Circulating Tumor DNA: Less Invasive, More Representative Method to Unveil the Genomic Landscape of Newly Diagnosed Multiple Myeloma Than Bone Marrow Aspirates
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Cohort and Sample Collection
2.2. Genomic DNA Extraction
2.3. Sequencing Library Preparation and Target Capture
2.4. Sequencing Data Analysis
2.5. Enrich Rare Mutation Sequencing (ER-Seq) of ctDNA
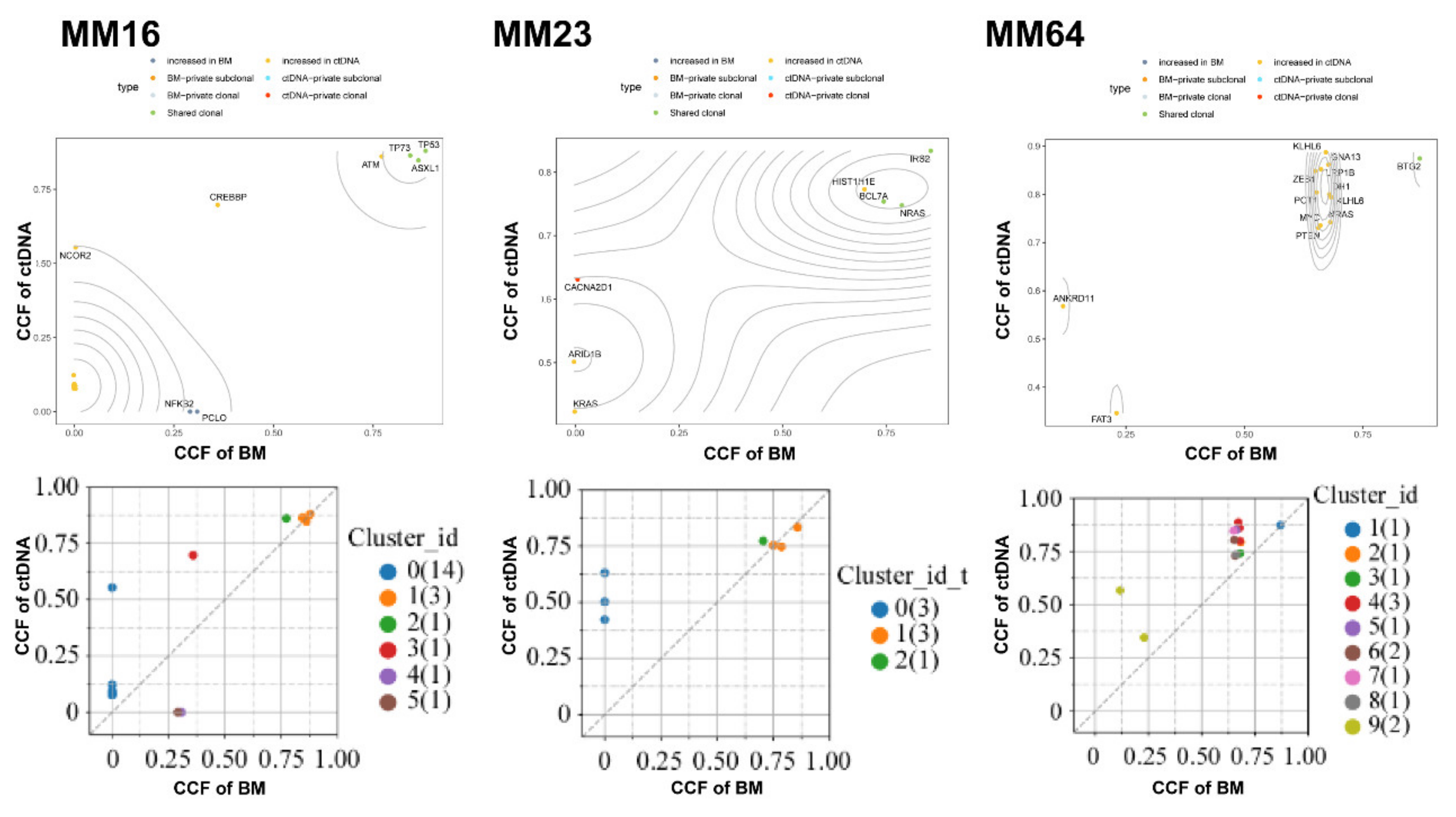
2.6. Clonal Population Structure Analysis
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Statistical Analysis
3. Results
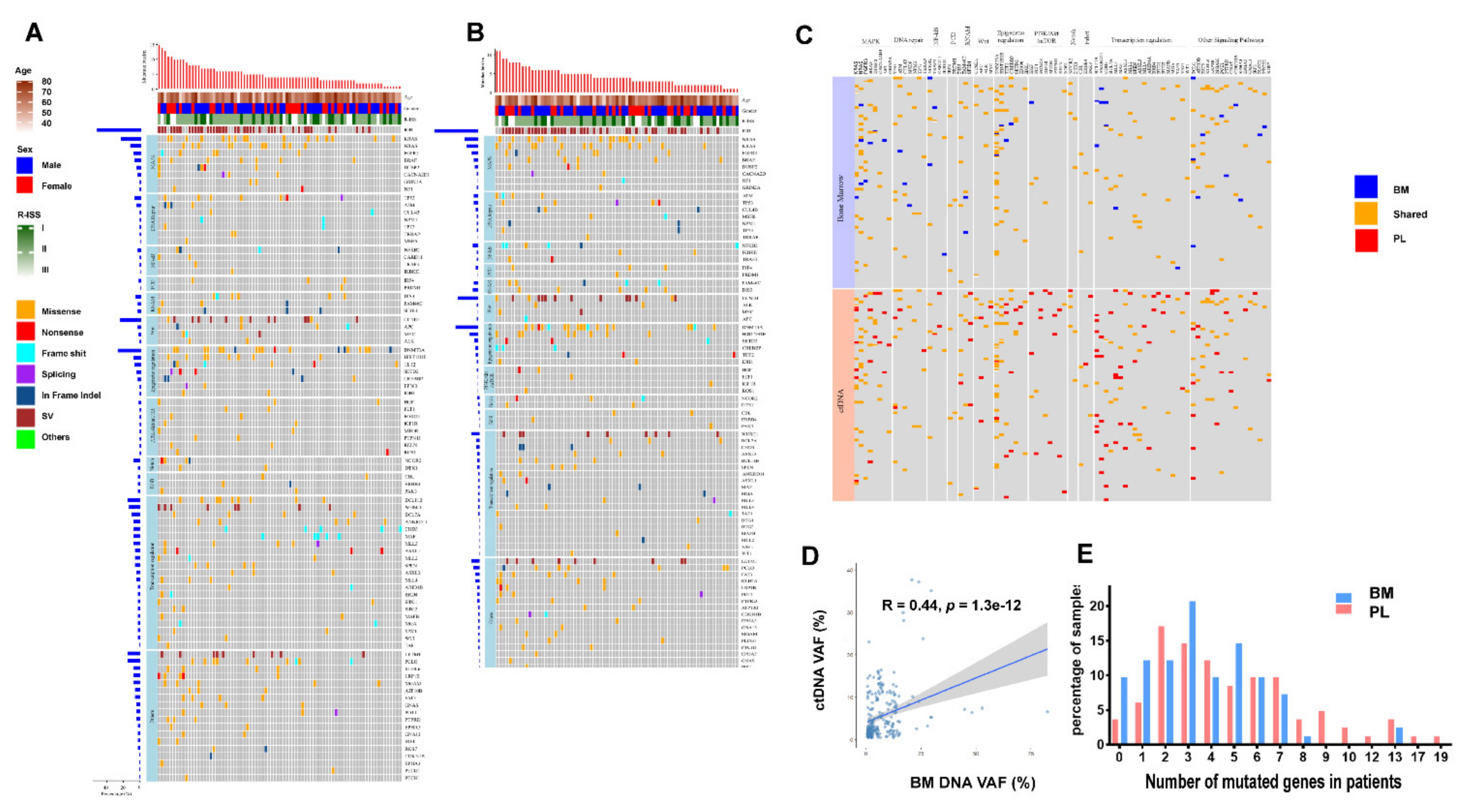
3.1. Higher Levels of Mutation Identified in ctDNA Than in BM
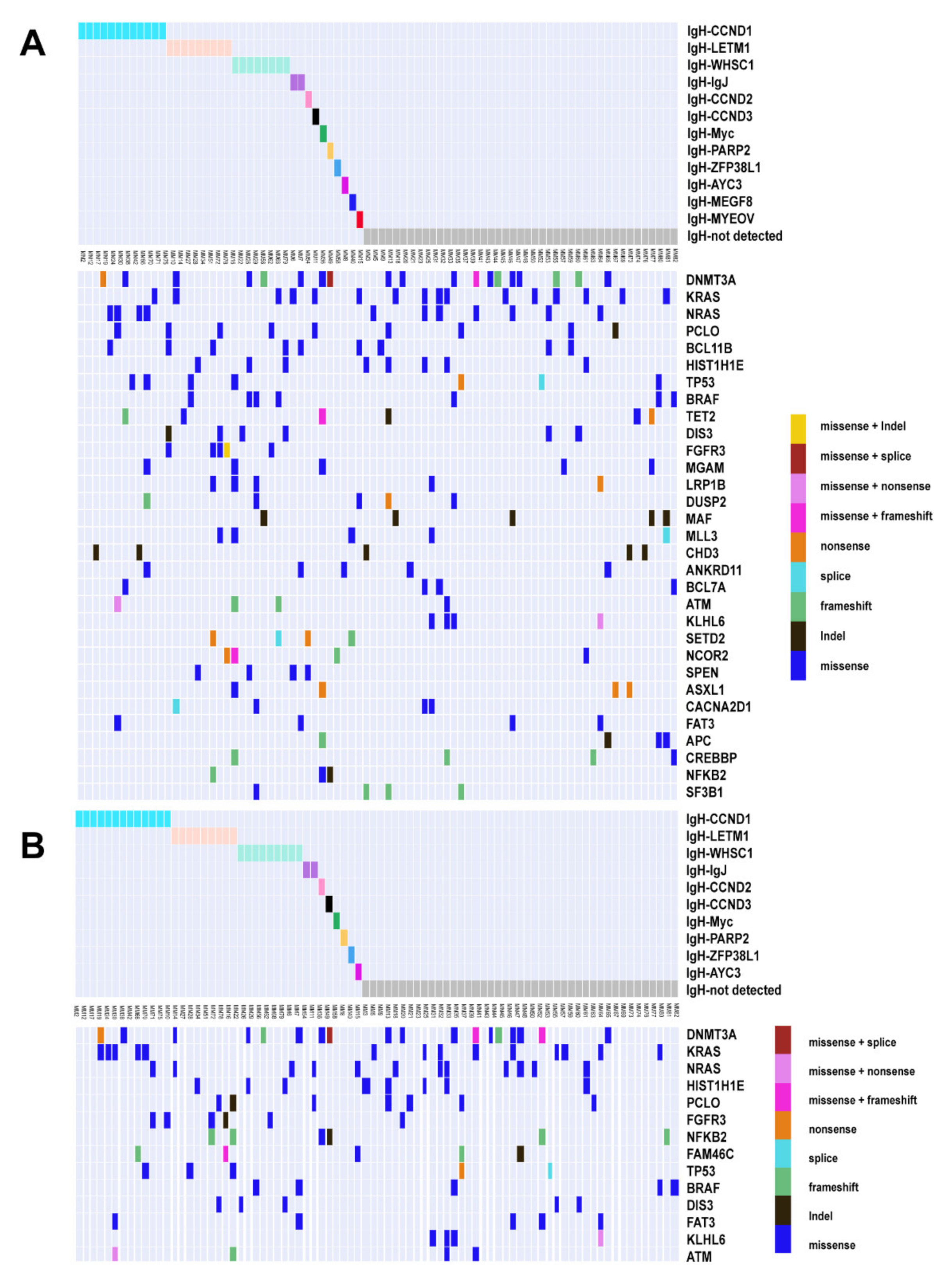
3.2. Unique Mutated Gene Profiles in ctDNA
3.3. High Sensitivity of Structural Variations (SV) and Association of IGH Translocation with Gene Mutations in ctDNA
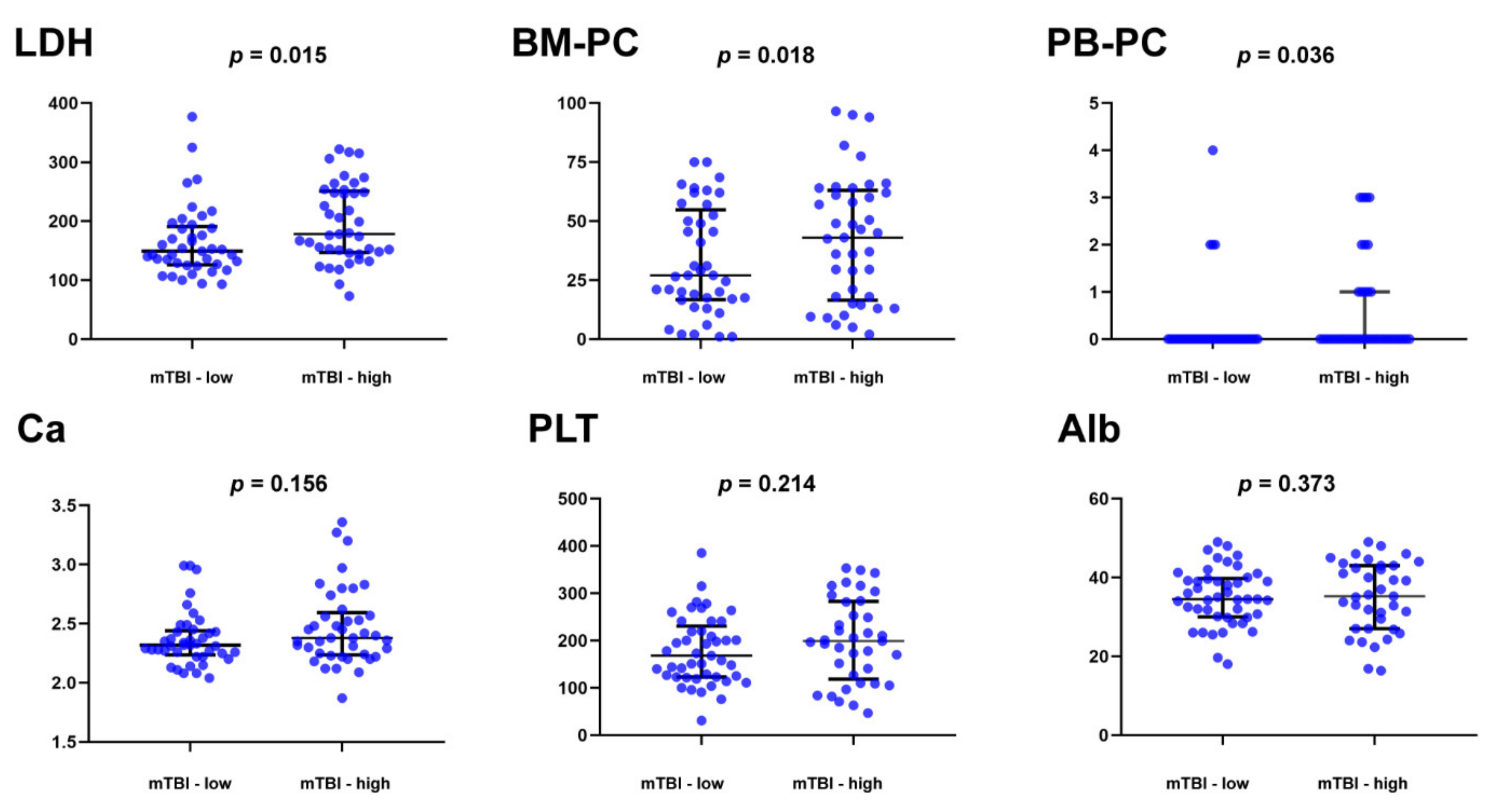
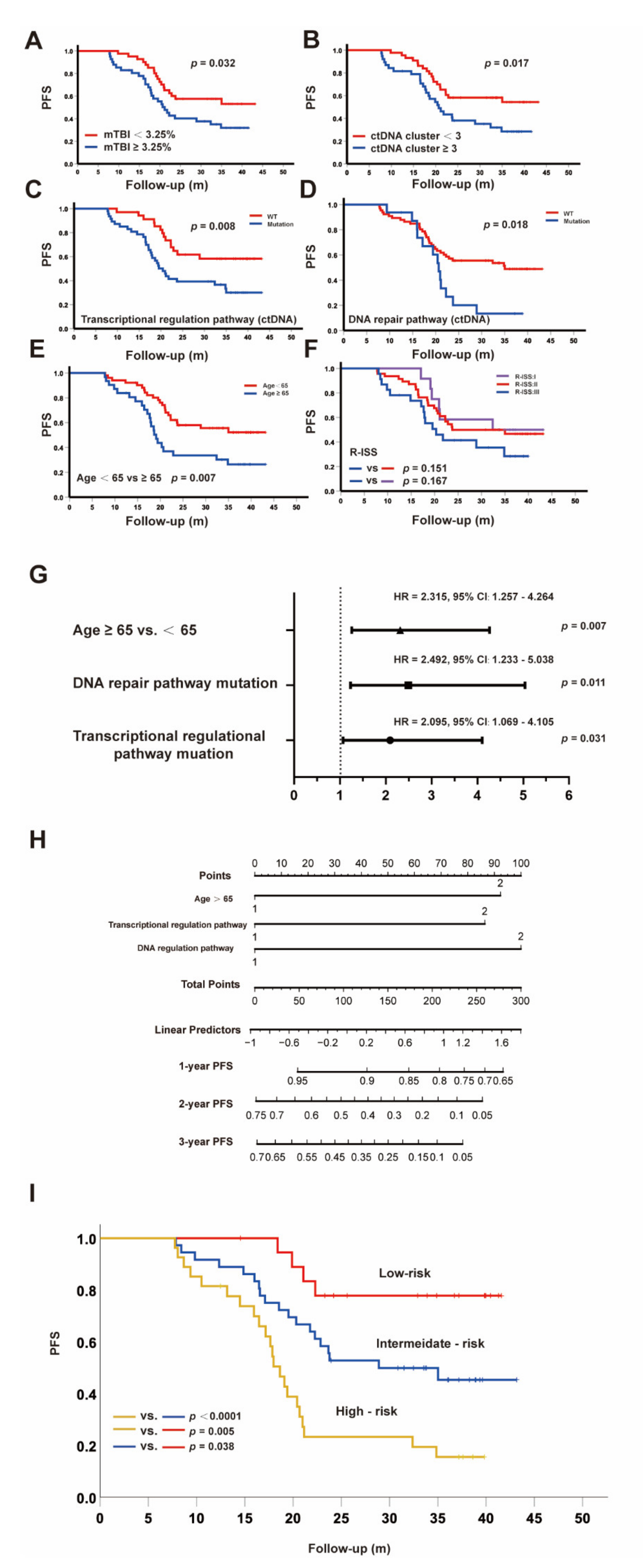
3.4. Clinical Significance of the Molecular Tumor Burden Index in ctDNA
3.5. ctDNA as a Promising Parameter to Predict Inferior Survival
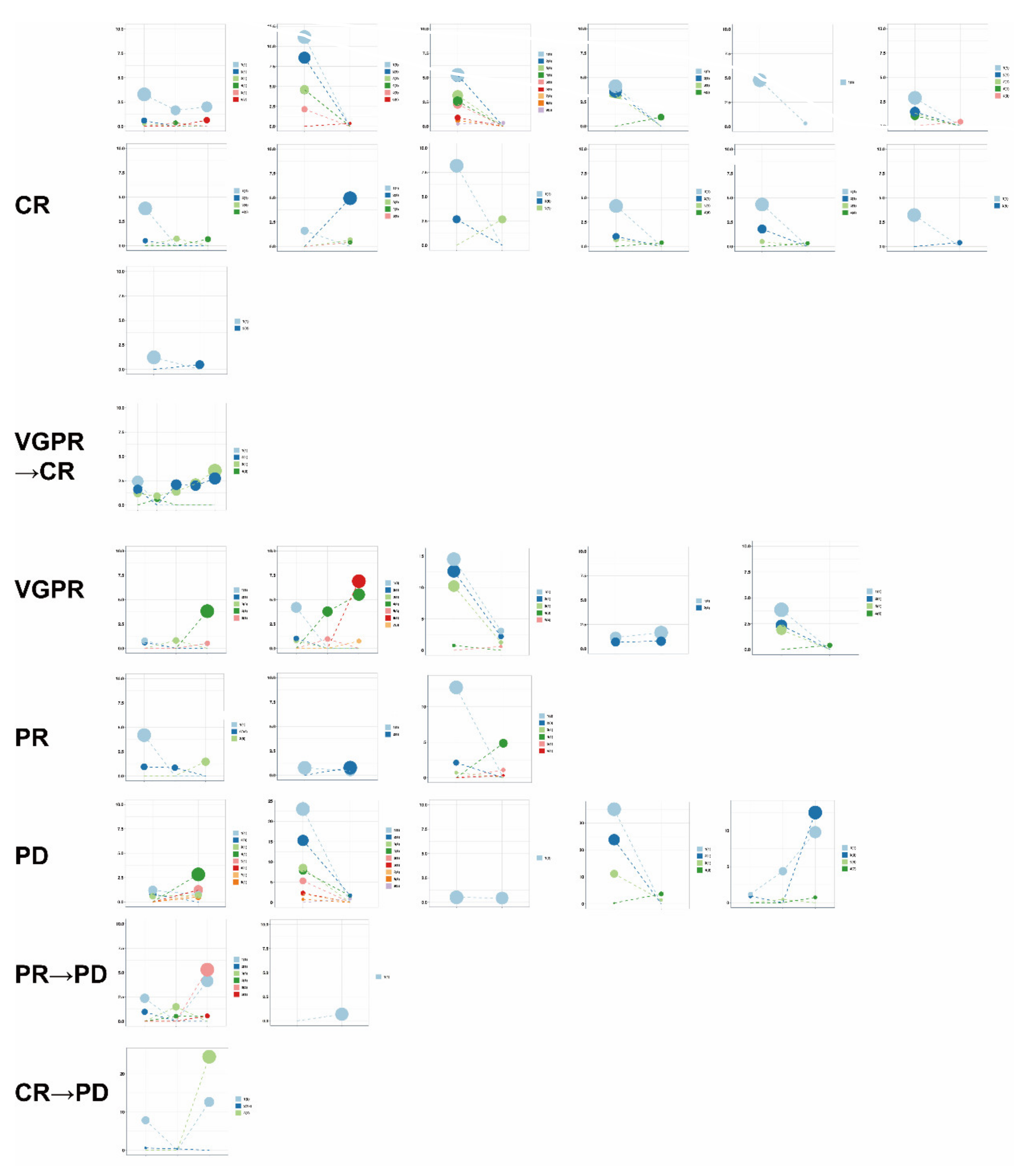
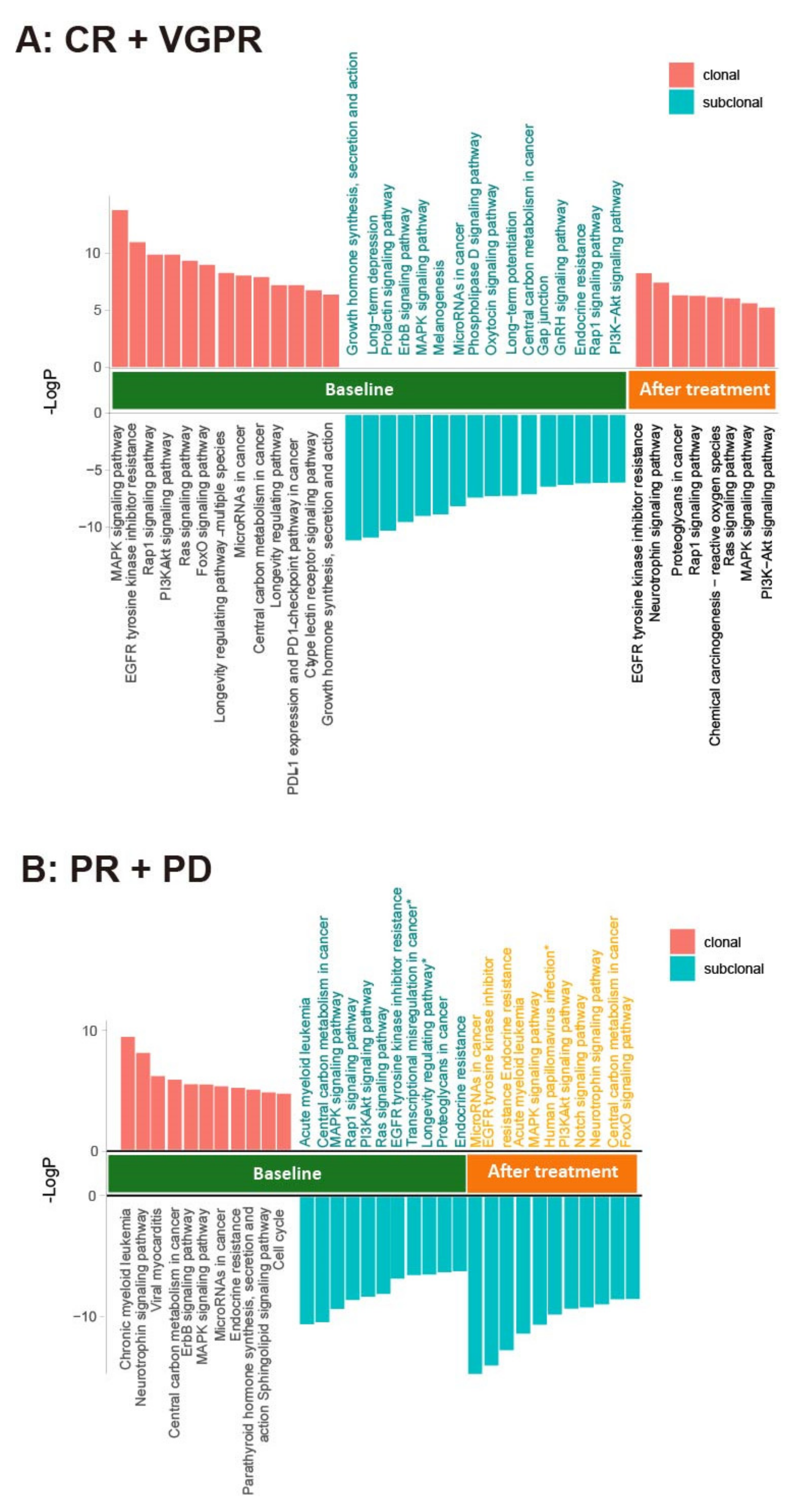
3.6. Use of Clonal Composition Analysis of ctDNA to Monitor Therapeutic Response
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gertz, M.A.; Lacy, M.Q.; Dispenzieri, A.; Greipp, P.R.; Litzow, M.R.; Henderson, K.J.; Van Wier, S.A.; Ahmann, G.J.; Fonseca, R. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood 2005, 106, 2837–2840. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.K.; Rajkumar, S.V.; Dispenzieri, A.; Lacy, M.Q.; Hayman, S.R.; Buadi, F.K.; Zeldenrust, S.R.; Dingli, D.; Russell, S.J.; Lust, J.A.; et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood 2008, 111, 2516–2520. [Google Scholar] [CrossRef] [PubMed]
- Chapman, M.A.; Lawrence, M.S.; Keats, J.J.; Cibulskis, K.; Sougnez, C.; Schinzel, A.C.; Harview, C.L.; Brunet, J.P.; Ahmann, G.J.; Adli, M.; et al. Initial genome sequencing and analysis of multiple myeloma. Nature 2011, 471, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Carter, S.L.; Cruz-Gordillo, P.; Lawrence, M.S.; Auclair, D.; Sougnez, C.; Knoechel, B.; Gould, J.; Saksena, G.; et al. Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell 2014, 25, 91–101. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.E.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 2018, 132, 587–597. [Google Scholar] [CrossRef]
- Manier, S.; Salem, K.Z.; Park, J.; Landau, D.A.; Getz, G.; Ghobrial, I.M. Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 100–113. [Google Scholar] [CrossRef]
- Merz, M.; Merz, A.M.A.; Wang, J.; Wei, L.; Hu, Q.; Hutson, N.; Rondeau, C.; Celotto, K.; Belal, A.; Alberico, R.; et al. Deciphering spatial genomic heterogeneity at a single cell resolution in multiple myeloma. Nat. Commun. 2022, 13, 807. [Google Scholar] [CrossRef]
- Bolli, N.; Genuardi, E.; Ziccheddu, B.; Martello, M.; Oliva, S.; Terragna, C. Next-Generation Sequencing for Clinical Management of Multiple Myeloma: Ready for Prime Time? Front. Oncol. 2020, 10, 189. [Google Scholar] [CrossRef]
- Munshi, N.C.; Avet-Loiseau, H.; Anderson, K.C.; Neri, P.; Paiva, B.; Samur, M.; Dimopoulos, M.; Kulakova, M.; Lam, A.; Hashim, M.; et al. A large meta-analysis establishes the role of MRD negativity in long-term survival outcomes in patients with multiple myeloma. Blood Adv. 2020, 4, 5988–5999. [Google Scholar] [CrossRef]
- Rasche, L.; Chavan, S.S.; Stephens, O.W.; Patel, P.H.; Tytarenko, R.; Ashby, C.; Bauer, M.; Stein, C.; Deshpande, S.; Wardell, C.; et al. Spatial genomic heterogeneity in multiple myeloma revealed by multi-region sequencing. Nat. Commun. 2017, 8, 268. [Google Scholar] [CrossRef]
- Lin, C.; Liu, X.; Zheng, B.; Ke, R.; Tzeng, C.M. Liquid Biopsy, ctDNA Diagnosis through NGS. Life 2021, 11, 890. [Google Scholar] [CrossRef]
- Manier, S.; Park, J.; Capelletti, M.; Bustoros, M.; Freeman, S.S.; Ha, G.; Rhoades, J.; Liu, C.J.; Huynh, D.; Reed, S.C.; et al. Whole-exome sequencing of cell-free DNA and circulating tumor cells in multiple myeloma. Nat. Commun. 2018, 9, 1691. [Google Scholar] [CrossRef]
- Deshpande, S.; Tytarenko, R.G.; Wang, Y.; Boyle, E.M.; Ashby, C.; Schinke, C.D.; Thanendrarajan, S.; Zangari, M.; Zhan, F.; Davies, F.E.; et al. Monitoring treatment response and disease progression in myeloma with circulating cell-free DNA. Eur. J. Haematol. 2021, 106, 230–240. [Google Scholar] [CrossRef]
- Kis, O.; Kaedbey, R.; Chow, S.; Danesh, A.; Dowar, M.; Li, T.; Li, Z.; Liu, J.; Mansour, M.; Masih-Khan, E.; et al. Circulating tumour DNA sequence analysis as an alternative to multiple myeloma bone marrow aspirates. Nat. Commun. 2017, 8, 15086. [Google Scholar] [CrossRef]
- Guo, G.; Raje, N.S.; Seifer, C.; Kloeber, J.; Isenhart, R.; Ha, G.; Yee, A.J.; O’Donnell, E.K.; Tai, Y.T.; Richardson, P.G.; et al. Genomic discovery and clonal tracking in multiple myeloma by cell-free DNA sequencing. Leukemia 2018, 32, 1838–1841. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Morley, R.; Khong, T.; Kalff, A.; Bergin, K.; Hocking, J.; Savvidou, I.; Bowen, K.M.; Ramachandran, M.; Choi, K.; et al. Monitoring tumour burden and therapeutic response through analysis of circulating tumour DNA and extracellular RNA in multiple myeloma patients. Leukemia 2019, 33, 2022–2033. [Google Scholar] [CrossRef]
- Mithraprabhu, S.; Khong, T.; Ramachandran, M.; Chow, A.; Klarica, D.; Mai, L.; Walsh, S.; Broemeling, D.; Marziali, A.; Wiggin, M.; et al. Circulating tumour DNA analysis demonstrates spatial mutational heterogeneity that coincides with disease relapse in myeloma. Leukemia 2017, 31, 1695–1705. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Li, J.; Lupat, R.; Amarasinghe, K.C.; Thompson, E.R.; Doyle, M.A.; Ryland, G.L.; Tothill, R.W.; Halgamuge, S.K.; Campbell, I.G.; Gorringe, K.L. CONTRA: Copy number analysis for targeted resequencing. Bioinformatics 2012, 28, 1307–1313. [Google Scholar] [CrossRef]
- Cowell, J.K.; Lo, K.C. Application of oligonucleotides arrays for coincident comparative genomic hybridization, ploidy status and loss of heterozygosity studies in human cancers. Methods Mol. Biol. 2009, 556, 47–65. [Google Scholar] [CrossRef]
- Zaretsky, J.M.; Garcia-Diaz, A.; Shin, D.S.; Escuin-Ordinas, H.; Hugo, W.; Hu-Lieskovan, S.; Torrejon, D.Y.; Abril-Rodriguez, G.; Sandoval, S.; Barthly, L.; et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N. Engl. J. Med. 2016, 375, 819–829. [Google Scholar] [CrossRef]
- Lv, X.; Zhao, M.; Yi, Y.; Zhang, L.; Guan, Y.; Liu, T.; Yang, L.; Chen, R.; Ma, J.; Yi, X. Detection of Rare Mutations in CtDNA Using Next Generation Sequencing. JoVE J. Vis. Exp. 2017, 24, e56342. [Google Scholar] [CrossRef]
- Roth, A.; Khattra, J.; Yap, D.; Wan, A.; Laks, E.; Biele, J.; Ha, G.; Aparicio, S.; Bouchard-Cote, A.; Shah, S.P. PyClone: Statistical inference of clonal population structure in cancer. Nat. Methods 2014, 11, 396–398. [Google Scholar] [CrossRef]
- Murtaza, M.; Dawson, S.J.; Pogrebniak, K.; Rueda, O.M.; Provenzano, E.; Grant, J.; Chin, S.F.; Tsui, D.W.Y.; Marass, F.; Gale, D.; et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat. Commun. 2015, 6, 8760. [Google Scholar] [CrossRef] [PubMed]
- Bolli, N.; Biancon, G.; Moarii, M.; Gimondi, S.; Li, Y.; de Philippis, C.; Maura, F.; Sathiaseelan, V.; Tai, Y.T.; Mudie, L.; et al. Analysis of the genomic landscape of multiple myeloma highlights novel prognostic markers and disease subgroups. Leukemia 2018, 32, 2604–2616. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, N.; Mi, L.; Shi, Y.; Liu, W.; Song, Y.; Shu, S.; Zhu, J. Correlation of mutational landscape and survival outcome of peripheral T-cell lymphomas. Exp. Hematol. Oncol. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Tsai, C.J.; Jang, H. Why Are Some Driver Mutations Rare? Trends Pharmacol. Sci. 2019, 40, 919–929. [Google Scholar] [CrossRef] [PubMed]
- Vendramini, E.; Bomben, R.; Pozzo, F.; Bittolo, T.; Tissino, E.; Gattei, V.; Zucchetto, A. KRAS and RAS-MAPK Pathway Deregulation in Mature B Cell Lymphoproliferative Disorders. Cancers 2022, 14, 666. [Google Scholar] [CrossRef] [PubMed]
- Smiech, M.; Leszczynski, P.; Kono, H.; Wardell, C.; Taniguchi, H. Emerging BRAF Mutations in Cancer Progression and Their Possible Effects on Transcriptional Networks. Genes 2020, 11, 1342. [Google Scholar] [CrossRef]
- Song, J.; Zhang, H.; Dong, N.; Zhang, X.; Hussaini, M.; Jain, A.; Moscinski, L.; Shain, K.; Baz, R.; Alsina, M.; et al. The Application of NextGen Sequencing in the Diagnosis of Myeloid Neoplasms in Myeloma Patients With Cytopenia. Clin. Lymphoma Myeloma Leuk. 2021, 22, e414–e426. [Google Scholar] [CrossRef]
- Huang, Q.S.; Wang, J.Z.; Qin, Y.Z.; Zeng, Q.Z.; Jiang, Q.; Jiang, H.; Lu, J.; Liu, H.X.; Liu, Y.; Wang, J.B.; et al. Overexpression of WT1 and PRAME predicts poor outcomes of patients with myelodysplastic syndromes with thrombocytopenia. Blood Adv. 2019, 3, 3406–3418. [Google Scholar] [CrossRef]
- Yang, L.; Wang, Y.Z.; Zhu, H.H.; Chang, Y.; Li, L.D.; Chen, W.M.; Long, L.Y.; Zhang, Y.H.; Liu, Y.R.; Lu, J.; et al. PRAME Gene Copy Number Variation Is Related to Its Expression in Multiple Myeloma. DNA Cell Biol. 2017, 36, 1099–1107. [Google Scholar] [CrossRef]
- Coccaro, N.; Tota, G.; Anelli, L.; Zagaria, A.; Casieri, P.; Cellamare, A.; Minervini, C.F.; Minervini, A.; Cumbo, C.; Impera, L.; et al. MYEOV gene overexpression in primary plasma cell leukemia with t(11;14)(q13;q32). Oncol. Lett. 2016, 12, 1460–1464. [Google Scholar] [CrossRef]
- Specht, K.; Haralambieva, E.; Bink, K.; Kremer, M.; Mandl-Weber, S.; Koch, I.; Tomer, R.; Hofler, H.; Schuuring, E.; Kluin, P.M.; et al. Different mechanisms of cyclin D1 overexpression in multiple myeloma revealed by fluorescence in situ hybridization and quantitative analysis of mRNA levels. Blood 2004, 104, 1120–1126. [Google Scholar] [CrossRef]
- Yi, Z.; Ma, F.; Rong, G.; Liu, B.; Guan, Y.; Li, J.; Sun, X.; Wang, W.; Guan, X.; Mo, H.; et al. The molecular tumor burden index as a response evaluation criterion in breast cancer. Signal Transduct. Target. Ther. 2021, 6, 251. [Google Scholar] [CrossRef]
- Cheng, Q.; Cai, L.; Zhang, Y.; Chen, L.; Hu, Y.; Sun, C. Circulating Plasma Cells as a Biomarker to Predict Newly Diagnosed Multiple Myeloma Prognosis: Developing Nomogram Prognostic Models. Front. Oncol. 2021, 11, 639528. [Google Scholar] [CrossRef]
- Tang, B.; Zhu, J.; Li, J.; Fan, K.; Gao, Y.; Cheng, S.; Kong, C.; Zheng, L.; Wu, F.; Weng, Q.; et al. The ferroptosis and iron-metabolism signature robustly predicts clinical diagnosis, prognosis and immune microenvironment for hepatocellular carcinoma. Cell Commun. Signal. 2020, 18, 174. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, W.; Wang, J. Mutations In Thirty Hotspot Genes In Newly Diagnosed Chinese Multiple Myeloma Patients. OncoTargets Ther. 2019, 12, 9999–10010. [Google Scholar] [CrossRef]
- Montefiori, L.E.; Mullighan, C.G. Redefining the biological basis of lineage-ambiguous leukemia through genomics: BCL11B deregulation in acute leukemias of ambiguous lineage. Best Pract. Res. Clin. Haematol. 2021, 34, 101329. [Google Scholar] [CrossRef]
- Lennon, M.J.; Jones, S.P.; Lovelace, M.D.; Guillemin, G.J.; Brew, B.J. Bcl11b-A Critical Neurodevelopmental Transcription Factor-Roles in Health and Disease. Front. Cell Neurosci. 2017, 11, 89. [Google Scholar] [CrossRef]
- Gutierrez, A.; Kentsis, A.; Sanda, T.; Holmfeldt, L.; Chen, S.C.; Zhang, J.; Protopopov, A.; Chin, L.; Dahlberg, S.E.; Neuberg, D.S.; et al. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood 2011, 118, 4169–4173. [Google Scholar] [CrossRef]
- Cardona-Benavides, I.J.; de Ramon, C.; Gutierrez, N.C. Genetic Abnormalities in Multiple Myeloma: Prognostic and Therapeutic Implications. Cells 2021, 10, 336. [Google Scholar] [CrossRef]
- Walker, B.A.; Boyle, E.M.; Wardell, C.P.; Murison, A.; Begum, D.B.; Dahir, N.M.; Proszek, P.Z.; Johnson, D.C.; Kaiser, M.F.; Melchor, L.; et al. Mutational Spectrum, Copy Number Changes, and Outcome: Results of a Sequencing Study of Patients With Newly Diagnosed Myeloma. J. Clin. Oncol. 2015, 33, 3911–3920. [Google Scholar] [CrossRef]
- Walker, B.A.; Mavrommatis, K.; Wardell, C.P.; Ashby, T.C.; Bauer, M.; Davies, F.; Rosenthal, A.; Wang, H.; Qu, P.; Hoering, A.; et al. A high-risk, Double-Hit, group of newly diagnosed myeloma identified by genomic analysis. Leukemia 2019, 33, 159–170. [Google Scholar] [CrossRef]
- Keats, J.J.; Fonseca, R.; Chesi, M.; Schop, R.; Baker, A.; Chng, W.J.; Van Wier, S.; Tiedemann, R.; Shi, C.X.; Sebag, M.; et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 2007, 12, 131–144. [Google Scholar] [CrossRef]
- Morgan, G.J.; Walker, B.A.; Davies, F.E. The genetic architecture of multiple myeloma. Nat. Rev. Cancer 2012, 12, 335–348. [Google Scholar] [CrossRef]
- Bertamini, L.; Oliva, S.; Rota-Scalabrini, D.; Paris, L.; More, S.; Corradini, P.; Ledda, A.; Gentile, M.; De Sabbata, G.; Pietrantuono, G.; et al. High Levels of Circulating Tumor Plasma Cells as a Key Hallmark of Aggressive Disease in Transplant-Eligible Patients With Newly Diagnosed Multiple Myeloma. J. Clin. Oncol. 2022, 40, 3120–3131. [Google Scholar] [CrossRef] [PubMed]

| Factors | Values |
|---|---|
| Sex, Male (%) | 51 (62.2%) |
| Age (Median, Range) | 62 (33–80) |
| DS (II/III) | 1/4/77 |
| ISS (I/II/III) | 19/21/42 |
| Renal insufficiency (SCr > 2 mg/dL) | 21 (25.6%) |
| Low PLT (<100 × 109/L) | 10 (12.2%) |
| High LDH | 18 (22.0%) |
| Del (17p) | 10 (12.2%) |
| Gain/Amplification of 1q21 | 44 (53.7%) |
| Del (RB1) | 38 (46.3%) |
| T(11;14) | 20 (24.4%) |
| T(4;14) | 18 (22.0%) |
| EMD | 25 (30.5%) |
| Response (CR/VGPR/PR/NR or PD) | 17/20/30/15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Guo, J.; Yi, Y.; Gao, X.; Wen, L.; Duan, W.; Wen, Z.; Liu, Y.; Guan, Y.; Xia, X.; et al. Circulating Tumor DNA: Less Invasive, More Representative Method to Unveil the Genomic Landscape of Newly Diagnosed Multiple Myeloma Than Bone Marrow Aspirates. Cancers 2022, 14, 4914. https://doi.org/10.3390/cancers14194914
Liu Y, Guo J, Yi Y, Gao X, Wen L, Duan W, Wen Z, Liu Y, Guan Y, Xia X, et al. Circulating Tumor DNA: Less Invasive, More Representative Method to Unveil the Genomic Landscape of Newly Diagnosed Multiple Myeloma Than Bone Marrow Aspirates. Cancers. 2022; 14(19):4914. https://doi.org/10.3390/cancers14194914
Chicago/Turabian StyleLiu, Yang, Jiapei Guo, Yuting Yi, Xuan Gao, Lei Wen, Wenbing Duan, Zhaohong Wen, Yaoyao Liu, Yanfang Guan, Xuefeng Xia, and et al. 2022. "Circulating Tumor DNA: Less Invasive, More Representative Method to Unveil the Genomic Landscape of Newly Diagnosed Multiple Myeloma Than Bone Marrow Aspirates" Cancers 14, no. 19: 4914. https://doi.org/10.3390/cancers14194914
APA StyleLiu, Y., Guo, J., Yi, Y., Gao, X., Wen, L., Duan, W., Wen, Z., Liu, Y., Guan, Y., Xia, X., Ma, L., Fu, R., Liu, L., Huang, X., Ge, Q., & Lu, J. (2022). Circulating Tumor DNA: Less Invasive, More Representative Method to Unveil the Genomic Landscape of Newly Diagnosed Multiple Myeloma Than Bone Marrow Aspirates. Cancers, 14(19), 4914. https://doi.org/10.3390/cancers14194914





