Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. In Vitro Modeling of the Human Brain with Induced-Pluripotent Stem Cells
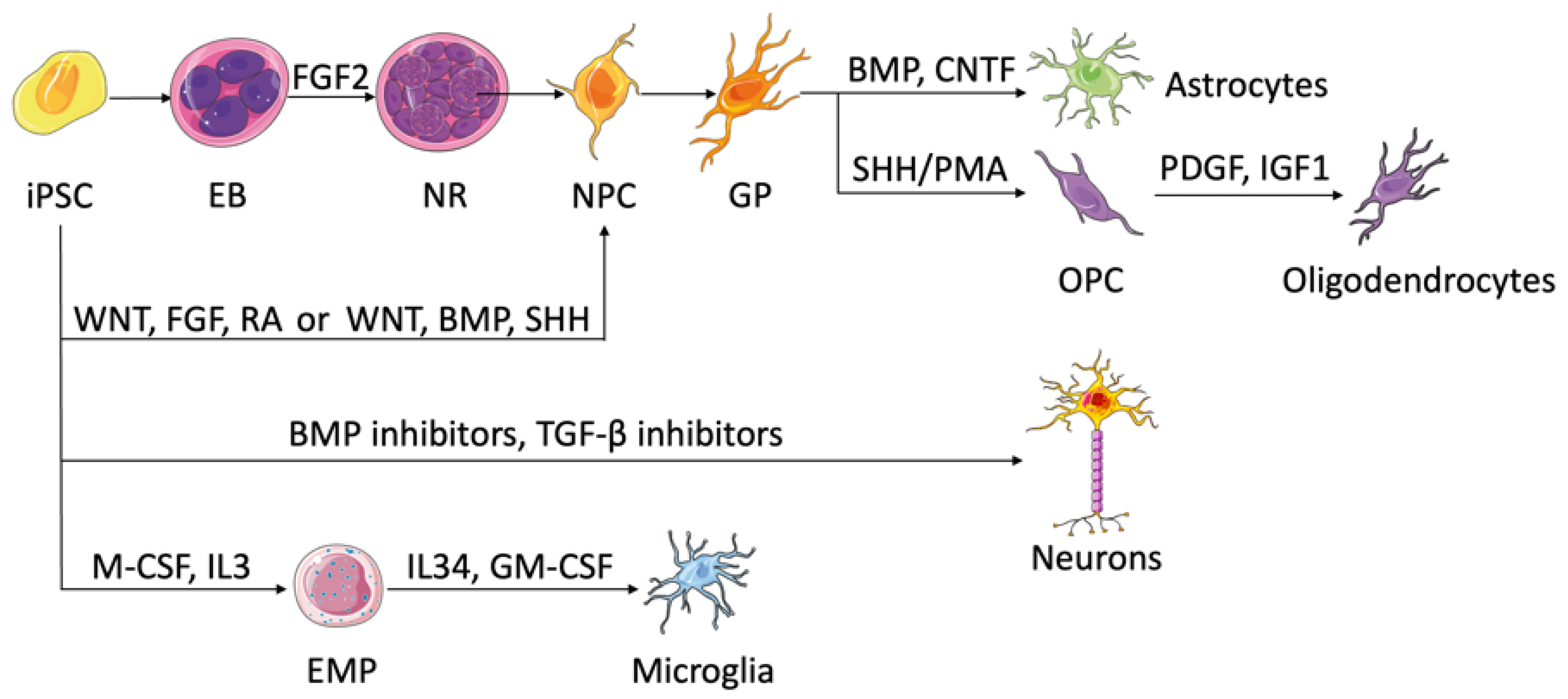
2.1. Deriving Brain Microenvironment Cells Using iPSCs
2.1.1. Neural Progenitor Cells and Neurons
2.1.2. Astrocytes
2.1.3. Oligodendrocytes
2.1.4. Microglia
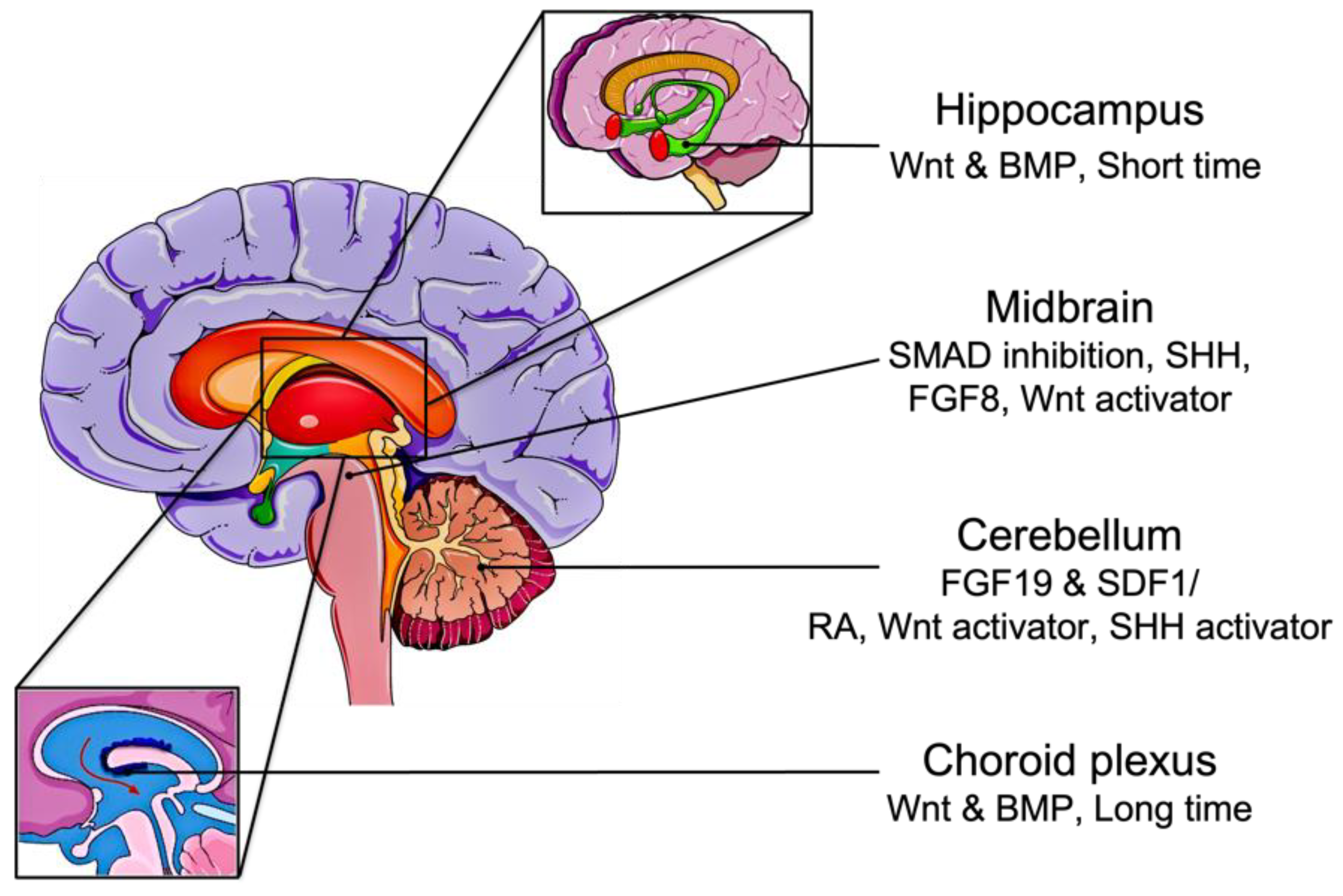
2.2. Derivation of Brain Organoids from iPSC
2.3. Induced PSC-Derived Blood–Brain Barrier Models
3. Induced Pluripotent Stem Cell-Derived Brain Tumor Models
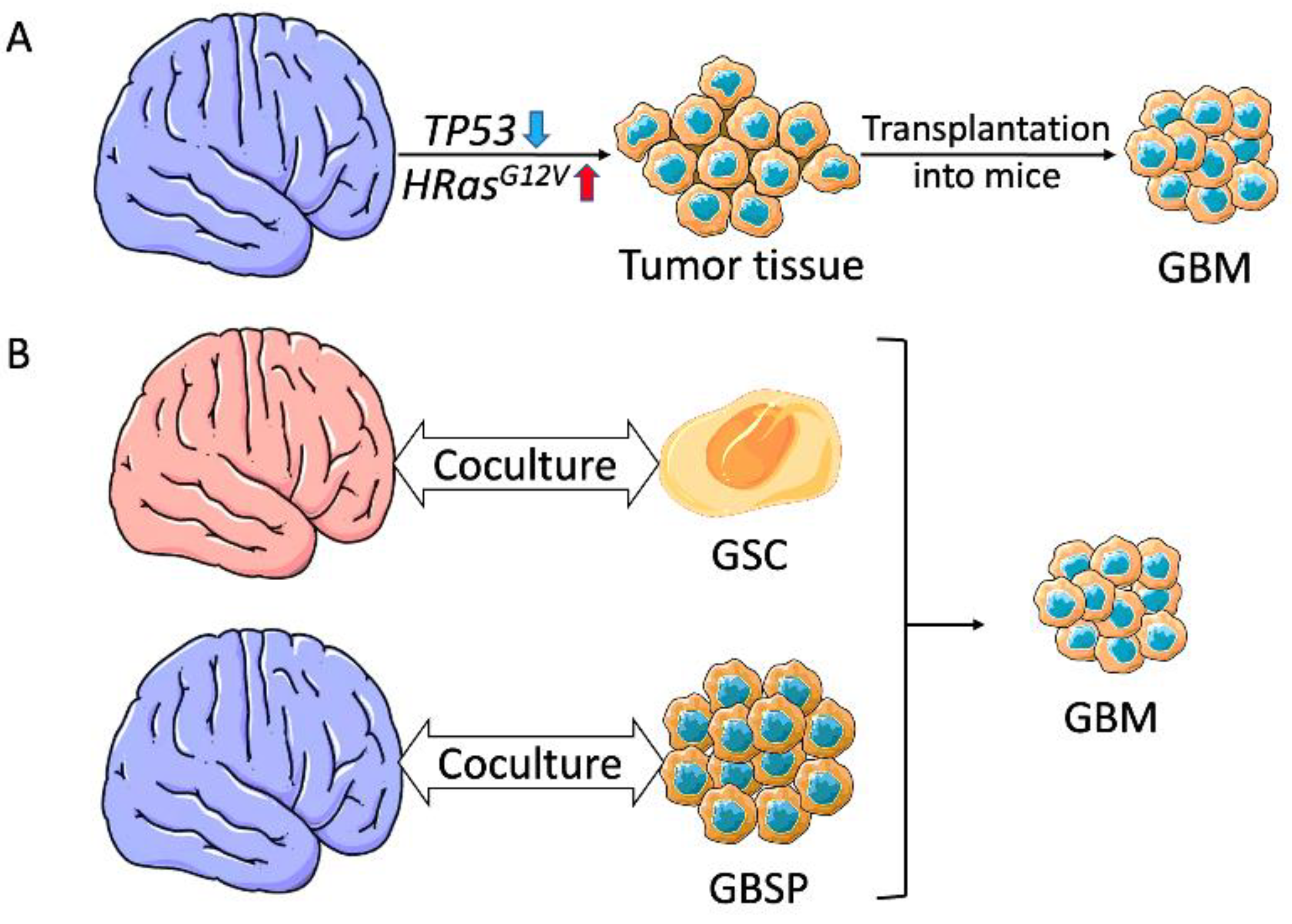
3.1. Induced PSC-Derived Glioblastoma Models
3.1.1. Glioblastoma Organoids Using Genetically Engineered Brain Organoids
3.1.2. Glioblastoma Organoids Using Coculture Organoids
3.1.3. Other iPSC-Derived Models for Glioblastoma
3.2. Induced PSC-Derived Medulloblastoma Models
3.2.1. SHH-Medulloblastoma Models
3.2.2. Group 3 Medulloblastoma Models
3.3. Other Brain Tumor Models
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mitalipov, S.; Wolf, D. Totipotency, Pluripotency and Nuclear Reprogramming. In Engineering of Stem Cells; Springer: Berlin/Heidelberg, Germany, 2009; pp. 185–199. [Google Scholar]
- Takahashi, K.; Yamanaka, S. Induced pluripotent stem cells in medicine and biology. Development 2013, 140, 2457–2461. [Google Scholar] [CrossRef] [PubMed]
- Hochedlinger, K.; Jaenisch, R. Nuclear reprogramming and pluripotency. Nature 2006, 441, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Induced pluripotent stem cells: Past, present, and future. Cell Stem Cell 2012, 10, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef]
- Park, I.-H.; Zhao, R.; West, J.A.; Yabuuchi, A.; Huo, H.; Ince, T.A.; Lerou, P.H.; Lensch, M.W.; Daley, G.Q. Reprogramming of human somatic cells to pluripotency with defined factors. Nature 2008, 451, 141–146. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Grskovic, M.; Javaherian, A.; Strulovici, B.; Daley, G.Q. Induced pluripotent stem cells—Opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 2011, 10, 915–929. [Google Scholar] [CrossRef]
- Luo, J.; Li, P. Human pluripotent stem cell-derived brain organoids as in vitro models for studying neural disorders and cancer. Cell Biosci. 2021, 11, 99. [Google Scholar] [CrossRef]
- Urbaniak, A.; Reed, M.R.; Heflin, B.; Gaydos, J.; Piña-Oviedo, S.; Jędrzejczyk, M.; Klejborowska, G.; Stępczyńska, N.; Chambers, T.C.; Tackett, A.J. Anti-glioblastoma activity of monensin and its analogs in an organoid model of cancer. Biomed. Pharmacother. 2022, 153, 113440. [Google Scholar] [CrossRef]
- Deng, W.-L.; Gao, M.-L.; Lei, X.-L.; Lv, J.-N.; Zhao, H.; He, K.-W.; Xia, X.-X.; Li, L.-Y.; Chen, Y.-C.; Li, Y.-P. Gene correction reverses ciliopathy and photoreceptor loss in iPSC-derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018, 10, 1267–1281. [Google Scholar] [CrossRef]
- Sugita, S.; Iwasaki, Y.; Makabe, K.; Kimura, T.; Futagami, T.; Suegami, S.; Takahashi, M. Lack of T cell response to iPSC-derived retinal pigment epithelial cells from HLA homozygous donors. Stem Cell Rep. 2016, 7, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Cooper, O.; Seo, H.; Andrabi, S.; Guardia-Laguarta, C.; Graziotto, J.; Sundberg, M.; McLean, J.R.; Carrillo-Reid, L.; Xie, Z.; Osborn, T. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson’s disease. Sci. Transl. Med. 2012, 4, 141ra190. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Okano, H. Cell transplantation therapies for spinal cord injury focusing on induced pluripotent stem cells. Cell Res. 2013, 23, 70–80. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, J.S.; Song, B.; Herrington, T.M.; Park, T.-Y.; Lee, N.; Ko, S.; Jeon, J.; Cha, Y.; Kim, K.; Li, Q. Personalized iPSC-derived dopamine progenitor cells for Parkinson’s disease. N. Engl. J. Med. 2020, 382, 1926–1932. [Google Scholar] [CrossRef] [PubMed]
- Madrid, M.; Sumen, C.; Aivio, S.; Saklayen, N. Autologous induced pluripotent stem cell–based cell therapies: Promise, progress, and challenges. Curr. Protoc. 2021, 1, e88. [Google Scholar] [CrossRef]
- Ito, T.; Kawai, Y.; Yasui, Y.; Iriguchi, S.; Minagawa, A.; Ishii, T.; Miyoshi, H.; Taketo, M.M.; Kawada, K.; Obama, K. The therapeutic potential of multiclonal tumoricidal T cells derived from tumor infiltrating lymphocyte-derived iPS cells. Commun. Biol. 2021, 4, 694. [Google Scholar] [CrossRef]
- Ueda, T.; Kumagai, A.; Iriguchi, S.; Yasui, Y.; Miyasaka, T.; Nakagoshi, K.; Nakane, K.; Saito, K.; Takahashi, M.; Sasaki, A. Non–clinical efficacy, safety and stable clinical cell processing of induced pluripotent stem cell-derived anti–glypican-3 chimeric antigen receptor-expressing natural killer/innate lymphoid cells. Cancer Sci. 2020, 111, 1478–1490. [Google Scholar] [CrossRef]
- Okano, H.; Morimoto, S. iPSC-based disease modeling and drug discovery in cardinal neurodegenerative disorders. Cell Stem Cell 2022, 29, 189–208. [Google Scholar] [CrossRef]
- Pasteuning-Vuhman, S.; de Jongh, R.; Timmers, A.; Pasterkamp, R.J. Towards advanced iPSC-based drug development for neurodegenerative disease. Trends Mol. Med. 2021, 27, 263–279. [Google Scholar] [CrossRef]
- Huse, J.T.; Holland, E.C. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat. Rev. Cancer 2010, 10, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Antonica, F.; Aiello, G.; Soldano, A.; Abballe, L.; Miele, E.; Tiberi, L. Modeling Brain Tumors: A Perspective Overview of in vivo and Organoid Models. Front. Mol. Neurosci. 2022, 15, 818696. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, J. The emergence of stem cell-based brain organoids: Trends and challenges. BioEssays 2019, 41, 1900011. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro-Oncology 2017, 19, v1–v88. [Google Scholar] [CrossRef]
- Udaka, Y.T.; Packer, R.J. Pediatric Brain Tumors. Neurol. Clin. 2018, 36, 533–556. [Google Scholar] [CrossRef]
- Krieger, T.G.; Tirier, S.M.; Park, J.; Jechow, K.; Eisemann, T.; Peterziel, H.; Angel, P.; Eils, R.; Conrad, C. Modeling glioblastoma invasion using human brain organoids and single-cell transcriptomics. Neuro-Oncology 2020, 22, 1138–1149. [Google Scholar] [CrossRef]
- Plummer, S.; Wallace, S.; Ball, G.; Lloyd, R.; Schiapparelli, P.; Quiñones-Hinojosa, A.; Hartung, T.; Pamies, D. A Human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. 2019, 9, 1407. [Google Scholar] [CrossRef]
- Sancho-Martinez, I.; Nivet, E.; Xia, Y.; Hishida, T.; Aguirre, A.; Ocampo, A.; Ma, L.; Morey, R.; Krause, M.N.; Zembrzycki, A.; et al. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat. Commun. 2016, 7, 10743. [Google Scholar] [CrossRef]
- Koga, T.; Chen, C.C.; Furnari, F.B. Genome engineering evolves brain tumor modeling. Neurol. Med.-Chir. 2020, 60, 329–336. [Google Scholar] [CrossRef]
- He, F.; Sun, Y.E. Glial cells more than support cells? Int. J. Biochem. Cell Biol. 2007, 39, 661–665. [Google Scholar] [CrossRef]
- Koo, B.; Choi, B.; Park, H.; Yoon, K.J. Past, Present, and Future of Brain Organoid Technology. Mol. Cells 2019, 42, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Logan, S.; Arzua, T.; Canfield, S.G.; Seminary, E.R.; Sison, S.L.; Ebert, A.D.; Bai, X. Studying Human Neurological Disorders Using Induced Pluripotent Stem Cells: From 2D Monolayer to 3D Organoid and Blood Brain Barrier Models. Compr. Physiol. 2019, 9, 565–611. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-C.; Wernig, M.; Duncan, I.D.; Thomson, J.A. In vitro differentiation of transplantable neural precursors from human embryonic stem cells. Nat. Biotechnol. 2001, 19, 1129–1133. [Google Scholar] [CrossRef] [PubMed]
- Boulting, G.L.; Kiskinis, E.; Croft, G.F.; Amoroso, M.W.; Oakley, D.H.; Wainger, B.J.; Williams, D.J.; Kahler, D.J.; Yamaki, M.; Davidow, L.; et al. resource A functionally characterized test set of human induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 279–286. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, S.C. Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 2016, 19, 573–586. [Google Scholar] [CrossRef]
- Rajan, P.; McKay, R.D.G. Multiple Routes to Astrocytic Differentiation in the CNS. J. Neurosci. 1998, 18, 3620–3629. [Google Scholar] [CrossRef]
- Krencik, R.; Weick, J.P.; Liu, Y.; Zhang, Z.J.; Zhang, S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011, 29, 528–534. [Google Scholar] [CrossRef]
- Leventoux, N.; Morimoto, S.; Imaizumi, K.; Sato, Y.; Takahashi, S.; Mashima, K.; Ishikawa, M.; Sonn, I.; Kondo, T.; Watanabe, H.; et al. Human Astrocytes Model Derived from Induced Pluripotent Stem Cells. Cells 2020, 9, 2680. [Google Scholar] [CrossRef]
- Douvaras, P.; Wang, J.; Zimmer, M.; Hanchuk, S.; O’Bara, M.A.; Sadiq, S.; Sim, F.J.; Goldman, J.; Fossati, V. Efficient Generation of Myelinating Oligodendrocytes from Primary Progressive Multiple Sclerosis Patients by Induced Pluripotent Stem Cells. Stem Cell Rep. 2014, 3, 250–259. [Google Scholar] [CrossRef]
- Wang, S.; Bates, J.; Li, X.; Schanz, S.; Chandler-Militello, D.; Levine, C.; Maherali, N.; Studer, L.; Hochedlinger, K.; Windrem, M.; et al. Human iPSC-Derived Oligodendrocyte Progenitor Cells Can Myelinate and Rescue a Mouse Model of Congenital Hypomyelination. Cell Stem Cell 2013, 12, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.R.; Sun, T.; Zhu, Z.; Ma, N.; Garcia, M.; Stiles, C.D.; Rowitch, D.H. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell 2002, 109, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Chanoumidou, K.; Mozafari, S.; Baron-Van Evercooren, A.; Kuhlmann, T. Stem cell derived oligodendrocytes to study myelin diseases. Glia 2020, 68, 705–720. [Google Scholar] [CrossRef] [PubMed]
- García-León, J.A.; Kumar, M.; Boon, R.; Chau, D.; One, J.; Wolfs, E.; Eggermont, K.; Berckmans, P.; Gunhanlar, N.; de Vrij, F.; et al. SOX10 Single Transcription Factor-Based Fast and Efficient Generation of Oligodendrocytes from Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 655–672. [Google Scholar] [CrossRef]
- Shaker, M.R.; Pietrogrande, G.; Martin, S.; Lee, J.H.; Sun, W.; Wolvetang, E.J. Rapid and Efficient Generation of Myelinating Human Oligodendrocytes in Organoids. Front. Cell. Neurosci. 2021, 15, 631548. [Google Scholar] [CrossRef]
- Hasselmann, J.; Blurton-Jones, M. Human iPSC-derived microglia: A growing toolset to study the brain’s innate immune cells. Glia 2020, 68, 721–739. [Google Scholar] [CrossRef]
- Haenseler, W.; Rajendran, L. Concise Review: Modeling Neurodegenerative Diseases with Human Pluripotent Stem Cell-Derived Microglia. Stem Cells 2019, 37, 724–730. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, S.N.; Xu, T.Y.; Miao, Z.W.; Su, D.F.; Miao, C.Y. Organoid technology for brain and therapeutics research. CNS Neurosci. Ther. 2017, 23, 771–778. [Google Scholar] [CrossRef]
- Benito-Kwiecinski, S.; Lancaster, M.A. Brain organoids: Human neurodevelopment in a dish. Cold Spring Harb. Perspect. Biol. 2020, 12, a035709. [Google Scholar] [CrossRef]
- Eiraku, M.; Watanabe, K.; Matsuo-Takasaki, M.; Kawada, M.; Yonemura, S.; Matsumura, M.; Wataya, T.; Nishiyama, A.; Muguruma, K.; Sasai, Y. Self-Organized Formation of Polarized Cortical Tissues from ESCs and Its Active Manipulation by Extrinsic Signals. Cell Stem Cell 2008, 3, 519–532. [Google Scholar] [CrossRef]
- Kleinman, H.K.; McGarvey, M.L.; Hassell, J.R.; Star, V.L.; Cannon, F.B.; Laurie, G.W.; Martin, G.R. Basement membrane complexes with biological activity. Biochemistry 1986, 25, 312–318. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Pasca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, H.; Kadoshima, T.; Soen, M.; Narii, N.; Ishida, Y.; Ohgushi, M.; Takahashi, J.; Eiraku, M.; Sasai, Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015, 6, 8896. [Google Scholar] [CrossRef]
- Qian, X.; Jacob, F.; Song, M.M.; Nguyen, H.N.; Song, H.; Ming, G.L. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018, 13, 565–580. [Google Scholar] [CrossRef]
- Muguruma, K.; Nishiyama, A.; Kawakami, H.; Hashimoto, K.; Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015, 10, 537–550. [Google Scholar] [CrossRef]
- Hua, T.; Bejoy, J.; Song, L.; Wang, Z.; Zeng, Z.; Zhou, Y.; Li, Y.; Sang, Q.-X.A. Cerebellar Differentiation from Human Stem Cells Through Retinoid, Wnt, and Sonic Hedgehog Pathways. Tissue Eng. Part A 2021, 27, 881–893. [Google Scholar] [CrossRef]
- Hua, T.; Liu, C.; Kiran, S.; Gray, K.; Jung, S.; Meckes, D.G.; Li, Y.; Sang, Q.-X.A. Phenotypic, metabolic, and biogenesis properties of human stem cell-derived cerebellar spheroids. Sci. Rep. 2022, 12, 12880. [Google Scholar] [CrossRef]
- Cederquist, G.Y.; Asciolla, J.J.; Tchieu, J.; Walsh, R.M.; Cornacchia, D.; Resh, M.D.; Studer, L. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019, 37, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Rifes, P.; Isaksson, M.; Rathore, G.S.; Aldrin-Kirk, P.; Møller, O.K.; Barzaghi, G.; Lee, J.; Egerod, K.L.; Rausch, D.M.; Parmar, M.; et al. Modeling neural tube development by differentiation of human embryonic stem cells in a microfluidic WNT gradient. Nat. Biotechnol. 2020, 38, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Bagley, J.A.; Reumann, D.; Bian, S.; Lévi-Strauss, J.; Knoblich, J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods 2017, 14, 743–751. [Google Scholar] [CrossRef] [PubMed]
- Birey, F.; Andersen, J.; Makinson, C.D.; Islam, S.; Wei, W.; Huber, N.; Fan, H.C.; Metzler, K.R.C.; Panagiotakos, G.; Thom, N. Assembly of functionally integrated human forebrain spheroids. Nature 2017, 545, 54–59. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Patterson, B.; Kang, Y.J.; Govindaiah, G.; Roselaar, N.; Cakir, B.; Kim, K.Y.; Lombroso, A.P.; Hwang, S.M.; et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017, 21, 383–398.e387. [Google Scholar] [CrossRef]
- Xiang, Y.; Tanaka, Y.; Cakir, B.; Patterson, B.; Kim, K.Y.; Sun, P.; Kang, Y.J.; Zhong, M.; Liu, X.; Patra, P.; et al. hESC-Derived Thalamic Organoids Form Reciprocal Projections When Fused with Cortical Organoids. Cell Stem Cell 2019, 24, 487–497.e487. [Google Scholar] [CrossRef] [PubMed]
- Ormel, P.R.; Vieira de Sá, R.; Van Bodegraven, E.J.; Karst, H.; Harschnitz, O.; Sneeboer, M.A.; Johansen, L.E.; van Dijk, R.E.; Scheefhals, N.; Berdenis van Berlekom, A. Microglia innately develop within cerebral organoids. Nat. Commun. 2018, 9, 4167. [Google Scholar] [CrossRef]
- Song, L.; Yuan, X.; Jones, Z.; Vied, C.; Miao, Y.; Marzano, M.; Hua, T.; Sang, Q.-X.A.; Guan, J.; Ma, T. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci. Rep. 2019, 9, 11055. [Google Scholar] [CrossRef]
- Ham, O.; Jin, Y.B.; Kim, J.; Lee, M.O. Blood vessel formation in cerebral organoids formed from human embryonic stem cells. Biochem. Biophys. Res. Commun. 2020, 521, 84–90. [Google Scholar] [CrossRef]
- Mansour, A.A.; Gonçalves, J.T.; Bloyd, C.W.; Li, H.; Fernandes, S.; Quang, D.; Johnston, S.; Parylak, S.L.; Jin, X.; Gage, F.H. An in vivo model of functional and vascularized human brain organoids. Nat. Biotechnol. 2018, 36, 432–441. [Google Scholar] [CrossRef]
- Qian, X.; Su, Y.; Adam, C.D.; Deutschmann, A.U.; Pather, S.R.; Goldberg, E.M.; Su, K.; Li, S.; Lu, L.; Jacob, F. Sliced human cortical organoids for modeling distinct cortical layer formation. Cell Stem Cell 2020, 26, 766–781. e769. [Google Scholar] [CrossRef]
- Fernández-López, D.; Faustino, J.; Daneman, R.; Zhou, L.; Lee, S.Y.; Derugin, N.; Wendland, M.F.; Vexler, Z.S. Neurobiology of Disease Blood-Brain Barrier Permeability Is Increased After Acute Adult Stroke but Not Neonatal Stroke in the Rat. J. Neurosci. 2012, 32, 9588–9600. [Google Scholar] [CrossRef] [PubMed]
- Benson, P.F.; Joseph, M.C. The Blood-Brain Barrier. Dev. Med. Child Neurol. 1961, 3, 510–514. [Google Scholar] [CrossRef] [PubMed]
- Bhalerao, A.; Sivandzade, F.; Archie, S.R.; Chowdhury, E.A.; Noorani, B.; Cucullo, L. In vitro modeling of the neurovascular unit: Advances in the field. Fluids Barriers CNS 2020, 17, 22. [Google Scholar] [CrossRef] [PubMed]
- Jarazo, J.; Barmpa, K.; Modamio, J.; Saraiva, C.; Sabaté-Soler, S.; Rosety, I.; Griesbeck, A.; Skwirblies, F.; Zaffaroni, G.; Smits, L.M.; et al. Parkinson’s Disease Phenotypes in Patient Neuronal Cultures and Brain Organoids Improved by 2-Hydroxypropyl-β-Cyclodextrin Treatment. Mov. Disord. 2021, 37, 80–94. [Google Scholar] [CrossRef] [PubMed]
- Lacalle-Aurioles, M.; Cassel de Camps, C.; Zorca, C.E.; Beitel, L.K.; Durcan, T.M. Applying hiPSCs and Biomaterials Towards an Understanding and Treatment of Traumatic Brain Injury. Front. Cell. Neurosci. 2020, 14, 594304. [Google Scholar] [CrossRef] [PubMed]
- Brown, L.S.; Foster, C.G.; Courtney, J.-M.; King, N.E.; Howells, D.W.; Sutherland, B.A. Pericytes and neurovascular function in the healthy and diseased brain. Front. Cell. Neurosci. 2019, 13, 282. [Google Scholar] [CrossRef] [PubMed]
- Lippmann, E.S.; Azarin, S.M.; Kay, J.E.; Nessler, R.A.; Wilson, H.K.; Al-Ahmad, A.; Palecek, S.P.; Shusta, E.V. Derivation of blood-brain barrier endothelial cells from human pluripotent stem cells. Nat. Biotechnol. 2012, 30, 783–791. [Google Scholar] [CrossRef]
- Workman, M.J.; Svendsen, C.N. Recent advances in human iPSC-derived models of the blood–brain barrier. Fluids Barriers CNS 2020, 17, 30. [Google Scholar] [CrossRef]
- Wilson, H.K.; Canfield, S.G.; Hjortness, M.K.; Palecek, S.P.; Shusta, E.V. Exploring the effects of cell seeding density on the differentiation of human pluripotent stem cells to brain microvascular endothelial cells. Fluids Barriers CNS 2015, 12, 1–12. [Google Scholar] [CrossRef]
- Hollmann, E.K.; Bailey, A.K.; Potharazu, A.V.; Neely, M.D.; Bowman, A.B.; Lippmann, E.S. Accelerated differentiation of human induced pluripotent stem cells to blood–brain barrier endothelial cells. Fluids Barriers CNS 2017, 14, 13. [Google Scholar] [CrossRef]
- Park, T.-E.; Mustafaoglu, N.; Herland, A.; Hasselkus, R.; Mannix, R.; FitzGerald, E.A.; Prantil-Baun, R.; Watters, A.; Henry, O.; Benz, M. Hypoxia-enhanced Blood-Brain Barrier Chip recapitulates human barrier function and shuttling of drugs and antibodies. Nat. Commun. 2019, 10, 2621. [Google Scholar] [CrossRef] [PubMed]
- Qian, T.; Maguire, S.E.; Canfield, S.G.; Bao, X.; Olson, W.R.; Shusta, E.V.; Palecek, S.P. Directed differentiation of human pluripotent stem cells to blood-brain barrier endothelial cells. Sci. Adv. 2017, 3, e1701679. [Google Scholar] [CrossRef] [PubMed]
- Neal, E.H.; Marinelli, N.A.; Shi, Y.; McClatchey, P.M.; Balotin, K.M.; Gullett, D.R.; Hagerla, K.A.; Bowman, A.B.; Ess, K.C.; Wikswo, J.P. A simplified, fully defined differentiation scheme for producing blood-brain barrier endothelial cells from human iPSCs. Stem Cell Rep. 2019, 12, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Praça, C.; Rosa, S.C.; Sevin, E.; Cecchelli, R.; Dehouck, M.-P.; Ferreira, L.S. Derivation of brain capillary-like endothelial cells from human pluripotent stem cell-derived endothelial progenitor cells. Stem Cell Rep. 2019, 13, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Pong, S.; Lizano, P.; Karmacharya, R. Derivation, expansion, cryopreservation and characterization of brain microvascular endothelial cells from human induced pluripotent stem cells. JoVE J. Vis. Exp. 2020, 165, e61629. [Google Scholar]
- Wilson, H.K.; Faubion, M.G.; Hjortness, M.K.; Palecek, S.P.; Shusta, E.V. Cryopreservation of brain endothelial cells derived from human induced pluripotent stem cells is enhanced by rho-associated coiled coil-containing kinase inhibition. Tissue Eng. Part C Methods 2016, 22, 1085–1094. [Google Scholar] [CrossRef]
- Lu, T.M.; Houghton, S.; Magdeldin, T.; Barcia Durán, J.G.; Minotti, A.P.; Snead, A.; Sproul, A.; Nguyen, D.H.T.; Xiang, J.; Fine, H.A.; et al. Pluripotent stem cell-derived epithelium misidentified as brain microvascular endothelium requires ETS factors to acquire vascular fate. Proc. Natl. Acad. Sci. USA 2021, 118, e2016950118. [Google Scholar] [CrossRef]
- Gastfriend, B.D.; Nishihara, H.; Canfield, S.G.; Foreman, K.L.; Engelhardt, B.; Palecek, S.P.; Shusta, E.V. Wnt signaling mediates acquisition of blood–brain barrier properties in naïve endothelium derived from human pluripotent stem cells. eLife 2021, 10, e70992. [Google Scholar] [CrossRef]
- Yamazaki, T.; Mukouyama, Y.-S. Tissue specific origin, development, and pathological perspectives of pericytes. Front. Cardiovasc. Med. 2018, 5, 78. [Google Scholar] [CrossRef]
- Chan, X.Y.; Black, R.; Dickerman, K.; Federico, J.; Levesque, M.; Mumm, J.; Gerecht, S. Three-Dimensional Vascular Network Assembly from Diabetic Patient-Derived Induced Pluripotent Stem Cells. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2677–2685. [Google Scholar] [CrossRef]
- Orlova, V.V.; Van Den Hil, F.E.; Petrus-Reurer, S.; Drabsch, Y.; Ten Dijke, P.; Mummery, C.L. Generation, expansion and functional analysis of endothelial cells and pericytes derived from human pluripotent stem cells. Nat. Protoc. 2014, 9, 1514–1531. [Google Scholar] [CrossRef] [PubMed]
- Colunga, T.; Hayworth, M.; Kreß, S.; Reynolds, D.M.; Chen, L.; Nazor, K.L.; Baur, J.; Singh, A.M.; Loring, J.F.; Metzger, M. Human pluripotent stem cell-derived multipotent vascular progenitors of the mesothelium lineage have utility in tissue engineering and repair. Cell Rep. 2019, 26, 2566–2579.e2510. [Google Scholar] [CrossRef] [PubMed]
- Faal, T.; Phan, D.T.; Davtyan, H.; Scarfone, V.M.; Varady, E.; Blurton-Jones, M.; Hughes, C.C.; Inlay, M.A. Induction of mesoderm and neural crest-derived pericytes from human pluripotent stem cells to study blood-brain barrier interactions. Stem Cell Rep. 2019, 12, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Stebbins, M.J.; Gastfriend, B.D.; Canfield, S.G.; Lee, M.-S.; Richards, D.; Faubion, M.G.; Li, W.-J.; Daneman, R.; Palecek, S.P.; Shusta, E.V. Human pluripotent stem cell–derived brain pericyte–like cells induce blood-brain barrier properties. Sci. Adv. 2019, 5, eaau7375. [Google Scholar] [CrossRef]
- Jeske, R.; Albo, J.; Marzano, M.; Bejoy, J.; Li, Y. Engineering Brain-Specific Pericytes from Human Pluripotent Stem Cells. Tissue Eng. Part B Rev. 2020, 26, 367–382. [Google Scholar] [CrossRef]
- Appelt-Menzel, A.; Cubukova, A.; Günther, K.; Edenhofer, F.; Piontek, J.; Krause, G.; Stüber, T.; Walles, H.; Neuhaus, W.; Metzger, M. Establishment of a human blood-brain barrier co-culture model mimicking the neurovascular unit using induced pluri-and multipotent stem cells. Stem Cell Rep. 2017, 8, 894–906. [Google Scholar] [CrossRef]
- Delsing, L.; Dönnes, P.; Sánchez, J.; Clausen, M.; Voulgaris, D.; Falk, A.; Herland, A.; Brolén, G.; Zetterberg, H.; Hicks, R. Barrier properties and transcriptome expression in human iPSC-derived models of the blood–brain barrier. Stem Cells 2018, 36, 1816–1827. [Google Scholar] [CrossRef]
- Lippmann, E.S.; Al-Ahmad, A.; Azarin, S.M.; Palecek, S.P.; Shusta, E.V. A retinoic acid-enhanced, multicellular human blood-brain barrier model derived from stem cell sources. Sci. Rep. 2014, 4, 4160. [Google Scholar] [CrossRef]
- Canfield, S.G.; Stebbins, M.J.; Faubion, M.G.; Gastfriend, B.D.; Palecek, S.P.; Shusta, E.V. An isogenic neurovascular unit model comprised of human induced pluripotent stem cell-derived brain microvascular endothelial cells, pericytes, astrocytes, and neurons. Fluids Barriers CNS 2019, 16, 25. [Google Scholar] [CrossRef]
- Canfield, S.G.; Stebbins, M.J.; Morales, B.S.; Asai, S.W.; Vatine, G.D.; Svendsen, C.N.; Palecek, S.P.; Shusta, E.V. An isogenic blood–brain barrier model comprising brain endothelial cells, astrocytes, and neurons derived from human induced pluripotent stem cells. J. Neurochem. 2017, 140, 874–888. [Google Scholar] [CrossRef]
- Delsing, L.; Kallur, T.; Zetterberg, H.; Hicks, R.; Synnergren, J. Enhanced xeno-free differentiation of hiPSC-derived astroglia applied in a blood–brain barrier model. Fluids Barriers CNS 2019, 16, 27. [Google Scholar] [CrossRef] [PubMed]
- Marzano, M.; Chen, X.; Russell, T.A.; Medina, A.; Wang, Z.; Hua, T.; Zeng, C.; Wang, X.; Sang, Q.-X.; Tang, H. Studying the Inflammatory Responses to Amyloid Beta Oligomers in Brain-Specific Pericyte and Endothelial Co-Culture from Human Stem Cells. Front. Chem. Eng. 2022, 4, 927188. [Google Scholar] [CrossRef] [PubMed]
- Griffin, K.; Bejoy, J.; Song, L.; Hua, T.; Marzano, M.; Jeske, R.; Sang, Q.-X.A.; Li, Y. Human stem cell-derived aggregates of forebrain astroglia respond to amyloid beta oligomers. Tissue Eng. Part A 2020, 26, 527–542. [Google Scholar] [CrossRef] [PubMed]
- Song, H.W.; Foreman, K.L.; Gastfriend, B.D.; Kuo, J.S.; Palecek, S.P.; Shusta, E.V. Transcriptomic comparison of human and mouse brain microvessels. Sci. Rep. 2020, 10, 12358. [Google Scholar] [CrossRef]
- Warren, M.S.; Zerangue, N.; Woodford, K.; Roberts, L.M.; Tate, E.H.; Feng, B.; Li, C.; Feuerstein, T.J.; Gibbs, J.; Smith, B.; et al. Comparative gene expression profiles of ABC transporters in brain microvessel endothelial cells and brain in five species including human. Pharmacol. Res. 2009, 59, 404–413. [Google Scholar] [CrossRef]
- Thosar, S.S.; Johnson, B.D.; Johnston, J.D.; Wallace, J.P. Sitting and endothelial dysfunction: The role of shear stress. Med. Sci. Monit. 2012, 18, REV173–REV180. [Google Scholar] [CrossRef]
- Song, H.-H.G.; Rumma, R.T.; Ozaki, C.K.; Edelman, E.R.; Chen, C.S. Vascular tissue engineering: Progress, challenges, and clinical promise. Cell Stem Cell 2018, 22, 340–354. [Google Scholar] [CrossRef]
- Vatine, G.D.; Barrile, R.; Workman, M.J.; Sances, S.; Barriga, B.K.; Rahnama, M.; Barthakur, S.; Kasendra, M.; Lucchesi, C.; Kerns, J. Human iPSC-derived blood-brain barrier chips enable disease modeling and personalized medicine applications. Cell Stem Cell 2019, 24, 995–1005.e1006. [Google Scholar] [CrossRef]
- Lee, C.T.; Bendriem, R.M.; Wu, W.W.; Shen, R.F. 3D brain Organoids derived from pluripotent stem cells: Promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017, 24, 59. [Google Scholar] [CrossRef]
- Khamis, Z.I.; Al-Akkary, N.; Hua, T.; Draughon, S.A.; Li, Y.; Sang, Q.-X.A. Clinical investigations of immunotherapy for human primary brain tumors. Neuroimmunol. Neuroinflamm. 2021, 8, 154–173. [Google Scholar] [CrossRef]
- Ogawa, J.; Pao, G.M.; Shokhirev, M.N.; Verma, I.M. Glioblastoma Model Using Human Cerebral Organoids. Cell Rep. 2018, 23, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.; Repic, M.; Guo, Z.; Kavirayani, A.; Burkard, T.; Bagley, J.A.; Krauditsch, C.; Knoblich, J.A. Genetically engineered cerebral organoids model brain tumour formation. Nat. Methods 2018, 15, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Azzarelli, R. Organoid models of glioblastoma to study brain tumor stem cells. Front. Cell Dev. Biol. 2020, 8, 220. [Google Scholar] [CrossRef] [PubMed]
- Linkous, A.; Balamatsias, D.; Snuderl, M.; Edwards, L.; Miyaguchi, K.; Milner, T.; Reich, B.; Cohen-Gould, L.; Storaska, A.; Nakayama, Y.; et al. Modeling Patient-Derived Glioblastoma with Cerebral Organoids. Cell Rep. 2019, 26, 3203–3211.e3205. [Google Scholar] [CrossRef] [PubMed]
- Pine, A.R.; Cirigliano, S.M.; Nicholson, J.G.; Hu, Y.; Linkous, A.; Miyaguchi, K.; Edwards, L.; Singhania, R.; Schwartz, T.H.; Ramakrishna, R.; et al. Tumor microenvironment is critical for the maintenance of cellular states found in primary glioblastomas. Cancer Discov. 2020, 10, 964–979. [Google Scholar] [CrossRef] [PubMed]
- Azzarelli, R.; Ori, M.; Philpott, A.; Simons, B.D. Three-dimensional model of glioblastoma by co-culturing tumor stem cells with human brain organoids. Biol. Open 2021, 10, bio056416. [Google Scholar] [CrossRef]
- Goranci-Buzhala, G.; Mariappan, A.; Gabriel, E.; Ramani, A.; Ricci-Vitiani, L.; Buccarelli, M.; D’Alessandris, Q.G.; Pallini, R.; Gopalakrishnan, J. Rapid and Efficient Invasion Assay of Glioblastoma in Human Brain Organoids. Cell Rep. 2020, 31, 107738. [Google Scholar] [CrossRef]
- Koga, T.; Chaim, I.A.; Benitez, J.A.; Markmiller, S.; Parisian, A.D.; Hevner, R.F.; Turner, K.M.; Hessenauer, F.M.; D’Antonio, M.; Nguyen, N.p.D.; et al. Longitudinal assessment of tumor development using cancer avatars derived from genetically engineered pluripotent stem cells. Nat. Commun. 2020, 11, 550. [Google Scholar] [CrossRef]
- Hwang, J.W.; Loisel-Duwattez, J.; Desterke, C.; Latsis, T.; Pagliaro, S.; Griscelli, F.; Bennaceur-Griscelli, A.; Turhan, A.G. A novel neuronal organoid model mimicking glioblastoma (GBM) features from induced pluripotent stem cells (iPSC). Biochim. Biophys. Acta Gen. Subj. 2020, 1864, 129540. [Google Scholar] [CrossRef]
- Taylor, M.D.; Northcott, P.A.; Korshunov, A.; Remke, M.; Cho, Y.J.; Clifford, S.C.; Eberhart, C.G.; Parsons, D.W.; Rutkowski, S.; Gajjar, A.; et al. Molecular subgroups of medulloblastoma: The current consensus. Acta Neuropathol. 2012, 123, 465–472. [Google Scholar] [CrossRef]
- Sharma, T.; Schwalbe, E.C.; Williamson, D.; Sill, M.; Hovestadt, V.; Mynarek, M.; Rutkowski, S.; Robinson, G.W.; Gajjar, A.; Cavalli, F.; et al. Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019, 138, 309–326. [Google Scholar] [CrossRef]
- Huang, M.; Tailor, J.; Zhen, Q.; Gillmor, A.H.; Miller, M.L.; Weishaupt, H.; Chen, J.; Zheng, T.; Nash, E.K.; McHenry, L.K.; et al. Engineering Genetic Predisposition in Human Neuroepithelial Stem Cells Recapitulates Medulloblastoma Tumorigenesis. Cell Stem Cell 2019, 25, 433–446.e437. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Prim. 2019, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Susanto, E.; Navarro, A.M.; Zhou, L.; Sundström, A.; van Bree, N.; Stantic, M.; Moslem, M.; Tailor, J.; Rietdijk, J.; Zubillaga, V.; et al. Modeling SHH-driven medulloblastoma with patient iPS cell-derived neural stem cells. Proc. Natl. Acad. Sci. USA 2020, 117, 20127–20138. [Google Scholar] [CrossRef] [PubMed]
- Čančer, M.; Hutter, S.; Holmberg, K.O.; Rosén, G.; Sundström, A.; Tailor, J.; Bergström, T.; Garancher, A.; Essand, M.; Wechsler-Reya, R.J.; et al. Humanized Stem Cell Models of Pediatric Medulloblastoma Reveal an Oct4/mTOR Axis that Promotes Malignancy. Cell Stem Cell 2019, 25, 855–870.e11. [Google Scholar] [CrossRef]
- Ikemoto, Y.; Miyashita, T.; Nasu, M.; Hatsuse, H.; Kajiwara, K.; Fujii, K.; Motojima, T.; Kokido, I.; Toyoda, M.; Umezawa, A. Gorlin syndrome-induced pluripotent stem cells form medulloblastoma with loss of heterozygosity in PTCH1. Aging 2020, 12, 9935. [Google Scholar] [CrossRef]
- Northcott, P.A.; Buchhalter, I.; Morrissy, A.S.; Hovestadt, V.; Weischenfeldt, J.; Ehrenberger, T.; Gröbner, S.; Segura-Wang, M.; Zichner, T.; Rudneva, V.A.; et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017, 547, 311–317. [Google Scholar] [CrossRef]
- Ballabio, C.; Anderle, M.; Gianesello, M.; Lago, C.; Miele, E.; Cardano, M.; Aiello, G.; Piazza, S.; Caron, D.; Gianno, F.; et al. Modeling medulloblastoma in vivo and with human cerebellar organoids. Nat. Commun. 2020, 11, 583. [Google Scholar] [CrossRef]
- Xue, Y.; Fu, Y.; Zhao, F.; Gui, G.; Li, Y.; Rivero-Hinojosa, S.; Liu, G.; Li, Y.; Xia, S.; Eberhart, C.G.; et al. Frondoside A Inhibits an MYC-Driven Medulloblastoma Model Derived from Human-Induced Pluripotent Stem Cells. Mol. Cancer Ther. 2021, 20, 1199–1209. [Google Scholar] [CrossRef]
- Parisian, A.D.; Koga, T.; Miki, S.; Johann, P.D.; Kool, M.; Crawford, J.R.; Furnari, F.B. SMARCB1 loss interacts with neuronal differentiation state to block maturation and impact cell stability. Genes Dev. 2020, 34, 1316–1329. [Google Scholar] [CrossRef]
- Chakrabarti, J.; Pandey, R.; Churko, J.M.; Eschbacher, J.; Mallick, S.; Chen, Y.; Hermes, B.; Mallick, P.; Stansfield, B.N.; Pond, K.W. Development of Human Pituitary Neuroendocrine Tumor Organoids to Facilitate Effective Targeted Treatments of Cushing’s Disease. Cells 2022, 11, 3344. [Google Scholar] [CrossRef] [PubMed]
- Kanton, S.; Boyle, M.J.; He, Z.; Santel, M.; Weigert, A.; Sanchís-Calleja, F.; Guijarro, P.; Sidow, L.; Fleck, J.S.; Han, D. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature 2019, 574, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Jovanovich, N.; Habib, A.; Kodavali, C.; Edwards, L.; Amankulor, N.; Zinn, P.O. The Evolving Role of Induced Pluripotent Stem Cells and Cerebral Organoids in Treating and Modeling Neurosurgical Diseases. World Neurosurg. 2021, 155, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, H.; Ming, G.-l. Brain organoids: Advances, applications and challenges. Development 2019, 146, dev166074. [Google Scholar] [CrossRef]
- Romero-Morales, A.I.; O’Grady, B.J.; Balotin, K.M.; Bellan, L.M.; Lippmann, E.S.; Gama, V. Spin∞: An updated miniaturized spinning bioreactor design for the generation of human cerebral organoids from pluripotent stem cells. HardwareX 2019, 6, e00084. [Google Scholar] [CrossRef]
- Hubert, C.G.; Rivera, M.; Spangler, L.C.; Wu, Q.; Mack, S.C.; Prager, B.C.; Couce, M.; McLendon, R.E.; Sloan, A.E.; Rich, J.N. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo brain cancer stem cell organoids. Cancer Res. 2016, 76, 2465–2477. [Google Scholar] [CrossRef]
- Jacob, F.; Salinas, R.D.; Zhang, D.Y.; Nguyen, P.T.; Schnoll, J.G.; Wong, S.Z.H.; Thokala, R.; Sheikh, S.; Saxena, D.; Prokop, S. A patient-derived glioblastoma organoid model and biobank recapitulates inter-and intra-tumoral heterogeneity. Cell 2020, 180, 188–204.e122. [Google Scholar] [CrossRef]

| Brain Tumor | Author | Model | Features | Key Findings |
|---|---|---|---|---|
| Glioblastoma | Sancho-Martinez et al. [29] | Orthotopic engraftment of engineered NPCs in mice | TP53-/-, RASOE/EGFROE/SRCOE | Engineered NPCs acquired glioma features. Transplantation yielded aggressive tumors. |
| Ogawa et al. [112] | Engineered tumor organoid orthotopically engrafted in mice | HRasG12V/TP53-/- | Engineered cells expressed GBM markers and invasive phenotype. | |
| Bian et al. [113] | Engineered tumor organoid | MYCOE, CDKN2A–/–/CDKN2B–/–/EGFROE/EGFRvIIIOE, NF1–/–/PTEN–/–/TP53–/–, EGFRvIIIOE/PTEN–/–/CDKN2A–/– | GBM organoids were capable of in vivo expansion and progression. | |
| Ogawa et al. [112] | Engineered tumor cell line/patient-derived glioblastoma cell line transplanted into cerebral organoid | HRasG12V/TP53-/- (Engineered tumor cell line) | Patient-derived cells showed different invasion potential than that of organoid-derived tumor cell spheres. | |
| Linkous et al. [115] | Organoid cocultured with GSCs | - | Patient-specific GBM could be studied ex-vivo. Scalable GLICO provided a reliable phenocopy of patient GBM. | |
| Goranci-Buzhala et al. [118] | Hybrid organoids by coculturing iPSCs with GSCs | - | 3D GSC invasion assays were developed. | |
| Hwang et al. [120] | Engineered tumor organoid | c-metOE | iPSC aggregates displayed genomic network and phenotype of primary human GBM. Organoids were sensitive to temozolomide. | |
| Koga et al. [119] | Orthotopic engraftment of engineered NPCs in mice | NF1–/–/PTEN–/–,TP53-/-/PDGFRAΔ8−9 | GBM model contained intra- and inter-tumor heterogeneity and provided platform for assessment of tumor development. | |
| Medulloblastoma | Ballabio et al. [129] | Engineered tumor organoid | GFI1OE /c-MYCOE, OTX2OE /c-MYCOE | OTX2 and c-MYC were identified as strong Group 3 MB drivers. SMARCA4 expression and tazemetostat negate OTX2/c-MYC tumorigenesis. |
| Huang et al. [123] | Orthotopic engraftment of engineered/Gorlin NESCs in mice | MYCNOE, PTCH1+/− | Engineered NESCs mimicked tumor subtype and epigenetic profile. Gorlin NESCs retained MB predisposition. | |
| Čančer et al. [126] | Orthotopic engraftment of engineered NESCs in mice | MYCNOE | Aggressive SHH-MB with mTOR activation and increased Oct4 | |
| Ikemoto et al. [127] | Gorlin iPSCs subcutaneously implanted in mice | Heterozygous PTCH1 mutations | Gorlin iPSCs developed MB with secondary somatic PTCH1 mutations. | |
| Susanto et al. [125] | Orthotopic engraftment of Gorlin NESCs in mice | PTCH1 1762insG | Gorlin NESCs formed tumors mimicking human SHH-MB. | |
| Xue et al. [130] | Orthotopic engraftment of engineered NPCs in mice | c-MYC/DNp53 coexpression | MYC-driven tumors recapitulated group 3 MB. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khamis, Z.I.; Sarker, D.B.; Xue, Y.; Al-Akkary, N.; James, V.D.; Zeng, C.; Li, Y.; Sang, Q.-X.A. Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells. Cancers 2023, 15, 1253. https://doi.org/10.3390/cancers15041253
Khamis ZI, Sarker DB, Xue Y, Al-Akkary N, James VD, Zeng C, Li Y, Sang Q-XA. Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells. Cancers. 2023; 15(4):1253. https://doi.org/10.3390/cancers15041253
Chicago/Turabian StyleKhamis, Zahraa I., Drishty B. Sarker, Yu Xue, Nancy Al-Akkary, Viviana D. James, Changchun Zeng, Yan Li, and Qing-Xiang Amy Sang. 2023. "Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells" Cancers 15, no. 4: 1253. https://doi.org/10.3390/cancers15041253
APA StyleKhamis, Z. I., Sarker, D. B., Xue, Y., Al-Akkary, N., James, V. D., Zeng, C., Li, Y., & Sang, Q.-X. A. (2023). Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells. Cancers, 15(4), 1253. https://doi.org/10.3390/cancers15041253






