainen, I.; Rappu, P.; Heino, J.; Ost, P.; et al. Unravelling the proteomic landscape of extracellular vesicles in prostate cancer by density-based fractionation of urine. J. Extracell. Vesicles 2020, 9, 1736935. [Google Scholar] [CrossRef]
- Intasqui, P.; Bertolla, R.P.; Sadi, M.V. Prostate cancer proteomics: Clinically useful protein biomarkers and future perspectives. Expert Rev. Proteom. 2017, 15, 65–79. [Google Scholar] [CrossRef]
- Cerrato, A.; Bedia, C.; Capriotti, A.L.; Cavaliere, C.; Gentile, V.; Maggi, M.; Montone, C.M.; Piovesana, S.; Sciarra, A.; Tauler, R.; et al. Untargeted metabolomics of prostate cancer zwitterionic and positively charged compounds in urine. Anal. Chim. Acta 2021, 1158, 338381. [Google Scholar] [CrossRef]
- Lima, A.R.; De Bastos, M.L.; Carvalho, M.; de Pinho, P.G. Biomarker discovery in human prostate cancer: An update in metabolomics studies. Transl. Oncol. 2016, 9, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Kdadra, M.; Höckner, S.; Leung, H.; Kremer, W.; Schiffer, E. Metabolomics biomarkers of prostate cancer: A systematic review. Diagnostics 2019, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Biol. Lipids 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2016, 70, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Mao, J.; Ai, J.; Deng, Y.; Roth, M.R.; Pound, C.; Henegar, J.; Welti, R.; Bigler, S.A. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS ONE 2012, 7, e48889. [Google Scholar] [CrossRef]
- Kornberg, Z.; Cooperberg, M.; Spratt, D.; Feng, F. Genomic biomarkers in prostate cancer. Transl. Androl. Urol. 2018, 7, 459–471. [Google Scholar] [CrossRef]
- Heidrich, I.; Deitert, B.; Werner, S.; Pantel, K. Liquid biopsy for monitoring of tumor dormancy and early detection of disease recurrence in solid tumors. Cancer Metastasis Rev. 2023, 1–22. [Google Scholar] [CrossRef]
- Yamamichi, G.; Kato, T.; Uemura, M.; Nonomura, N. Diagnosing and Prognosing Bone Metastasis in Prostate Cancer: Clinical Utility of Blood Biomarkers. Anticancer. Res. 2023, 43, 283–290. [Google Scholar] [CrossRef]
- Rzhevskiy, A.S.; Kapitannikova, A.Y.; Butnaru, D.V.; Shpot, E.V.; Joosse, S.A.; Zvyagin, A.V.; Warkiani, M.E. Liquid Biopsy in Diagnosis and Prognosis of Non-Metastatic Prostate Cancer. Biomedicines 2022, 10, 3115. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Zappavigna, S.; Crocetto, F.; Giuliano, M.; Ribera, D.; Morra, R.; Scafuri, L.; Verde, A.; Bruzzese, D.; Iaccarino, S.; et al. Assessment of Total, PTEN(-), and AR-V7(+) Circulating Tumor Cell Count by Flow Cytometry in Patients with Metastatic Castration-Resistant Prostate Cancer Receiving Enzalutamide. Clin. Genitourin. Cancer 2021, 19, e286–e298. [Google Scholar] [CrossRef]
- Li, S.L.; An, N.; Liu, B.; Wang, S.Y.; Wang, J.J.; Ye, Y. Exosomes from LNCaP cells promote osteoblast activity through miR-375 transfer. Oncol. Lett. 2019, 17, 4463–4473. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Li, S.-L.; Ma, Y.-Y.; Diao, Y.-J.; Yang, L.; Su, M.-Q.; Li, Z.; Ji, Y.; Wang, J.; Lei, L.; et al. Exosomal miR-141-3p regulates osteoblast activity to promote the osteoblastic metastasis of prostate cancer. Oncotarget 2017, 8, 94834–94849. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Ding, M.; Xu, K.; Yang, C.; Mao, L.J. Exosomes in diagnosis and therapy of prostate cancer. Oncotarget 2017, 8, 97693–97700. [Google Scholar] [CrossRef] [PubMed]
- Taverna, G.; Tidu, L.; Grizzi, F.; Torri, V.; Mandressi, A.; Sardella, P.; La Torre, G.; Cocciolone, G.; Seveso, M.; Giusti, G.; et al. Olfactory system of highly trained dogs detects prostate cancer in urine samples. J. Urol. 2015, 193, 1382–1387. [Google Scholar] [CrossRef] [PubMed]
- Khalid, T.; Aggio, R.; White, P.; De Lacy Costello, B.; Persad, R.; Alkateb, H.; Jones, P.; Probert, C.S.; Ratcliffe, N. Urinary Volatile Organic Compounds for the Detection of Prostate Cancer. PLoS ONE 2015, 10, e0143283. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.; Pinto, J.; Azevedo, A.I.; Barros-Silva, D.; Jerónimo, C.; Henrique, R.; de Lourdes Bastos, M.; Guedes de Pinho, P.; Carvalho, M. Identification of a biomarker panel for improvement of prostate cancer diagnosis by volatile metabolic profiling of urine. Br. J. Cancer 2019, 121, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Lima, A.R.; Pinto, J.; Carvalho-Maia, C.; Jerónimo, C.; Henrique, R.; de Bastos, M.L.; Carvalho, M.; Guedes de Pinho, P. A Panel of Urinary Volatile Biomarkers for Differential Diagnosis of Prostate Cancer from Other Urological Cancers. Cancers 2020, 12, 2017. [Google Scholar] [CrossRef] [PubMed]
- Ashour, N.; Angulo, J.; Andrés, G.; Alelú, R.; González-Corpas, A.; Toledo, M.; Rodríguez-Barbero, J.; López, J.; Sánchez-Chapado, M.; Ropero, S. A DNA hypermethylation profile reveals new potential biomarkers for prostate cancer diagnosis and prognosis. Prostate 2014, 74, 1171–1182. [Google Scholar] [CrossRef]
- Payne, S.R.; Serth, J.; Schostak, M.; Kamradt, J.; Strauss, A.; Thelen, P.; Model, F.; Day, J.K.; Liebenberg, V.; Morotti, A.; et al. DNA methylation biomarkers of prostate cancer: Confirmation of candidates and evidence urine is the most sensitive body fluid for non-invasive detection. Prostate 2009, 69, 1257–1269. [Google Scholar] [CrossRef]
- Sunami, E.; Shinozaki, M.; Higano, C.S.; Wollman, R.; Dorff, T.B.; Tucker, S.J.; Martinez, S.R.; Singer, F.R.; Hoon, D.S. Multimarker Circulating DNA Assay for Assessing Blood of Prostate Cancer Patients. Clin. Chem. 2009, 55, 559–567. [Google Scholar] [CrossRef]
- Buszewska-Forajta, M.; Pomastowski, P.; Monedeiro, F.; Walczak-Skierska, J.; Markuszewski, M.; Matuszewski, M.; Markuszewski, M.; Buszewski, B. Lipidomics as a Diagnostic Tool for Prostate Cancer. Cancers 2021, 13, 2000. [Google Scholar] [CrossRef] [PubMed]
- Min, H.K.; Lim, S.; Chung, B.C.; Moon, M.H. Shotgun lipidomics for candidate biomarkers of urinary phospholipids in prostate cancer. Anal. Bioanal. Chem. 2011, 399, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Nakayama, K.; Goto, T.; Kimura, H.; Akamatsu, S.; Hayashi, Y.; Fujita, K.; Kobayashi, T.; Shimizu, K.; Nonomura, N.; et al. High level of phosphatidylcholines/lysophosphatidylcholine ratio in urine is associated with prostate cancer. Cancer Sci. 2021, 112, 4292–4302. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; et al. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar] [CrossRef]
- Jentzmik, F.; Stephan, C.; Miller, K.; Schrader, M.; Erbersdobler, A.; Kristiansen, G.; Lein, M.; Jung, K. Sarcosine in Urine after Digital Rectal Examination Fails as a Marker in Prostate Cancer Detection and Identification of Aggressive Tumours. Eur. Urol. 2010, 58, 12–18. [Google Scholar] [CrossRef]
- Zhang, T.; Watson, D.G.; Wang, L.; Abbas, M.; Murdoch, L.; Bashford, L.; Ahmad, I.; Lam, N.-Y.; Ng, A.C.F.; Leung, H.Y. Application of Holistic Liquid Chromatography-High Resolution Mass Spectrometry Based Urinary Metabolomics for Prostate Cancer Detection and Biomarker Discovery. PLoS ONE 2013, 8, e65880. [Google Scholar] [CrossRef]
- Pinto, F.G.; Mahmud, I.; Harmon, T.A.; Rubio, V.Y.; Garrett, T.J. Rapid Prostate Cancer Noninvasive Biomarker Screening Using Segmented Flow Mass Spectrometry-Based Untargeted Metabolomics. J. Proteome Res. 2020, 19, 2080–2091. [Google Scholar] [CrossRef]
- Jones, A.L.; Dhanapala, L.; Baldo, T.A.; Sharafeldin, M.; Krause, C.E.; Shen, M.; Moghaddam, S.; Faria, R.C.; Dey, D.K.; Watson, R.W.; et al. Prostate Cancer Diagnosis in the Clinic Using an 8-Protein Biomarker Panel. Anal. Chem. 2021, 93, 1059–1067. [Google Scholar] [CrossRef]
- Song, J.; Ma, S.; Sokoll, L.J.; Eguez, R.V.; Höti, N.; Zhang, H.; Mohr, P.; Dua, R.; Patil, D.; May, K.D.; et al. A panel of selected serum protein biomarkers for the detection of aggressive prostate cancer. Theranostics 2021, 11, 6214–6224. [Google Scholar] [CrossRef]
- Al-Ruwaili, A.J.; Larkin, S.E.T.; Zeidan, A.B.; Taylor, M.G.; Adra, C.N.; Aukim-Hastie, C.L.; Townsend, A.P. Discovery of Serum Protein Biomarkers for Prostate Cancer Progression by Proteomic Analysis. Cancer Genom.-Proteom. 2010, 7, 93–103. [Google Scholar]
- A Haj-Ahmad, T.; Abdalla, M.A.; Haj-Ahmad, Y. Potential Urinary Protein Biomarker Candidates for the Accurate Detection of Prostate Cancer among Benign Prostatic Hyperplasia Patients. J. Cancer 2014, 5, 103–114. [Google Scholar] [CrossRef]
- Daniel, R.; Wu, Q.; Williams, V.; Clark, G.; Guruli, G.; Zehner, Z. A Panel of MicroRNAs as Diagnostic Biomarkers for the Identification of Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 1281. [Google Scholar] [CrossRef]
- Giglio, S.; De Nunzio, C.; Cirombella, R.; Stoppacciaro, A.; Faruq, O.; Volinia, S.; Baldassarre, G.; Tubaro, A.; Ishii, H.; Croce, C.M.; et al. A preliminary study of micro-RNAs as minimally invasive biomarkers for the diagnosis of prostate cancer patients. J. Exp. Clin. Cancer Res. 2021, 40, 79. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.J.; Richards, R.S.; Chow, C.W.K.; Doi, S.A.R.; Schirra, H.J.; Buck, M.; Samaratunga, H.; Perry-Keene, J.; Payton, D.; Yaxley, J.; et al. Prostate-based biofluids for the detection of prostate cancer: A comparative study of the diagnostic performance of cell-sourced RNA biomarkers. Prostate Int. 2016, 4, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Van Neste, L.; Hendriks, R.J.; Dijkstra, S.; Trooskens, G.; Cornel, E.B.; Jannink, S.A.; de Jong, H.; Hessels, D.; Smit, F.P.; Melchers, W.J.G.; et al. Detection of High-grade Prostate Cancer Using a Urinary Molecular Biomarker–Based Risk Score. Eur. Urol. 2016, 70, 740–748. [Google Scholar] [CrossRef] [PubMed]
- Capelli, L.; Bax, C.; Grizzi, F.; Taverna, G. Optimization of training and measurement protocol for eNose analysis of urine headspace aimed at prostate cancer diagnosis. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Woollam, M.; Wang, L.; Grocki, P.; Liu, S.; Siegel, A.; Kalra, M.; Goodpaster, J.; Yokota, H.; Agarwal, M. Tracking the Progression of Triple Negative Mammary Tumors over Time by Chemometric Analysis of Urinary Volatile Organic Compounds. Cancers 2021, 13, 1462. [Google Scholar] [CrossRef] [PubMed]
- Woollam, M.; Teli, M.; Liu, S.; Daneshkhah, A.; Siegel, A.P.; Yokota, H.; Agarwal, M. Urinary Volatile Terpenes Analyzed by Gas Chromatography–Mass Spectrometry to Monitor Breast Cancer Treatment Efficacy in Mice. J. Proteome Res. 2020, 19, 1913–1922. [Google Scholar] [CrossRef]
- Woollam, M.; Teli, M.; Angarita-Rivera, P.; Liu, S.; Siegel, A.P.; Yokota, H.; Agarwal, M. Detection of volatile organic compounds (VOCs) in urine via gas chromatography-mass spectrometry QTOF to differentiate between localized and metastatic models of breast cancer. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Silva, C.; Passos, M.; Câmara, J. Solid phase microextraction, mass spectrometry and metabolomic approaches for detection of potential urinary cancer biomarkers—A powerful strategy for breast cancer diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef]
- Kwak, J.; Grigsby, C.C.; Rizki, M.M.; Preti, G.; Köksal, M.; Josue, J.; Yamazaki, K.; Beauchamp, G.K. Differential binding between volatile ligands and major urinary proteins due to genetic variation in mice. Physiol. Behav. 2012, 107, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dong, R.; Wang, X.; Lian, A.; Chi, C.; Ke, C.; Guo, L.; Liu, S.; Zhao, W.; Xu, G.; et al. Exhaled volatile organic compounds as lung cancer biomarkers during one-lung ventilation. Sci. Rep. 2014, 4, 7312. [Google Scholar] [CrossRef] [PubMed]
- Siegel, A.P.; Daneshkhah, A.; Hardin, D.S.; Shrestha, S.; Varahramyan, K.; Agarwal, M. Analyzing breath samples of hypoglycemic events in type 1 diabetes patients: Towards developing an alternative to diabetes alert dogs. J. Breath Res. 2017, 11, 026007. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Broadhurst, D.I.; Wilson, M.; Wishart, D.S. Translational biomarker discovery in clinical metabolomics: An introductory tutorial. Metabolomics 2012, 9, 280–299. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, C.V.; Pereira, F.; Pereira, J.A.M.; Câmara, J.S. Volatilomics: An Emerging and Promising Avenue for the Detection of Potential Prostate Cancer Biomarkers. Cancers 2022, 14, 3982. [Google Scholar] [CrossRef]
- Wen, Q.; Boshier, P.; Myridakis, A.; Belluomo, I.; Hanna, G.B. Urinary Volatile Organic Compound Analysis for the Diagnosis of Cancer: A Systematic Literature Review and Quality Assessment. Metabolites 2020, 11, 17. [Google Scholar] [CrossRef]
- Buljubasic, F.; Buchbauer, G. The scent of human diseases: A review on specific volatile organic compounds as diagnostic biomarkers. Flavour Fragr. J. 2015, 30, 5–25. [Google Scholar] [CrossRef]
- Grocki, P.; Woollam, M.; Wang, L.; Liu, S.; Kalra, M.; Siegel, A.P.; Li, B.-Y.; Li, H.; Yokota, H.; Agarwal, M. Chemometric Analysis of Urinary Volatile Organic Compounds to Monitor the Efficacy of Pitavastatin Treatments on Mammary Tumor Progression over Time. Molecules 2022, 27, 4277. [Google Scholar] [CrossRef]
- Silva, C.L.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Implementing a central composite design for the optimization of solid phase microextraction to establish the urinary volatomic expression: A first approach for breast cancer. Metabolomics 2019, 15, 64. [Google Scholar] [CrossRef]
- Kure, S.; Satoi, S.; Kitayama, T.; Nagase, Y.; Nakano, N.; Yamada, M.; Uchiyama, N.; Miyashita, S.; Iida, S.; Takei, H.; et al. A prediction model using 2-propanol and 2-butanone in urine distinguishes breast cancer. Sci. Rep. 2021, 11, 19801. [Google Scholar] [CrossRef]
- Filipiak, W.; Sponring, A.; Mikoviny, T.; Ager, C.; Schubert, J.; Miekisch, W.; Amann, A.; Troppmair, J. Release of volatile organic compounds (VOCs) from the lung cancer cell line CALU-1 in vitro. Cancer Cell Int. 2008, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Passos, M.; Câmara, J.S. Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Br. J. Cancer 2011, 105, 1894–1904. [Google Scholar] [CrossRef] [PubMed]
- Saccenti, E.; Hoefsloot, H.C.J.; Smilde, A.K.; Westerhuis, J.A.; Hendriks, M.M.W.B. Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics 2013, 10, 361–374. [Google Scholar] [CrossRef]
- Ying, X. An Overview of Overfitting and its Solutions. J. Phys. Conf. Ser. 2019, 1168, 022022. [Google Scholar] [CrossRef]
- Janfaza, S.; Khorsand, B.; Nikkhah, M.; Zahiri, J. Digging deeper into volatile organic compounds associated with cancer. Biol. Methods Protoc. 2019, 4, bpz014. [Google Scholar] [CrossRef]
- Liu, Y. Fatty acid oxidation is a dominant bioenergetic pathway in prostate cancer. Prostate Cancer Prostatic Dis. 2006, 9, 230–234. [Google Scholar] [CrossRef]
- Aiderus, A.; Black, M.A.; Dunbier, A.K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer 2018, 18, 805. [Google Scholar] [CrossRef]
- Singh, S.; Rajendran, R.; Kuroda, K.; Isogai, E.; Krstic-Demonacos, M.; Demonacos, C. Oxidative stress and breast cancer biomarkers: The case of the cytochrome P450 2E1. J. Cancer Metastasis Treat. 2016, 2, 268–276. [Google Scholar] [CrossRef]
- Gonzalez, F.J. Role of cytochromes P450 in chemical toxicity and oxidative stress: Studies with CYP2E1. Mutat. Res. 2005, 569, 101–110. [Google Scholar] [CrossRef]
- Mullen, P.J.; Yu, R.; Longo, J.; Archer, M.C.; Penn, L.Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 2016, 16, 718–731. [Google Scholar] [CrossRef]
- Basso, A.; Kirschmeier, P.; Bishop, W. Lipid posttranslational modifications. Farnesyl transferase inhibitors. J. Lipid Res. 2006, 47, 15–31. [Google Scholar] [CrossRef] [PubMed]
- Ashida, S.; Kawada, C.; Inoue, K. Stromal regulation of prostate cancer cell growth by mevalonate pathway enzymes HMGCS1 and HMGCR. Oncol. Lett. 2017, 14, 6533–6542. [Google Scholar] [CrossRef] [PubMed]
- Göbel, A.; Rauner, M.; Hofbauer, L.C.; Rachner, T.D. Cholesterol and beyond—The role of the mevalonate pathway in cancer biology. Biochim. Et Biophys. Acta (BBA)—Rev. Cancer 2020, 1873, 188351. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Recio, C.; Aranda-Tavío, H.; Guerra-Rodríguez, M.; García-Castellano, J.; Fernández-Pérez, L. The Mevalonate Pathway, a Metabolic Target in Cancer Therapy. Front. Oncol. 2021, 11, 626971. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, H.; Daulton, E.; Bannaga, A.S.; Arasaradnam, R.P.; Covington, J.A. Urinary Volatiles and Chemical Characterisation for the Non-Invasive Detection of Prostate and Bladder Cancers. Biosensors 2021, 11, 437. [Google Scholar] [CrossRef]
- Saalberg, Y.; Wolff, M. VOC breath biomarkers in lung cancer. Clin. Chim. Acta 2016, 459, 5–9. [Google Scholar] [CrossRef]
- Guest, C.; Harris, R.; Sfanos, K.S.; Shrestha, E.; Partin, A.W.; Trock, B.; Mangold, L.; Bader, R.; Kozak, A.; Mclean, S.; et al. Feasibility of integrating canine olfaction with chemical and microbial profiling of urine to detect lethal prostate cancer. PLoS ONE 2021, 16, e0245530. [Google Scholar] [CrossRef]
- da Costa, B.; De Martinis, B. Analysis of urinary VOCs using mass spectrometric methods to diagnose cancer: A review. Clin. Mass Spectrom. 2020, 18, 27–37. [Google Scholar] [CrossRef]
- Guo, L.; Qiu, Z.; Wang, Y.; Yu, K.; Zheng, X.; Li, Y.; Liu, M.; Wang, G.; Guo, N.; Yang, M.; et al. Volatile Organic Compounds to Identify Infectious (Bacteria/Viruses) Diseases of the Central Nervous System: A Pilot Study. Eur. Neurol. 2021, 84, 325–332. [Google Scholar] [CrossRef]
- El Qader, A.A.; Lieberman, D.; Avni, Y.S.; Svobodin, N.; Lazarovitch, T.; Sagi, O.; Zeiri, Y. Volatile organic compounds generated by cultures of bacteria and viruses associated with respiratory infections. Biomed. Chromatogr. 2015, 29, 1783–1790. [Google Scholar] [CrossRef]
- Woollam, M.; Angarita-Rivera, P.; Siegel, A.P.; Kalra, V.; Kapoor, R.; Agarwal, M. Exhaled VOCs can discriminate subjects with COVID-19 from healthy controls. J. Breath Res. 2022, 16, 036002. [Google Scholar] [CrossRef] [PubMed]
- Sanmukh, S.G.; Dos Santos, N.J.; Barquilha, C.N.; De Carvalho, M.; Dos Reis, P.P.; Delella, F.K.; Carvalho, H.F.; Latek, D.; Fehér, T.; Felisbino, S.L. Bacterial RNA virus MS2 exposure increases the expression of cancer progression genes in the LNCaP prostate cancer cell line. Oncol. Lett. 2023, 25, 86. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woollam, M.; Siegel, A.P.; Munshi, A.; Liu, S.; Tholpady, S.; Gardner, T.; Li, B.-Y.; Yokota, H.; Agarwal, M. Canine-Inspired Chemometric Analysis of Volatile Organic Compounds in Urine Headspace to Distinguish Prostate Cancer in Mice and Men. Cancers 2023, 15, 1352. https://doi.org/10.3390/cancers15041352
Woollam M, Siegel AP, Munshi A, Liu S, Tholpady S, Gardner T, Li B-Y, Yokota H, Agarwal M. Canine-Inspired Chemometric Analysis of Volatile Organic Compounds in Urine Headspace to Distinguish Prostate Cancer in Mice and Men. Cancers. 2023; 15(4):1352. https://doi.org/10.3390/cancers15041352
Chicago/Turabian StyleWoollam, Mark, Amanda P. Siegel, Adam Munshi, Shengzhi Liu, Sunil Tholpady, Thomas Gardner, Bai-Yan Li, Hiroki Yokota, and Mangilal Agarwal. 2023. "Canine-Inspired Chemometric Analysis of Volatile Organic Compounds in Urine Headspace to Distinguish Prostate Cancer in Mice and Men" Cancers 15, no. 4: 1352. https://doi.org/10.3390/cancers15041352
APA StyleWoollam, M., Siegel, A. P., Munshi, A., Liu, S., Tholpady, S., Gardner, T., Li, B.-Y., Yokota, H., & Agarwal, M. (2023). Canine-Inspired Chemometric Analysis of Volatile Organic Compounds in Urine Headspace to Distinguish Prostate Cancer in Mice and Men. Cancers, 15(4), 1352. https://doi.org/10.3390/cancers15041352





